Abstract
Altered glycosylation on the surfaces or secreted proteins of tumor cells is common in pancreatic cancer and is thought to promote cancer progression, but the factors leading to the changes in carbohydrate structures are incompletely understood. We hypothesized that pro-inflammatory conditions can lead to alterations in cancer-associated glycans on mucins produced by pancreatic-cancer cells. Using a novel antibody-glycan microarray method, we measured the effects of pro-inflammatory stimuli (oxidative stress and treatment with the cytokines IFNγ, IL-1α, and TNFα) on the expression and glycosylation of the mucins MUC1, MUC5AC, and MUC16 in multiple pancreatic cancer cell lines. Mucin glycosylation was significantly affected in specific cell lines, particularly in structures involving terminal galactose or N-acetylgalactosamine. In addition, the responses of the cell lines grouped according to the expression of cell-surface markers that are associated with tumorigenicity, as cell lines bearing minimal surface markers showed evidence of increased O-glycan extension and decreased presentation of terminal β1,4-linked galactose, opposite to cell lines bearing multiple markers. These results suggest mechanisms whereby inflammation might influence tumor behavior in a cell-type specific manner through modulating the presentation of cancer-associated glycans.
Keywords: mucin, glycosylation, pro-inflammatory cytokines, antibody array, pancreatic cancer
INTRODUCTION
A common feature of pancreatic cancer is alterations to the glycan chains on cell-surface and secreted proteins. These alterations are thought to be involved in cancer progression through affecting cell adhesion, migration, metastatic capability, and immune invasion1, 2. Cancer-associated glycans are often found on a class of proteins called mucins3, which are large, highly O-glycosylated proteins involved in the protection and control of signaling at epithelial surfaces. Mucin themselves exert profound tumor-promoting effects, in part mediated through interactions with their glycan moieties4–6. Therefore, understanding the cancer biology of glycan alterations on mucins is important for developing strategies for combating pancreatic cancer.
The causes of cancer-associated glycosylation alterations on mucins are not well understood. The assembly of O-glycan chains is determined by a complex integration of factors such as monosaccharide precursor availability, flux of the protein substrates, and glycosyltransferase activities. Changes to glycosyltransferase activities are likely important in cancer, as suggested by the ability of certain glycosyltransferases to control the formation of cancer-associated glycans7–10. Glycosyltransferase activities and the resulting glycan structures are known to be regulated by cytokine signaling in systems such as leukocyte11, 12 and endothelial13 activation, inflammatory responses14, 15, and tumor cells16, 17. This regulation of glycosyltransferase activity by cytokines suggests a role for external signaling from the tumor microenvironment in the appearance of cancer-associated glycosylation on cancer cells.
Pancreatic cancer cells typically are part of a pro-inflammatory microenvironment, which contributes negatively to overall outcomes18, 19. Inflammation has been shown to be involved in pancreatic cancer development and progression in a variety of ways, such as by creating a high oxidative stress and mutagenic environment, by supplying tumor-promoting growth factors and chemokines, and by modulating immune responses20, 21. Such an environment also could provide the conditions necessary to stimulate changes in the glycosyltransferases and other factors that produce cancer-associated glycans.
We therefore hypothesized that a pro-inflammatory environment could lead to altered glycosylation in pancreatic-cancer cells. This hypothesis was tested using antibody-lectin sandwich microarrays, which enable reproducible measurements of glycan levels on multiple proteins captured directly from biological samples22. Conventional methods for looking at glycosylation on specific proteins in biological samples, such as immunoprecipitation followed by enzymatic or mass spectrometry analysis, do not have the throughput or the precision to allow comparisons between multiple samples. Antibody-lectin sandwich microarrays, which build on previous lectin-based methods for studying the glycobiology of proteins and the roles of glycans in diseases23–26, are complementary to those methods, in that they yield less-detailed structural information but allow the investigation of changes to protein abundance and glycosylation for multiple proteins.
In addition to testing the hypothesis that pro-inflammatory stimuli affect glycosylation, we investigated the heterogeneity between the cell types in their alterations to mucin expression and glycosylation. Populations of transformed cells in tumors can be highly heterogeneous. A recent study demonstrated that a minority of cells within pancreatic tumors, defined by high levels of the surface markers CD24, CD44, and ESA, is much more tumorigenic than the other cells27. These tumorigenic cells, sometimes termed “cancer stem cells,” have the ability to self renew and form new, heterogeneous tumors. Similar observations have been made in tumors from other tissues28–30. Since mucin expression and glycosylation are implicated in cancer cell growth and metastasis, it is possible that highly tumorigenic cancer cells have a higher propensity to activate altered expression or glycosylation of mucins. Therefore we also tested the hypothesis that tumorigenic pancreatic cancer cells and non-tumorigenic cancer cells display distinct changes to mucin expression or glycosylation in response to pro-inflammatory stimuli.
MATERIALS AND METHODS
Antibodies
Antibodies and biotinylated lectins were purchased from various sources (Table 1). The antibodies used for cell surface marker detection in flow cytometry analysis were anti-CD44-allophycocyanin, anti-CD24-phycoerythrin, and anti-ESA-FITC (Biomeda, Foster City, CA). To prepare antibodies for printing in arrays, they were purified by dialysis (Slide-A-lyzer, Pierce Biotechnology, Rockford, IL) against phosphate-buffered saline (PBS) and ultracentrifugation, and the concentrations were measured (MicroBCA, Pierce Biotechnology, Rockford, IL) and adjusted to 0.5 mg/ml. The integrity of each antibody was confirmed by reducing and non-reducing acrylamide gel electrophoresis. Biotinylation of the antibodies was performed using EZ-Link sulfo-NHS-LC-biotin (Pierce Biotechnology, Rockford, IL), using mild conditions (pH 7.4 on ice for one hour) to minimize possible disruptions to antibody affinities through over-labeling. The reaction mixtures were quenched with a tenth volume of 1M Tris (pH 7.5) for 20 minutes, and the antibodies were purified using size-exclusion gel filtration (Biospin P6 columns, BioRad, Hercules, CA).
Table 1.
| Antibodies | Clone No. | Target | Company | Cat. No. | Species |
|---|---|---|---|---|---|
| Anti CA19-9 | 1.B.844 | Sialyl Lewisa | USBio | C0075-07 | Mouse |
| Anti-MUC1-Ab1 | 1.B.831 | MUC1 core protein | USBio | C0050-23 | Mouse |
| Anti-MUC1-Ab2 | VU-11D1 | MUC1 core protein | Biogenesis | 6378-0155 | Mouse |
| Anti-MUC5AC-Ab1 | CLH2 | MUC5AC core protein | Chemicon | MAB2011 | Mouse |
| Anti-MUC5AC-Ab2 | 45M1 | MUC5AC core protein | Biogenesis | 1695-0128 | Mouse |
| Anti-MUC16-Ab1 | 1.B.821 | MUC16 core protein | USBio | C0050-01 | Mouse |
| Anti-MUC16-Ab2 | X325 | MUC16 core protein | AbCam | ab10033 | Mouse |
| Anti-MUC16-Ab3 | 653F | MUC16 core protein | Neoclone | - | Mouse |
| Anti-MUC16-Ab4 | 618F | MUC16 core protein | Neoclone | - | Mouse |
| Lectins | |||||
| GSL-I | Terminal GalNAcα, terminal Fucα1,2 | Vector Laboratories | B-1105 | Griffonia simplicifolia | |
| AAL | Terminal Fucα1,2, Fucα1,3, and Fucα1,4 | Vector Laboratories | B-1395 | Aleuria aurantia (mushroom) | |
| BPL | Terminal Galβ1,3 and Galβ1,4, terminal lactosamine | Vector Laboratories | B-1285 | Bauhinia purpurea (alba) | |
| ConA | N-glycan high mannose, terminal Manα | Vector Laboratories | BK-1000 | Canavalia ensiformis (Jack bean) | |
| SBA | Terminal GlcNAcβ, GalNAcα,GalNAcβ, lactosamine | Vector Laboratories | BK-1000 | Glycine max (soybean) | |
| WGA | Internal lactosamine, terminal GlcNAcβ | Vector Laboratories | BK-1000 | Triticum vulgaris (wheat germ) | |
| UEA | Terminal Fucα1,2 | Vector Laboratories | BK-1000 | Ulex europaeus (Furze gorse) | |
| RCA-I | Terminal Galβ1,4, terminal lactosamine | Vector Laboratories | BK-1000 | Ricinus communis (castor bean) | |
| PNA | O-glycan core 1 (T-antigen), terminal Galβ1,3 | Vector Laboratories | BK-1000 | Arachis hypogaea (peanuts) | |
| GSL-II | Terminal GlcNAcβ, terminal Manα | Vector Laboratories | BK-3000 | Griffonia simplicifolia | |
| ECL | Galβ-1,4GlcNAc | Vector Laboratories | BK-3000 | Erythrina cristagalli | |
| Jacalin | O-glycan core 1 (T-antigen) | Vector Laboratories | BK-3000 | Artocarpus integrifolia (Jackfruit) | |
| LEL | Internal and terminal lactosamine | Vector Laboratories | BK-3000 | Lycopersicon esculentum (Tomato) | |
| STL | Internal and terminal lactosamine | Vector Laboratories | BK-3000 | Solanum tuberosum (Potato) | |
| LCA | N-glycan high mannose, terminal Manα | Vector Laboratories | B-1045 | Lens culinaris (lentil) | |
| VVL | Terminal GalNAcα, terminal GalNAcβ | Vector Laboratories | B-1235 | Vicia villosa (vetch) | |
| HPA | Terminal GalNAcα (Tn antigen) | Sigma | L-6512 | Helix pomatia (Roman snail) | |
| DSL | Internal lactosamine | Vector Laboratories | BK-3000 | Datura stramonium | |
Cell culture
Human pancreatic cancer cell lines (Supplementary Table 1) were from American Type Culture Collection (ATCC). AsPC-1, BxPC3 and SU.86.86 cells were maintained in RPMI 1640 medium; Hs-766T, MIAPaCa, PANC-1, SW1990 and MPanc-96 cells were maintained in DMEM medium; Capan-2 cells were maintained in McCOY’s 5A medium; HPAF-II and L3-3 cells were maintained in a 1:1 mixture of MEM and DMEM medium. All the media were supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Capan-1 and CFPAC cells were maintained in IMDM medium supplemented with 20% FBS. HPAC cells were maintained in a 1:1 mixture of DMEM and Ham’s F12 medium supplemented with 5% FBS. The normal human pancreatic ductal epithelial cells (HPDE) were kindly provided by Dr. Tsao (Ontario Cancer Institute, Toronto, Ontario, Canada) and were maintained in Keratinocyte SFM medium supplemented with EGF, bovine pituitary extract, and antibiotics (Invitrogen, Carlsbad, CA).
Preparation of cell lysates and conditioned media
Pancreatic cancer cell lines were grown in appropriate media containing 10% FBS until reaching 70–80% confluence. The cells were washed with PBS buffer and incubated with 10 μM H2O2 (Sigma, St. Louis, MO), 10 ng/ml interferon-γ (IFNγ), 10 ng/ml interleukin-1α (IL-1α), or 10 ng/ml tumor necrosis factor α (TNFα, R&D Systems, Minneapolis, MN) in a serum-free medium for 24 hr. The culture supernatants were collected, and cell debris was removed by centrifugation. For protein lysate preparation, cell pellets were lysed in buffer containing 50 mM HEPES, pH7.5, 0.5 mM EDTA, 50 mM NaCl, 1% CHAPS and protease inhibitor cocktail (Roche, Indianapolis, IN). Proteins were extracted for 30 min on a rocking platform at 4°C, and the insoluble cell debris was removed by centrifugation at 15,000 × g for 10 minutes, followed by determination of the protein concentrations (MicroBCA, Pierce Biotechnology, Rockford, IL).
Microarray fabrication and preparation
Antibody microarrays were prepared as described previously22, 31. A piezoelectric non-contact printer (Biochip Arrayer, PerkinElmer Life Sciences, Waltham, MA) was used to spot approximately 350 pl of each antibody solution on the surfaces of ultrathin nitrocellulose-coated microscope slides (PATH slides, GenTel Biosciences, Madison, WI). Forty-eight identical arrays were printed on each slide, spaced by 4.5 mm, arranged with four arrays across the width of the slide and 12 arrays along the length. A wax border was precisely imprinted around each of the arrays to define hydrophobic boundaries (SlideImprinter, The Gel Company, San Francisco, CA). The slides were stored in a vacuum-sealed container with desiccant at 4°C. When ready to use, the slides were restored to room temperature before breaking the vacuum seal (to prevent condensation on the slides) and washed in PBST0.5 (PBS buffer containing 0.5% Tween-20) three times, three minutes each, to remove unbound antibodies and to clean the surfaces.
Antibody microarray assays
The arrays were used either to detect core proteins, in a sandwich immunoassay, or to detect glycan structures on captured proteins. The antibody sandwich assays were performed as previously described with a few modifications22, 32. The slides printed with antibody arrays were blocked in a 1% BSA solution in PBS containing 0.5% Tween-20 (PBST0.5) for 1 hr, after which they were washed in PBST0.5 three times, three minutes each, and dried by centrifugation at 160 × g for 1 min. When conditioned media were analyzed, culture supernatants were mixed with 1/10 volume of 10X protease inhibitor cocktail, and 6 μl of sample was applied to each array. When total cell lysates were analyzed, the lysates were diluted with PBS buffer to a concentration of 1.5 mg/ml and 6 μl of sample was applied to each array. After a 1-hr incubation, the slides were washed with PBST0.1 and dried as before. The arrays were incubated with biotin-labeled antibodies at a concentration of 1~10 μg/ml for 1 hr, washed and dried, and incubated with 2.5 μg/ml streptavidin-phycoerythrin (Invitrogen, Carlsbad, CA) for another hour. The slides were washed, dried, and scanned for fluorescence emission at 570 nm using a microarray scanner (LS Reloaded, TECAN, Durham, NC). All arrays probed with the same detection antibody were scanned in one batch at a single laser power and detector gain setting.
The slides used for lectin detection were first chemically derivatized as described before22. Briefly, the slides were incubated in a coupling buffer (0.1M sodium acetate, pH5.5) with 0.1% Tween-20 for 10 min, transferred to a 200 mM NaIO4 solution in coupling buffer (Pierce Biotechnology, Rockford, IL) and incubated at 4°C for 5 hr in the dark to oxidize the glycan groups on printed antibodies. The slides were rinsed in coupling buffer and incubated with a solution containing 1mM MPBH (4-(4-N-Maleimidophenyl) butyric acid hydrazide hydrochloride) and 1 mM Cysteine-Glycine dipeptide (Pierce Biotechnology, Rockford, IL) in coupling buffer for 2 hr at room temperature to derivatize the carbonyl groups. The slides were rinsed with PBST0.1 and incubated with 1 mM Cysteine-Glycine in PBST0.1 overnight at 4°C. After the slides were rinsed thoroughly with PBST0.1 and dried by centrifugation, they were processed as above. Biotin-labeled lectins (at a concentration of 10 μg/ml in PBST0.1 buffer containing 0.1% BSA) were used as the detection reagents.
Microarray data analysis
The image data were quantified using microarray image processing software (GenePix Pro 5.1, Axon Instruments, Union City, CA). The local backgrounds were subtracted from the median intensity of each spot, yielding the net signal. Spots with defects or highly abnormal backgrounds were eliminated from the analysis. The signal intensities from replicate antibody measurements were averaged (geometric mean). The Student’s t-test in Microsoft Excel was used to compare data between groups.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis, Immunoprecipitations, and immunoblotting
RESULTS
Characterization of pancreatic cancer cell lines
The goals of this work were to examine whether mucin glycosylation can be affected by pro-inflammatory stimuli and to determine if these changes are related to particular cell-surface marker phenotypes. To characterize the phenotypes, the basal expression of three cell surface markers that are associated with tumorigenicity in pancreatic cancer (cancer-stem-cell markers), CD44, ESA, and CD24, were determined in 15 cell lines by flow cytometry (Figure 1A). All 15 cell lines expressed CD44; all but four expressed CD24; and two of the four CD24-negative lines also did not express ESA. The receptor-tyrosine-kinase MET has been implicated as a marker for stem-cell characteristics in pancreatic cancer33. Most of the cell lines showed high MET expression (Figure 1A), except for AsPC-1 and MIAPaCa, which were also negative in CD24 and ESA expression.
Figure 1.
Characterization of pancreatic cell lines. (A) Expression profile of pancreatic tumorigenic markers and the tyrosine kinase MET. CD24, CD44, and ESA cell surface levels were determined by flow cytometry, and MET total protein levels were measured by Western blot. * MET expression in HPDE cells was not analyzed. (B) Mucin expression patterns. The columns indicate the protein levels measured by antibody microarrays, using the indicated capture and detection antibodies. The values from the conditioned media and cell lysate samples were normalized to set the maximum values at the same level. The bands in the bottom panels represent the transcript levels of the corresponding mucins as measured by RT-PCR analysis.
In the same cell lines we characterized the basal expression of three cancer-associated mucins that are found in the blood of a high percentage of pancreatic cancer patients: MUC1, MUC5AC, and MUC16. The expression of the three mucins was measured at the transcript level by RT-PCR and at the protein level by antibody arrays. All the cell lines expressed some MUC1; nearly all expressed MUC5AC; and a few expressed a high level of MUC16 (Fig. 1B). The RNA and protein expression levels generally correlated well, which supports the reliability of the results and indicates that these protein levels are largely transcriptionally regulated. The protein measurements also were compared between antibody arrays and Western blots (Supplementary Figures 1 and 2). The assays agreed well, confirming the accuracy and specificity of these antibody array assays for measuring protein levels. Some differences between the RT-PCR, Western blot, and sandwich assays are expected since they are measuring divergent molecular forms.
The protein levels in the cell lysates generally correlated with the levels in the culture supernatants (Fig 1B), indicating that the release from the cell surface is proportional to the total levels. MUC1 and MUC16 are transmembrane proteins that can be released from the surface, and MUC5AC is a secreted mucin that can also be found in the cell lysates because of the formation of surface-associated gels. No strong correlations were observed between mucin expression and tumorigenic-marker status, although AsPC-1 and MIAPaCa, the two cell lines expressing only one marker (CD44), also expressed low basal levels of all three mucins. Therefore, based on the expression patterns of the tumorigenic markers, MET kinase, and the tumor-associated mucins, AsPC-1 and MIAPaCa have a different phenotype than the other pancreatic cell lines.
Profiling mucin expression and glycosylation changes
We selected six cell lines that would represent different types of surface-marker status. Two cell lines were chosen that express all three surface markers (BxPC3 and MPanc-96); two that express two markers (PANC-1 and SW1990); and two that express just one marker (AsPC-1 and MIAPaCa). In order to conveniently measure changes in both the core protein levels and glycosylation of mucins, we used antibody microarrays that can be run to capture either type of information (Figures 2A and 2B)22. The six cell lines were cultured and treated with IFNγ, IL-1α, TNFα, H2O2, or normal media (as a control), and the culture supernatant from each condition was collected for analysis after 24 hours. The expression of the receptors for these cytokines was confirmed in each cell line by RT-PCR (Supplementary Figure 3), simply to confirm that the cell lines would be able to respond to these stimuli. Each sample was run multiple times on successive antibody arrays, with each array probed with a different detection reagent: either an antibody targeting one of the core proteins (to obtain the protein levels at the corresponding capture antibodies); an antibody targeting the carbohydrate antigen CA19-9; or one of 15 different lectins targeting various glycans on the captured proteins. The use of 15 different lectins provided a broad screen for potential glycosylation changes. These capture and detection reagents are listed in Table 1.
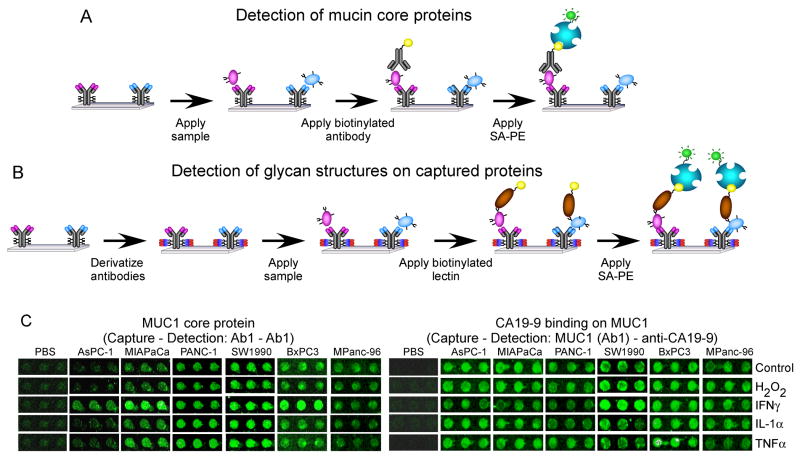
Figure 2.
Detection of mucin core proteins and glycans on antibody arrays. (A) Experimental scheme of mucin core protein detection. (B) Experimental scheme of glycan detection on captured proteins. (C) Scanned fluorescence images of portions of antibody microarrays. Triplicate anti-MUC1 (Ab1) capture antibody spots are shown from arrays incubated with various samples and probed with either anti-MUC1 (Ab1) or the CA 19-9 antibody.
Representative antibody spots from the arrays are shown in Figure 2C. The spot intensities were compared between conditions to assess changes in the protein and glycan levels. The entire data set is presented in Figure 3A. Several protein and glycan levels greatly increased or decreased (up to about eight-fold) after treatment, relative to the control. The largest changes were MUC1 core protein elevations and the elevated binding of the BPL, RCA, and SBA lectins to captured MUC1, MUC5AC and MUC16.
Figure 3.
Clusters of mucin expression and glycosylation changes in response to various treatments. (A) Complete data from the initial screening experiments. The column labels indicate the cell line and the treatment, and the row labels indicate the detection reagent and the capture antibody. The column labels are color-coded by the surface-marker status of the cell line: red indicates three markers, black indicates two markers, and green indicates one marker. The values from each treatment were divided by the values from the corresponding control, and each square represents those ratios, after log transformation (base 2) and using the scale indicated by the color bar. Yellow squares indicate elevations relative to control, and blue squares indicate reductions relative to control. The red boxes indicate groups of measurements showing the greatest changes relative to the controls. (B) Results from repeat data sets. The data from three independent experiment sets were treated as above and averaged prior to log transformation. The red boxes highlight results from the SBA, BPL, and RCA detection reagents, as well as the MUC1 core protein.
Based on the results from the above experiment set, we sought to confirm the reproducibility of selected assays and more precisely evaluate the glycosylation and mucin alterations. Each of the six cell lines was treated as before in three independent cultures, and the supernatants were probed with the same detection antibodies as before (anti-MUC1, anti-MUC5AC, anti-MUC16, and anti-CA19-9), but with a limited set of lectins (BPL, RCA, and SBA). The averaged data from the replicate experiments are shown in Figure 3B. (The raw data from each replicate set are available in the supplementary materials.) The patterns of mucin and glycan alteration were similar to the first set of experiments, indicating good reproducibility in the responses and in the analytical method. The average coefficient of variation (CV) between the three replicates (calculated individually for each measurement and averaged over the measurements) was 14.9%, also indicating a good overall reproducibility in the biological responses and the assay. The significance of individual measurements in the differences between conditions was tested using a t test, as described below.
Induced changes to mucin protein levels
We first determined the extent of induced changes to mucin core protein levels, as determined by the binding of the mucin detection antibodies on their corresponding capture antibodies (a sandwich immunoassay). MUC1 showed the greatest number of significant changes, relative to the control treatments (Fig. 4A and Supplementary Fig. 4). The observation of cytokine-induced control of MUC1 levels is consistent with previous findings in which MUC1 transcript levels were induced by IFNγ and TNFα in breast, airway, and hematopoietic systems34–36. Oxidative stress induced no significant protein changes. The MUC1 transcript levels, as determined by RT-PCR, correlated well with the protein levels (Supplementary Fig. 4), indicating that, in most cases, the changes in MUC1 secretion were likely due to changes in MUC1 gene expression. The levels of MUC5AC transcript and protein were not significantly influenced by the stimuli (Fig. 4A and Supplementary Fig. 5), suggesting that these cytokines are not strong regulators for MUC5AC expression in these cell lines. Secreted MUC16 protein levels were induced only in BxPC3 and SW1990, most strikingly by TNFα (with P-values 0.05 and 0.02, respectively). The MUC16 transcript levels also correlated well with the protein levels, as confirmed by RT-PCR in BxPC3 cells (Supplementary Fig. 5). These results indicate independent regulation of the different mucins, with MUC1 being the most regulated, MUC16 being induced only in specific cell lines and conditions, and MUC5AC rarely controlled by these stimuli.
Figure 4.
Comparisons of protein levels, glycan levels, and glycan-to-protein ratios. Mucin protein levels (A), glycan levels (B), and glycan-to-protein ratios (C) were clustered by hierarchical clustering. The cell lines and treatments are indicated by the column labels, which are color-coded by the surface-marker status of the cell line: red indicates three markers, black indicates two markers, and green indicates one marker. The detection and capture reagents are indicated by the row labels. The measurements from each treatment condition were averaged over the three replicate experiments and divided by the averaged values from the corresponding control conditions, resulting in ratios indicating changes relative to control (using the scale on the color bar). White boxes indicate significant changes (Student t-test, P<0.01) between treated and untreated.
Induced changes to mucin glycosylation
Next we investigated whether the glycosylation of the mucins can be modulated by inflammatory stimuli. We normalized each core protein measurement or each glycan measurement to the corresponding control measurements and clustered the resulting values (Figures 4A and 4B). The glycan levels on all three mucins showed some alterations (Fig. 4B), suggesting that inflammation-induced changes to glycosylation are more frequent than core protein changes, since only MUC1 showed frequent changes to core protein levels (Fig. 4A).
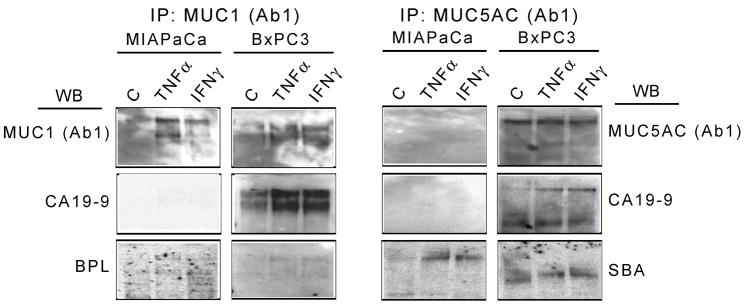
To evaluate the extent that the glycans were changing in relation to their underlying core proteins, we calculated the ratio of glycan-to-core-protein for each measurement and clustered the resulting ratios (Fig. 4C). Each ratio was normalized to its corresponding control measurement. Many of these glycan/protein ratios were significantly altered relative to the control conditions, supporting the concept that glycosylation on mucins indeed can be regulated by pro-inflammatory stimuli. The possibility exists that for some of the observed changes, it is not the glycans that are changing but rather simply the accessibility of the glycans, for example due to protein conformational changes or protein-protein interactions. To determine if indeed certain glycan structures were altered, we performed immunoprecipitations of the proteins followed by Western blots to detect the protein and glycan levels. For these selected measurements, the results agreed very well with the antibody microarray results (Fig. 5). For example, SBA binding shows selective elevation on MUC5AC from MIAPaCa; and MUC1 protein is elevated while MUC5AC protein is relatively constant. The CA 19-9 levels were up-regulated on MUC1 and MUC5AC in BxPC3, consistent with the microarray data, but were not detectable on both mucins in MIAPaCa by Western blot, perhaps because of reduced sensitivity of immunoblotting relative to antibody microarrays (unpublished observations). These results confirm that the glycans are changing, rather than being obscured somehow. Furthermore, the correspondence of the bands detected for glycans to the bands detected for proteins confirms that these glycans are attached to MUC1 and MUC5AC rather than on associated proteins.
Figure 5.
Assessment of glycan changes by immunoprecipitation and Western blotting. MIAPaCa and BxPC3 cells were left untreated (control) or treated with TNFα or IFNγ for 24 hours. Conditioned media were collected, immunoprecipitated with anti-MUC1 (Ab-1) or anti-MUC5AC (Ab-1) antibodies, and immunoblotted with the indicated antibodies or lectins. IP, immunoprecipitation; WB, Western blot; C, control.
Cell-type specific responses to pro-inflammatory stimuli
We next evaluated whether the responses to inflammatory stimuli were associated with cell phenotype, as defined by the expression of cell-surface markers. The grouping by cell type in the clusters of Figures 3 and 4 indicated that the induced glycan changes were often similar within a particular cell line and different between cell lines. Furthermore, the cell lines appeared to group according to surface-marker status, with the one -marker lines, AsPC-1 and MIAPaCa, showing different responses than the other cell lines, both in gains and losses of particular glycan structures (Figures 3 and 4).
We examined the significance of the differences between the high-marker cell lines and the low-marker cell lines in their responses to stimuli. The three replicate measurements from both the AsPC-1 and MIAPaCa cells (one-marker cell lines) were compared to the replicate measurements from both the BxPC3 and MPanc-96 cells (three-marker cell lines) using a t-test. Twenty-two significant differences were found, some with high significance (Supplementary Table 3). To investigate the contribution of chance associations, the groupings of cell lines were rearranged and the comparisons repeated. When we combined a one-marker cell line and a three-marker cell line together in each group, no significant differences (p > 0.01 for all measurements) occurred for one of the two combinations, and the other combination showed just two differences at a significance of p = 0.01 (data not shown). The highly-increased significance using the proper grouping suggests a real difference between the cells expressing three markers and those expressing one marker. In general, SBA binding increased in the one-marker cells and decreased in the three-marker cells, while the opposite was true for BPL, RCA, and CA 19-9 binding.
In order to gain more insights about what structures might define the differences between some of the cell lines, we combined the binding information from all the lectins, with reference to their particular specificities. We selected the changes occurring on MUC5AC secreted from the MIAPaCa and BxPC3 cell lines after stimulation with TNFα. Based on the determination of which lectins showed increased, equivalent, or reduced binding after stimulation (using the data from Fig. 4C and 3A), combined with knowledge of the specificities of the lectins (Table 1), we constructed a table of which glycan motifs showed up or down regulation (Fig. 6). (Data from the lectins Vicia Villosa and Helix Pomatia were obtained subsequent to the initial screen in order to gain additional insights into possible changes to terminal GalNAc, which is targeted by those glycans.) Since multiple structures may contribute to the binding levels of each lectin, conclusions cannot be made about particular structures, but this view can give information on the total levels of certain terminal groups. Looking at the greatest differences between the cell lines, and which motifs show consistency between the lectins that bind them, it appears that MIAPaCa could be showing increased chain lengths (increased polylactosamine) but a loss in terminal β1,4-linked Gal (loss in RCA binding). This loss in terminal Gal could be due to sialylation of the Gal (RCA binding is inhibited by sialylation) or to termination instead with GlcNAc, which is indicated by the strong SBA binding induction. BxPC3, on the other hand, shows decreased chain lengths but a gain in terminal β1,4-linked Gal. BPL binding patterns correlated strongly with those of RCA (Figs. 3 and 5). BPL binds both β1,3-linked and β1,4-linked terminal Gal, and RCA binds primarily β1-4-linked terminal Gal, so their correspondence supports primarily alterations to β1-4-linked terminal Gal.
Figure 6.
Changes to glycan motifs bound by lectin panels. Each square in the clusters represents the glycan/protein ratio (normalized to the control ratios) at the MUC5AC capture antibody detected with a particular lectin (indicated in the column labels) using media from either MIAPaCa (left cluster) or BxPC3 (right cluster) after treatment with TNFα. The ratios from Fig. 4C were used for the lectins SBA, RCA, and BPL, and for the CA 19-9 antibody. For the other lectins, the glycan/protein ratios were calculated from the data of Fig. 3A. Yellow indicates elevated relative to control, and blue indicates decreased relative to control. The highlighted lectins and glycans indicated major differences between the cell lines.
Therefore, divergent and seemingly complementary alterations are observed between these representative 1-marker and 3-marker cell lines. Other changes are common between the cell lines, such as loss of the TF antigen. Further experimentation will be required to define which structures are responsible for the lectin-binding patterns.
DISCUSSION
This work explored the question of the origin of the altered mucin glycosylation that is often seen in cancer. We demonstrated that pro-inflammatory signaling can affect both mucin protein expression and glycosylation and that glycosylation alterations may be cell type-dependent. The fact that glycans on tumor cells are remodeled in response to cytokine signaling has implications for understanding the origin and functions of cancer-associated glycans. Since pancreatic tumor cells are usually part of an inflammatory environment, they are exposed to a variety of cytokines and growth factors. The emergence of particular glycan structures on those cells, some of which have been shown to be functionally important in cancer progression, may be in response to that environment. A detailed study exploring induced glycosylation changes on multiple proteins was not previously possible due to limitations in the available technologies. Antibody-lectin sandwich arrays allowed broad profiling of changes in many different glycan structures on multiple proteins.
Clearly much diversity was present between the cell lines in their glycan alterations. Tumors are indeed heterogeneous, comprising sub-populations of transformed cells with divergent behaviors. The variability in tumorigenicity associated with cell-surface markers is a prominent example of that heterogeneity28–30. This study provided evidence that the cell surface markers that define tumorgenic potential in pancreatic cancer may be associated with particular types of glycan remodeling in response to cytokine stimulation. Further work with primary tumor cells would provide more direct evidence for this conclusion.
Detailed information about structures cannot be drawn from lectin binding patterns, although some conclusions can be drawn from panels of lectins regarding primarily terminal structures. Other methods are needed to characterize complete structures, which could be the goal of a future studies. It appears that the changes most associated with a representative one-marker cell line, MIAPaCa, are increased chain extensions with reduced terminal β1,4-linked Gal, along with increased terminal GlcNAc or GalNAc (Fig. 6). The representative three-marker cell line, BxPC3, showed largely the opposite changes, although both showed reductions in the Core-1 TF antigen. These changes could relate to several glycan alterations that have been shown to be important in human cancer. For example, alterations to the amount of lactosamine chains, as observed here, is largely controlled by glycan branching, which has been associated with cancer both on N-glycans37, 38 and O-glycans39. Lactosamine extension also is associated with the formation of the Lewis blood group structures (such as the CA 19-9 antigen), since those structures are found on the termini of lactosamine chains10, 39–41. Most cancer samples show an increase in the TF antigen42, rather than a decrease as observed here, but this study does show that the TF antigen can be modulated by cytokines, so the particular direction of that modulation might be different in our conditions.
These types of glycan alterations could have functional consequences and also related to the behavioral differences between tumorigenic and non-tumorigenic cancer cells. The Lewis blood group structures that are increased with O-glycan extension and branching are ligands for selectin receptors found on endothelial cells and lymphocytes43, and increased presentation of this structure on pancreatic cells leads to increased metastases44–46 and reduced survival in pancreatic cancer47, 48. Increased presentation of the TF antigen, resulting from halted O-glycan extension and the exposure of the Core-1 structure, can be mitogenic49, 50 or pro-metastatic through interations with the protein galectin-351. In cancer patients, both the truncated and extended O-glycans are present on mucins in varying degrees, which is consistent with different cell populations, perhaps a tumorigenic and non-tumorigenic population, preferentially producing either kind. It will be important to determine whether the cell-type-specific variation in inflammatory response observed in this work also occurs in primary tumor cells, which would help to explain behavioral variation and facilitate strategies to target highly tumorigenic cells.
A potential source of ambiguity in this method is that glycosylation can affect the affinities of antibodies, which would mean that the ability to capture the core protein of particular glycoforms would be compromised. The main antibody we used to detect MUC5AC, the 45M1 clone, binds an epitope in one of the cysteine-rich domains of the molecule52, which is not heavily glycosylated. The binding of 45M1 is sensitive to denaturation52, but no data has been presented on the effects of glycosylation. The other anti-MUC5AC antibody we used, CLH2, binds in the tandem repeat domain53, and again its sensitivity to glycosylation is not known. The protein levels and glycan levels detected on these two antibodies correlate extremely well for multiple detection reagents (Fig. 3a). Therefore, it is unlikely that glycoforms greatly affect the binding of these antibodies, since they bind very different regions of the molecule yet correlate so well. One of the clones we used to detect MUC1, VU-11D1, binds in the tandem repeat region but does not show strong variability based on glycosylation, as reported from a workshop on the properties of anti-MUC1 monoclonal antibodies54. Information on the glycosylation sensitivity of our other anti-MUC1 antibody, 1.B.831, is not available. However, the close correlation between the clones in the overall protein and glycosylation profiles (Fig. 3a) indicates that they are probably not greatly affected by glycosylation, unless they are all affected in the same way.
We did not address the mechanisms linking the external stimuli to the glycosylation alterations, but it may be valuable in future studies to explore the signaling pathways and transcriptional responses involved. Previous studies have shown that mucins are transcriptionally-regulated by pro-inflammatory cytokines and hormones6, 34, 35, 55, 56, and that those responses act through the transcription factors NF-κB/SP1/STAT1 and the MAPK and PKC signaling pathways. The MAPK pathway also was shown to be involved in the transcriptional regulation of the glycosyltransferase C2GnT16, which may link the MAPK pathway to the regulation of both mucin expression and glycosylation. Another mechanism of inducing glycan alterations would be through effects on the availability of particular monosaccharides. Determining the mechanism of glycan alteration may provide information on the metabolic state of the cell and the resulting behavioral responses to inflammation.
In conclusion, this study demonstrates that glycans on mucins secreted by cancer cells can be altered in response to pro-inflammatory signaling, and that particular phenotypes of cancer cells may have divergent glycan alterations. This relationship offers a possible molecular link between inflammation and the behavioral characteristics of sub-populations of pancreatic tumor cells. Antibody-lectin sandwich arrays provided a convenient means to profile changes in glycosylation among multiple proteins and cell lines. Future work will further define the nature and function of glycan structures that are associated with adaptive responses of subpopulations of cancer cells.
Supplementary Material
Acknowledgments
We thank Dr. Brian Cao (VARI) for kindly providing the anti-MET monoclonal antibody; Andrew Porter (VARI) and Adam Granger (University of California, San Francisco) for laboratory support; and Rich West (VARI) for the flow cytometry analysis. We gratefully acknowledge support of this work by the Michigan Technology Tri-Corridor (Grant 687 to the Michigan Proteomics Alliance for Cancer Research, to G.O., B.H., and Y.W.); the NCI (R21 CA122890, to B.H. and Y.W.); and the Van Andel Research Institute.
References
- 1.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–88. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 2.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochimica et biophysica acta. 1999;1473(1):21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 3.Burdick MD, Harris A, Reid CJ, Iwamura T, Hollingsworth MA. Oligosaccharides expressed on MUC1 produced by pancreatic and colon tumor cell lines. The Journal of biological chemistry. 1997;272(39):24198–202. doi: 10.1074/jbc.272.39.24198. [DOI] [PubMed] [Google Scholar]
- 4.Moniaux N, Andrianifahanana M, Brand RE, Batra SK. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. Br J Cancer. 2004;91(9):1633–8. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4(1):45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 6.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochimica et biophysica acta. 2006;1765(2):189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Dalziel M, Whitehouse C, McFarlane I, Brockhausen I, Gschmeissner S, Schwientek T, Clausen H, Burchell JM, Taylor-Papadimitriou J. The relative activities of the C2GnT1 and ST3Gal-I glycosyltransferases determine O-glycan structure and expression of a tumor-associated epitope on MUC1. The Journal of biological chemistry. 2001;276(14):11007–15. doi: 10.1074/jbc.M006523200. [DOI] [PubMed] [Google Scholar]
- 8.Sewell R, Backstrom M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, Gatgens J, Clausen H, Hansson GC, Burchell J, Taylor-Papadimitriou J. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. The Journal of biological chemistry. 2006;281(6):3586–94. doi: 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- 9.Beum PV, Singh J, Burdick M, Hollingsworth MA, Cheng PW. Expression of core 2 beta-1,6-N-acetylglucosaminyltransferase in a human pancreatic cancer cell line results in altered expression of MUC1 tumor-associated epitopes. The Journal of biological chemistry. 1999;274(35):24641–8. doi: 10.1074/jbc.274.35.24641. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi J, Shinohara H, Nonomura C, Ando H, Takaku S, Nojiri H, Nakamura M. Not core 2 beta 1,6-N-acetylglucosaminyltransferase-2 or -3 but -1 regulates sialyl-Lewis x expression in human precursor B cells. Glycobiology. 2005;15(3):271–80. doi: 10.1093/glycob/cwi005. [DOI] [PubMed] [Google Scholar]
- 11.Lim YC, Henault L, Wagers AJ, Kansas GS, Luscinskas FW, Lichtman AH. Expression of functional selectin ligands on Th cells is differentially regulated by IL-12 and IL-4. J Immunol. 1999;162(6):3193–201. [PubMed] [Google Scholar]
- 12.Tjew SL, Brown KL, Kannagi R, Johnson P. Expression of N-acetylglucosamine 6-O-sulfotransferases (GlcNAc6STs)-1 and -4 in human monocytes: GlcNAc6ST-1 is implicated in the generation of the 6-sulfo N-acetyllactosamine/Lewis x epitope on CD44 and is induced by TNF-alpha. Glycobiology. 2005;15(7):7C–13C. doi: 10.1093/glycob/cwi050. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Vallejo JJ, Van Dijk W, Van Het Hof B, Van Die I, Engelse MA, Van Hinsbergh VW, Gringhuis SI. Activation of human endothelial cells by tumor necrosis factor-alpha results in profound changes in the expression of glycosylation-related genes. Journal of cellular physiology. 2006;206(1):203–10. doi: 10.1002/jcp.20458. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Lehotay M, Anastassiades T, Harrison M, Brockhausen I. The effect of TNF-alpha on glycosylation pathways in bovine synoviocytes Biochemistry and cell biology. Biochimie et biologie cellulaire. 2004;82(5):559–68. doi: 10.1139/o04-058. [DOI] [PubMed] [Google Scholar]
- 15.Delmotte P, Degroote S, Merten MD, Van Seuningen I, Bernigaud A, Figarella C, Roussel P, Perini JM. Influence of TNFalpha on the sialylation of mucins produced by a transformed cell line MM-39 derived from human tracheal gland cells. Glycoconjugate journal. 2001;18(6):487–97. doi: 10.1023/a:1016038219183. [DOI] [PubMed] [Google Scholar]
- 16.Beum PV, Bastola DR, Cheng PW. Mucin biosynthesis: epidermal growth factor downregulates core 2 enzymes in a human airway adenocarcinoma cell line. American journal of respiratory cell and molecular biology. 2003;29(1):48–56. doi: 10.1165/rcmb.2002-0147OC. [DOI] [PubMed] [Google Scholar]
- 17.Higai K, Miyazaki N, Azuma Y, Matsumoto K. Interleukin-1beta induces sialyl Lewis X on hepatocellular carcinoma HuH-7 cells via enhanced expression of ST3Gal IV and FUT VI gene. FEBS letters. 2006;580(26):6069–75. doi: 10.1016/j.febslet.2006.09.073. [DOI] [PubMed] [Google Scholar]
- 18.Barber MD, Ross JA, Fearon KC. Changes in nutritional, functional, and inflammatory markers in advanced pancreatic cancer. Nutr Cancer. 1999;35(2):106–10. doi: 10.1207/S15327914NC352_2. [DOI] [PubMed] [Google Scholar]
- 19.Jamieson NB, Glen P, McMillan DC, McKay CJ, Foulis AK, Carter R, Imrie CW. Systemic inflammatory response predicts outcome in patients undergoing resection for ductal adenocarcinoma head of pancreas. Br J Cancer. 2005;92(1):21–3. doi: 10.1038/sj.bjc.6602305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrow B, Evers BM. Inflammation and the development of pancreatic cancer. Surg Oncol. 2002;10(4):153–69. doi: 10.1016/s0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 21.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Laroche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4(5):437–44. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 23.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6(6):985–9. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 24.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2(11):851–6. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 25.Zheng T, Peelen D, Smith LM. Lectin arrays for profiling cell surface carbohydrate expression. J Am Chem Soc. 2005;127(28):9982–3. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 26.Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconjugate journal. 2004;21(1–2):35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer research. 2007;67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 28.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 29.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 31.Orchekowski R, Hamelinck D, Li L, Gliwa E, van Brocklin M, Marrero JA, Vande Woude GF, Feng Z, Brand R, Haab BB. Antibody microarray profiling reveals individual and combined serum proteins associated with pancreatic cancer. Cancer research. 2005;65(23):11193–202. doi: 10.1158/0008-5472.CAN-05-1436. [DOI] [PubMed] [Google Scholar]
- 32.Forrester S, Kuick R, Hung KE, Kucherlapati R, Haab BB. Low-volume, high-throughput sandwich immunoassays for profiling plasma proteins in mice: identification of early-stage systemic inflammation in a mouse model of intestinal cancer. Molecular Oncology. 2007 doi: 10.1016/j.molonc.2007.06.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nature reviews. 2006;6(8):637–45. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 34.Lagow EL, Carson DD. Synergistic stimulation of MUC1 expression in normal breast epithelia and breast cancer cells by interferon-gamma and tumor necrosis factor-alpha. J Cell Biochem. 2002;86(4):759–72. doi: 10.1002/jcb.10261. [DOI] [PubMed] [Google Scholar]
- 35.Reddy PK, Gold DV, Cardillo TM, Goldenberg DM, Li H, Burton JD. Interferon-gamma upregulates MUC1 expression in haematopoietic and epithelial cancer cell lines, an effect associated with MUC1 mRNA induction. Eur J Cancer. 2003;39(3):397–404. doi: 10.1016/s0959-8049(02)00700-1. [DOI] [PubMed] [Google Scholar]
- 36.Thathiah A, Brayman M, Dharmaraj N, Julian JJ, Lagow EL, Carson DD. Tumor necrosis factor alpha stimulates MUC1 synthesis and ectodomain release in a human uterine epithelial cell line. Endocrinology. 2004;145(9):4192–203. doi: 10.1210/en.2004-0399. [DOI] [PubMed] [Google Scholar]
- 37.Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunology and cell biology. 2005;83(4):429–39. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 38.Saito H, Nishikawa A, Gu J, Ihara Y, Soejima H, Wada Y, Sekiya C, Niikawa N, Taniguchi N. cDNA cloning and chromosomal mapping of human N-acetylglucosaminyltransferase V+ Biochemical and biophysical research communications. 1994;198(1):318–27. doi: 10.1006/bbrc.1994.1045. [DOI] [PubMed] [Google Scholar]
- 39.Shimodaira K, Nakayama J, Nakamura N, Hasebe O, Katsuyama T, Fukuda M. Carcinoma-associated expression of core 2 beta-1,6-N-acetylglucosaminyltransferase gene in human colorectal cancer: role of O-glycans in tumor progression. Cancer research. 1997;57(23):5201–6. [PubMed] [Google Scholar]
- 40.Kim YS, Itzkowitz SH, Yuan M, Chung Y, Satake K, Umeyama K, Hakomori S. Lex and Ley antigen expression in human pancreatic cancer. Cancer research. 1988;48(2):475–82. [PubMed] [Google Scholar]
- 41.Maemura K, Fukuda M. Poly-N-acetyllactosaminyl O-glycans attached to leukosialin. The presence of sialyl Le(x) structures in O-glycans. J Biol Chem. 1992;267(34):24379–86. [PubMed] [Google Scholar]
- 42.Yu LG. The oncofetal Thomsen-Friedenreich carbohydrate antigen in cancer progression. Glycoconjugate journal. 2007;24(8):411–20. doi: 10.1007/s10719-007-9034-3. [DOI] [PubMed] [Google Scholar]
- 43.McEver RP. Selectin-carbohydrate interactions during inflammation and metastasis. Glycoconjugate journal. 1997;14(5):585–91. doi: 10.1023/a:1018584425879. [DOI] [PubMed] [Google Scholar]
- 44.Kishimoto T, Ishikura H, Kimura C, Takahashi T, Kato H, Yoshiki T. Phenotypes correlating to metastatic properties of pancreas adenocarcinoma in vivo: the importance of surface sialyl Lewis(a) antigen. Int J Cancer. 1996;69(4):290–4. doi: 10.1002/(SICI)1097-0215(19960822)69:4<290::AID-IJC9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 45.Iwai K, Ishikura H, Kaji M, Sugiura H, Ishizu A, Takahashi C, Kato H, Tanabe T, Yoshiki T. Importance of E-selectin (ELAM-1) and sialyl Lewis(a) in the adhesion of pancreatic carcinoma cells to activated endothelium. Int J Cancer. 1993;54(6):972–7. doi: 10.1002/ijc.2910540618. [DOI] [PubMed] [Google Scholar]
- 46.Kawarada Y, Ishikura H, Kishimoto T, Kato H, Yano T, Kato H, Yoshiki T. The role of sialylated Lewis antigens on hematogenous metastases of human pancreas carcinoma cell lines in vivo. Pathol Res Pract. 2000;196(4):259–63. doi: 10.1016/s0344-0338(00)80075-8. [DOI] [PubMed] [Google Scholar]
- 47.Ferrone CR, Finkelstein DM, Thayer SP, Muzikansky A, Fernandez-delCastillo C, Warshaw AL. Perioperative CA19-9 levels can predict stage and survival in patients with resectable pancreatic adenocarcinoma. J Clin Oncol. 2006;24(18):2897–902. doi: 10.1200/JCO.2005.05.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi S, Oda T, Hasebe T, Sasaki S, Kinoshita T, Konishi M, Ueda T, Nakahashi C, Ochiai T, Ochiai A. Overexpression of sialyl Lewis x antigen is associated with formation of extratumoral venous invasion and predicts postoperative development of massive hepatic metastasis in cases with pancreatic ductal adenocarcinoma. Pathobiology. 2001;69(3):127–35. doi: 10.1159/000048767. [DOI] [PubMed] [Google Scholar]
- 49.Campbell BJ, Finnie IA, Hounsell EF, Rhodes JM. Direct demonstration of increased expression of Thomsen-Friedenreich (TF) antigen in colonic adenocarcinoma and ulcerative colitis mucin and its concealment in normal mucin. The Journal of clinical investigation. 1995;95(2):571–6. doi: 10.1172/JCI117700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell BJ, Yu LG, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconjugate journal. 2001;18(11–12):851–8. doi: 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- 51.Yu LG, Andrews N, Zhao Q, McKean D, Williams JF, Connor LJ, Gerasimenko OV, Hilkens J, Hirabayashi J, Kasai K, Rhodes JM. Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. The Journal of biological chemistry. 2007;282(1):773–81. doi: 10.1074/jbc.M606862200. [DOI] [PubMed] [Google Scholar]
- 52.Lidell ME, Bara J, Hansson GC. Mapping of the 45M1 epitope to the C-terminal cysteine-rich part of the human MUC5AC mucin. The FEBS journal. 2008;275(3):481–9. doi: 10.1111/j.1742-4658.2007.06215.x. [DOI] [PubMed] [Google Scholar]
- 53.Reis CA, David L, Nielsen PA, Clausen H, Mirgorodskaya K, Roepstorff P, Sobrinho-Simoes M. Immunohistochemical study of MUC5AC expression in human gastric carcinomas using a novel monoclonal antibody. International journal of cancer. 1997;74(1):112–21. doi: 10.1002/(sici)1097-0215(19970220)74:1<112::aid-ijc19>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 54.Price M, Rye P, Petrakou E, Murray A, Brady K, Imai S, Haga S, Kiyozuka Y, Schol D, Meulenbroek M, Snijdewint F, von Mensdorff-Pouilly S, Verstraeten R, Kenemans P, Blockzjil A, Nilsson K, Nilsson O, Reddish M, Suresh M, Koganty R, Fortier S, Baronic L, Berg A, Longenecker M, Hilgers J, et al. Summary report on the ISOBM TD-4 Workshop: analysis of 56 monoclonal antibodies against the MUC1 mucin. San Diego, Calif. November 17–23, 1996. Tumour Biol. 1998;19(Suppl 1):1–20. doi: 10.1159/000056500. [DOI] [PubMed] [Google Scholar]
- 55.Theodoropoulos G, Carraway KL. Molecular signaling in the regulation of mucins. J Cell Biochem. 2007;102(5):1103–16. doi: 10.1002/jcb.21539. [DOI] [PubMed] [Google Scholar]
- 56.Andrianifahanana M, Singh AP, Nemos C, Ponnusamy MP, Moniaux N, Mehta PP, Varshney GC, Batra SK. IFN-gamma-induced expression of MUC4 in pancreatic cancer cells is mediated by STAT-1 upregulation: a novel mechanism for IFN-gamma response. Oncogene. 2007 doi: 10.1038/sj.onc.1210532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.