Summary
BMapBuilder builds maps of pairwise linkage disequilibrium (LD) in either two or three dimensions. The optimized resolution allows for graphical display of LD for single nucleotide polymorphisms (SNPs) in a whole chromosome.
One of the current goals to measure genome-wide LD is to estimate recombination in a population and its implication for gene mapping and association studies. This is, for example, one of the aims of the International HapMap Project (IHP) [6].
Because of their simplicity, pairwise measures of LD, such as D′ or r2, are the most popular measures used to capture the strength of association between pairs of SNPs [7]. Visual inspection of genome-wide LD requires graphical methods able to display large-scale patterns of LD. The commonly used graphical displays —sliding windows [4] and LD decay plots [8]— display average pairwise LD inside a region. The first one displays average LD over regions determined by a window of constant size, the second one displays patterns of LD decays for increasing physical or genetic distances. These plots however do not show pairwise LD but compress information into a one dimensional plot. Therefore a tool able to display a bidemensional LD along a whole chromosome would allow researchers to examine recombination, as well as distribution of haplotype blocks [6] across a whole chromosome, without information loss.
A two-dimensional display of LD is available in the software Haploview [2], but its resolution –more than 50 times greater than the optimal resolution– is suboptimal. Furthermore, in our experience this software can generate LD maps only for a region with at most about 6000 SNPs. The suboptimal resolution of Haploview implies that a map created to display LD of 1000 pairs of SNPs would require 50,912 pixels and hence 50 windows in a common screen with resolution 1024×768 to be displayed. The optimal resolution map would require only 1000 pixels and be displayed in only one window.
To the best of our knowledge, there is no software able to build pairwise LD maps for a fast screening of large data sets such as those produced by the IHP. To address this limitation, we have developed BMapBuilder: a program that can generate bitmap images to represent genome-wide pairwise LD, and can build LD maps over an entire chromosome. The program takes as input a text file, tab, blank or comma-delimited, with a row for each pair of SNPs and at least three columns — positions i, j of each SNP pair and a corresponding measure of LD, and creates an LD map that is saved as a png file. BMapBuilder provides users with a wide choice of resolutions. A resolution equal to s means that, for each pairwise estimate of LD between SNPs at loci i and j, i < j, a square of s × s pixels is plotted. The color intensity of each square follows the legend in Figure 1, when the selected color for the map is red. Equivalent scales are used for the other two color options of BMapBuilder: green and blue.
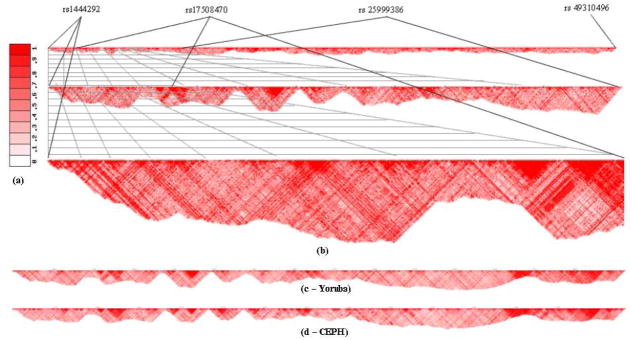
Fig. 1.
Examples of maps built by BMapBuilder using the ML estimates of D′. Panel a: color legend. Panel b-back: LD map based on all 19,854 SNPs that were genotyped in chromosome 22 of the Yoruba population. Panel b-middle: LD map based on the first 6050 SNPs. Panel b-front: LD map based on first 1200 SNPs. Each cell (i, j) represents the magnitude of LD between SNPs in position i, j, with i < j by the intensity described in the legend in panel a. Panel c: LD map based on the first 8,000 SNPs typed in chromosome 22 of the Yoruba population. The last SNP is in position 29437558. Panel d: LD map based based on the 7569 SNPs typed in the same physical region of chromosome 22 for the CEPH population.
To optimize the resolution without loosing information, BMap-Builder can use only one pixel to represent the estimation of LD between a pair of SNPs such as D′ or r2. Higher resolution can also be chosen to magnify the map for smaller DNA regions, and the current implementation allows a maximum resolution of 20. If, besides the estimate of a measure of LD for each pair of SNPs, the user provides two columns with allele frequencies, the program builds maps with different thresholds on the Minor Allele Frequencies (MAF) to reduce the bias towards disequilibrium of the Maximum Likelihood (ML) estimator of D′ [5, 9]. We emphasize that BmapBuilder allows users to use any pairwise measure of LD, including the standard D′ and r2 regardless of how they are computed. The input file can be generated with programs such as Haploview or BLink [1]. While Haploview uses ML to compute D′ and r2, BLink uses a Bayesian approach with a proper prior that reduces the bias toward disequilibrium of D′ in small samples.
Figure 1 (back image, panel b) shows a reduced image of a map generated by BMapBuilder by using a genotype dataset published by the IHP. The data consists of 19,854 SNPs genotyped in chromosome 22 using samples from 30 trios of the Yoruba population. The data are available in post-makeped format at http://bios.ugr.es/BMapBuilder/supplementary/data.html. To build the map we used the traditional ML estimates of D′ (available from the IHP website). The supplementary material contains also maps that were built using the ML estimates of r2, as well as the novel Bayesian estimator of D′ that is implemented in BLink [1]. The web site reports maps with resolution s = 1, so that only one pixel is used to graphically display the magnitude of LD. Larger resolutions maps with s = 4 can be examined by double-clicking on each map. If one wants to focus attention on smaller DNA regions, BMapBuilder can build maps with higher resolutions. As an example, Figure 1 (middle and front images, panel b) shows two zoomed maps of LD using overlapping subsets of SNPs. While the chomosome-wide map displays LD for all 19,854 SNPs, the middle image displays LD for 6050 SNPs and the front image displays LD for 1200 SNPs.
The figure highlights the LD landscape of the whole chromosome so that regions of higher LD can be identified by visual inspection, and patterns of LD can be compared across different populations. The maps in Figure 1, panels c and d, display LD for the first strand of chromosome 22, up to the physical positions 29437558. There are 8000 SNPs genotyped in the IHP for this strand in the Yoruba population (panel c) and 7569 in the CEPH population (panel d). Wider blocks are evident in the LD map of the CEPH population, in agreement with results showing a more reduced haplotype diversity in the European-descent population [8, 3].
The software also includes a 3D display option for smaller sets of SNPs. As an example, 3D maps can be created to visually examine the effect of other factors influencing LD, such as allele frequencies or types of polymorphisms. In BMapBuilder, different polymorphisms can be coded with different colors chosen by the user, so that colored 3D maps would be created. By using transparency features proportional to the estimator, 3D maps can be projected into 2D maps in which colors represent a types of polymorphisms and estimator values are represented by the transparency. Compared to standard 2-dimension LD maps, these projections include more information through different color choices. These images constitute a novel visual tool for a first step in the identification of patterns of LD that include other factors and open a new range of options.
Supplementary Material
Acknowledgments
This work was supported by NHLBI grants R21 HL080463-01 and 1R01DK069646-01A1 and the Spanish Research Program under projects TIN2004-07672-C03-02 and TIN2005-09098-C05-03. We thank Marco Ramoni who inspired us to write this paper.
Footnotes
Supplementary information: Maps displaying chomosome-wide LD are available at http://bios.ugr.es/BMapBuilder/supplementary.
Availability: The program is coded in Java, which runs on all relevant operating systems, including Windows, Mac and Unix/Linux, and is available from http://bios.ugr.es/BMapBuilder.
For Permissions, please journals.permissions@oxfordjournals.org
References
- 1.Abad-Grau MM, Sebastiani P. Bayesian correction for snp ascertainment bias: Third international conference, mdai 2006, tarragona, spain, april 3–5, 2006. proceedings. In: Torra Aida Valls Vicenc, Narukawa Yasuo, Domingo-Ferrer Josep., editors. LNCS. Vol. 3885. Springer; Berlin/Heidelberg: 2006. pp. 262–273. Modeling Decisions for Artificial Intelligence. [Google Scholar]
- 2.Barret JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 3.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 4.Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, Carter D, Papaspyridonos M, Livingstone S, Ganskek R, Lohmussaar E, Zernant J, Tonisson N, Remm M, Magi R, Puurand T, Vilo J, Kurg A, Rice K, Deloukas P, Mott R, Metspalu A, Bentley DR, Cardon LR, Dunham I. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418:544–548. doi: 10.1038/nature00864. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel S, Schaffner S, Nguyen H, Moore J, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Neen Liu-Cordero S, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander E, Daly M, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002:296. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 6.The International HapMap-Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott J. Analysis of human genetic linkage. John Hopkins; Baltimore, MD: 1999. [Google Scholar]
- 8.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 9.Teare MD, Dunning AM, Durocher F, Rennart G, Easton DF. Sampling distribution of summary linkage disequilibrium measures. Ann Hum Genet. 2002;66:223–233. doi: 10.1017/S0003480002001082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.