Abstract
The DNA of all organisms is constantly damaged by exogenous and endogenous agents. Base excision repair (BER) is important for the removal of several non-bulky lesions from the DNA, however not much is known about the contributions of other DNA repair pathways to the processing of non-bulky lesions. Here we utilized a luciferase reporter system to assess the contributions of transcription-coupled repair (TCR), BER and nucleotide excision repair (NER) to the repair of two non-bulky lesions, 8-oxoguanine (8OG) and uracil (U), in vivo under non-growth conditions. We demonstrate that both TCR and NER are utilized by Escherichia coli to repair 8OG and U. Additionally, the relative level of recognition of these lesions by BER and NER suggests that TCR can utilize components of either pathway for lesion removal, depending upon their availability. These findings indicate a dynamic flexibility of DNA repair pathways in the removal of non-bulky DNA lesions in prokaryotes, and reveal their respective contributions to the repair of 8OG and U in vivo.
Keywords: uracil, 8-oxoguanine, TCR, NER
1. Introduction
Most cells in nature are not in a constant growth state and are not engaged in continuous rounds of replication [1]. Thus, the functional viability of most cell populations likely depends more on the fidelity of transcription and translation than on replication. Maintenance of active regions of the genome is a biological priority as cells have evolved transcription-coupled repair (TCR), a system that preferentially targets repair of bulky DNA damage on the template strand of actively transcribed genes. TCR is mediated in E. coli by Mfd, which initiates DNA repair by recognizing a stalled RNA polymerase (RNAP), with subsequent targeting of nucleotide excision repair (NER) components [2].
Previous work from our laboratory demonstrated that non-bulky DNA damage such as 8-oxoguanine (8OG) and uracil (U) can be bypassed by RNAP in a mutagenic manner in E. coli leading to transcriptional mutagenesis (TM). Interestingly, it was also observed that the TM caused by 8OG was significantly elevated in the absence of Mfd, indicating that TCR acts on such oxidative DNA base damage in vivo [3]. Although base excision repair (BER) is the major pathway for transcription-independent repair of 8OG, a synergistic increase in TM in a TCR, BER-defective mutant reveals that Mfd utilizes components outside of BER to facilitate repair. While nucleotide excision repair (NER) proteins are known to interact with Mfd for repair of bulky DNA damage, they are not known to directly act on 8OG [4].
These above studies suggested a dynamic flexibility of DNA repair pathways for the repair of certain types of DNA damage. Such flexibility of pathway utilization could confer an ability for cells to adapt to changing environments and exposures to genotoxic agents. Genotoxic agents can introduce a spectrum of DNA damage at various locations throughout an organism’s genome [5]. If the majority of this DNA damage is handled by a single DNA repair pathway, genotoxic agent exposure could overwhelm single pathways, reducing expression of critical housekeeping genes, leading to cell death. It would be beneficial to the viability of an organism, therefore, if it were able to fall back on secondary, overlapping repair pathways to maximize DNA lesion removal. This repair flexibility would restore normal cell function quickly and reduce the likelihood of cell death due to genotoxic agent exposure. Indeed, it has been observed in the yeast Saccharomyces cerevisiae that there is overlap of DNA damage handling by BER, NER, recombination and translesion synthesis in replicating cells, and that compromising multiple pathways simultaneously is much more detrimental to the integrity of the genome than corruption of any pathway individually [6, 7]. While these previous studies did not examine which lesions specifically are subject to repair by such multiple pathways, they suggested that many oxidative DNA base damages could be accommodated by BER, NER and damage tolerance pathways. In the present study, we addressed whether flexibility of DNA repair pathways can be observed in E. coli for two spontaneous, frequently occurring non-bulky lesions, U and 8OG.
Since oxidative DNA base damage (8OG) is subject to TCR [3] in E. coli, it was of interest to expand our studies to other non-bulky lesions primarily repaired by BER. To address this issue, we chose to examine whether U was also repaired by TCR in vivo under non-growth conditions. Both U and 8OG are known to be mutagenic for both DNA polymerases and RNAPs [5, 8]. Additionally, repair by NER of non-bulky DNA damage was examined both as a possible component of TCR, and as a direct repair mechanism for these lesions. An important goal of this study was to determine whether these non-bulky lesions, which arise by very different mechanisms, are both subject to DNA repair flexibility under non-growth conditions.
2. Materials and Methods
2.1. Strain Constructions/Growth Conditions
The genotypes and sources of the E. coli strains used in this study are listed in Table 1. Cell growth media was liquid LB supplemented with the following antibiotics, when appropriate: kanamycin (50 μg/ml), tetracycline (15 μg/ml) and ampicillin (50 μg/ml). For newly constructed strains, generalized transduction was performed with phage P1 Δdam rev6 as described previously [9] with the donor and recipient designated P1(donor) x recipient in Table 1. Ultraviolet sensitivity of uvrA strains was confirmed for several tetracycline (BW1743 donor) or kanamycin (JW4019-2 donor) resistant transductants. ung strains were confirmed using λvir grown on dut-1 ung-1 strain, BW313 [10, 11].
Table 1.
Strains and Primers
| Strains | Description | Source |
|---|---|---|
| AB1157 | thr-1, araC14, leuB6 (Am), Δ(gpt-pro) lacY1, tsx-33, qsr’-0, glnV44 (AS), galK2 (Oc), Rac-0, hisG4 (Oc), rfbD1, mgl-51, rpoS396 (Am), rpsL31, kdgK51, xylA5, myl-1, argE3 (Oc), thi-1 | B. Weiss [30] |
| BD2008 | ung-151::Tn10 | [31] |
| BW313 | dut-1, ung-1 | B. Weiss |
| BW1743 | AB1157 uvrA::Tn10 | B. Weiss |
| DH12S | ϕ80dlacZM15, mcrA, Δ(mrr-hsdRMS-mcrBC), araD139, Δ(ara, leu)7697, ΔlacX74, galU, galK, rpsL, deoR, nupG, recA1 [F′proAB+ lacIqZΔM15] | P. Doetsch collection |
| JW4019-2 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), LAM-, rph-1, Δ(rhaD-rhaB)568, ΔuvrA753::kan, hsdR514 | Coli Genetic Stock Center, Yale University [32] |
| Z078 | AB1157 mutM::mini- Tn10 | [3] |
| Z079 | AB1157 mfd98::Tn5 | [3] |
| Z102 | AB1157 mfd98::Tn5, uvrA::Tn10 | P1(BW1743) × BW1603 |
| Z105 | AB1157 ung-151::Tn10 | P1(BD2008) × AB1157 |
| Z106 | AB1157 ung-151::Tn10, mfd98::Tn5 | P1(BD2008) × BW1603 |
| Z109 | AB1157 ΔuvrA753::kan | P1(JW4019-2) × AB1157 |
| Z110 | AB1157 ΔuvrA753::kan, mutM::mini- Tn10 | P1(JW4019-2) × Z078 |
| Z111 | AB1157 ΔuvrA753::kan, ung-151::Tn10 | P1(JW4019-2) × Z105 |
| Primers | Sequence | |
| Const-Normal | 5′phos-CGATTCCAATTAAGCGGGGGCCACCTGATATCCTTTGTATTTAAT-3′ | |
| Const-Stop | 5′phos-CGATTCCAATTAAGCGGGGGCCACCTGATATCCTTAGTATTTAAT-3′ | |
| Const-Ura | 5′phos-CGATTCCAATTAAGCGGGGGCCACCTGATATCCTTUGTATTTAAT-3′ | |
| Const-8oG | 5′phos-CGATTCCAATTAAGCGGGGGCCACCTGATATCCTT*GGTATTTAAT-3′ | |
2.2. Generation of Damage-Containing Construct
Constructs were prepared as previously described [3, 12]. The four constructs used in this study are designated as Normal/Normal, Stop/Stop, U/Stop and 8OG/Stop. The first word of the construct name refers to the primer used in the DNA polymerization reaction (Table 2), and the second word refers to the plus strand of the plasmids pBESTluc-f1-Normal or pBESTluc-f1-Stop.
Table 2.
Luciferase Activities 2 hr Following Induction in 8OG and U Repair-Proficient and Deficient Cells
| Straina | RLUb/106 cells ± SEMc | Mutant/Wild-Typed | p-Valuee |
|---|---|---|---|
| Normal/Normal | NTSf 5′ TAC AAA GGA 3′ TSg 3′ ATG TTT CCT 5′ |
||
| AB1157 | 228898 ± 155629h | 1.00 | |
| AB1157 ung | 124427 ± 15318 | 0.54 | 0.7494 |
| AB1157 mutM | 110627 ± 7749 | 0.48 | 0.6041 |
| AB1157 mfd | 134399 ± 29939 | 0.59 | 0.5738 |
| AB1157 uvrA(Tn10) | 218286 ± 45841h | 0.95 | 0.9484 |
| AB1157 uvrA(Tn10) mfd | 244658 ± 5075 | 1.07 | 0.9615 |
| AB1157 ung mfd | 76864 ± 15981 | 0.34 | 0.6428 |
| AB1157 uvrA(kan) | 8671 ± 1863 | 0.04 | 0.5036 |
| AB1157 uvrA(kan) mutM | 75942 ± 16282 | 0.33 | 0.6408 |
| AB1157 uvrA(kan) ung | 15990 ± 9225 | 0.07 | 0.5176 |
| Stop/Stop | NTSf 5′ TAC TAA GGA 3′ TSg 3′ ATG ATT CCT 5′ |
||
| AB1157 | 120 ± 39h | 1.00 | |
| AB1157 ung | 342 ± 138 | 2.86 | 0.3288 |
| AB1157 mutM | 112 ± 58 | 0.94 | 0.8813 |
| AB1157 mfd | 306 ± 76 | 2.56 | 0.1147 |
| AB1157 uvrA(Tn10) | 213 ± 73h | 1.78 | 0.2861 |
| AB1157 uvrA(Tn10) mfd | 110 ± 31 | 0.92 | 0.8420 |
| AB1157 ung mfd | 252 ± 153 | 2.11 | 0.4204 |
| AB1157 uvrA(kan) | 32 ± 5 | 0.27 | 0.1678 |
| AB1157 uvrA(kan) mutM | 126 ± 95 | 1.05 | 0.9443 |
| AB1157 uvrA(kan) ung | 134 ± 36 | 1.12 | 0.8288 |
| Ura/Stop | NTSf 5′ TAC TAA GGA 3′ TSg 3′ ATG UTT CCT 5′ |
||
| AB1157 | 193 ± 32h | 1.00 | |
| AB1157 ung | 11730 ± 711 | 60.69 | 0.0000000000017 |
| AB1157 mfd | 289 ± 73 | 1.50 | 0.1771 |
| AB1157 uvrA(Tn10) | 983 ± 90 | 5.09 | 0.000047 |
| AB1157 uvrA(Tn10) mfd | 1062 ± 83 | 5.50 | 0.0000000058 |
| AB1157 ung mfd | 55513 ± 10229 | 287.22 | 0.0000018 |
| AB1157 uvrA(kan) | 1928 ± 296 | 9.98 | 0.00000087 |
| AB1157 uvrA(kan) ung | 19448 ± 6784 | 100.62 | 0.0055 |
| 8OG/Stop | NTSf 5′ TAC TAA GGA 3′ TSg 3′ ATG 8TT CCT 5′ |
||
| AB1157 | 406 ± 89h | 1.00 | |
| AB1157 mutM | 19805 ± 7650 | 48.80 | 0.0029 |
| AB1157 mfd | 4848 ± 644 | 11.94 | 0.0000040 |
| AB1157 uvrA(Tn10) | 2267 ± 350 | 5.59 | 0.00010 |
| AB1157 uvrA(Tn10) mfd | 6035 ± 874 | 14.87 | 0.00000077 |
| AB1157 uvrA(kan) | 503 ± 78 | 1.24 | 0.4202 |
| AB1157 uvrA(kan) mutM | 57970 ± 8615 | 142.83 | 0.0000014 |
Full strain descriptions in Table 1.
Relative light units.
Each value is the average of at least three replicate samples ± standard error of the mean.
Ratio of mutant luciferase activity over repair-proficient cells luciferase activity for the same construct.
p-values for Student’s t-test comparison between mutant and repair-proficient cells luciferase activity for the same construct. Distributions were considered to be significantly different when p < 0.05.
Nontranscribed strand
Transcribed strand; U=uracil, 8=8-oxoguanine
This data included in [23]
2.3. TM Luciferase Reporter System (TM-LAS)
TM-LAS is illustrated in Supplementary Fig. 1. Competent cells were prepared as described previously [13] using a 1mM, pH 7.0 HEPES first wash. Luciferase assay was carried out as described previously [3, 14].
3. Results and Discussion
To investigate the dynamic flexibility of different DNA repair pathways to repair non-bulky DNA lesions in E. coli, we utilized DNA damage-tailored plasmid constructs and employed the transcriptional mutagenesis luciferase assay system (TM-LAS) (detailed in Supplementary Fig. 1) as a tool for the measurement of repair. In this system, the configuration of the damage-containing constructs places specific, known base damage across from a Stop codon sequence, enabling a measurement of transcriptional mutagenesis through production of a full-length luciferase protein product only when an unrepaired lesion is bypassed by RNAP (Supplementary Fig. 1B) [3]. Therefore, two factors can affect the total amount of active luciferase produced: (i) the rate of repair of the lesion by the available repair proteins in a cell, which will convert the lesion site to a Stop codon during repair synthesis, and (ii) the level of bypass by the RNAP for the lesion, which affects the total amount of full-length RNA produced from the luciferase gene. By comparing a single lesion (8OG or U) between strains with different DNA repair backgrounds, we were able to determine the relative roles of DNA repair proteins in the removal of this lesion. Thus TM caused by a U or 8OG is a probe for delineating the flexibility of DNA repair pathways.
By utilizing isogenic E. coli strains (Table 1) deficient in various components of DNA repair pathways, it is possible to mimic a state in which a DNA repair pathway is overwhelmed, such as would occur following exposure to a genotoxic agent, and investigate the repair of a specific base lesion under these conditions.
Several recent reports have addressed whether certain non-bulky DNA lesions are handled by TCR in bacteria [15], or mammals [16–20]. However, most of these studies have been performed in vitro [16, 18, 20]. Additionally, the few studies utilizing in vivo systems employed cells that were not under any growth restriction [15, 17, 19, 20]. Unrestricted continuous growth represents a living state rarely experienced by most cell types. It is conceivable that cells alter their priorities for DNA damage handling depending on whether they are dividing or not. These studies were carried out with E. coli under growth restriction, through use of the DNA gyrase inhibitor novobiocin. Novobiocin prevents DNA replication, and its use in these experiments provides a system that more closely resembles a natural non-growth state of bacteria [21].
3.1. TCR initiates repair of U in vivo
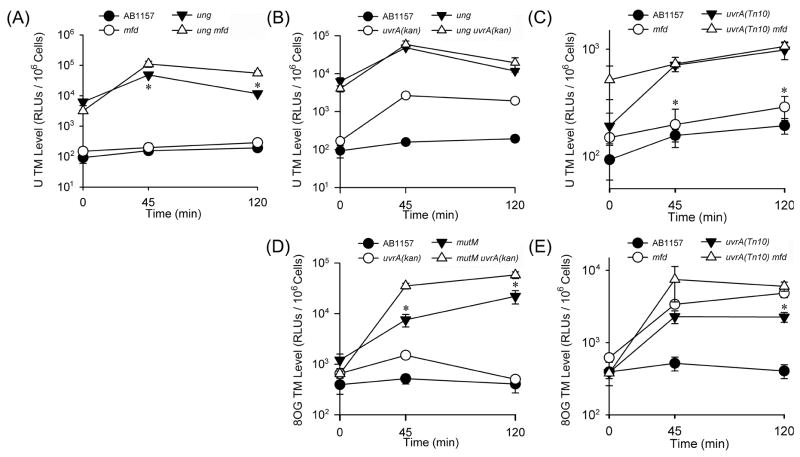
To determine whether TCR acts on non-bulky base lesions in E. coli, we assessed U-mediated TM in mfd mutants. Unlike what was previously observed for 8OG [3], we did not observe any significant increase in TM in the mfd single mutant (Fig. 1A, 1C and Table 2). Therefore, we examined TM in a double mutant, ung mfd. At 45 and 120 minutes following induction of transcription, U-mediated TM in the ung mfd double mutant was exacerbated 2- to 4-fold compared to the ung single mutant (Fig. 1A, Table 2). This result confirms a role for TCR in the repair of U in vivo. Therefore, despite the fact that both U and 8OG are primarily repaired by BER in replicating and non-replicating cells, these results indicate that TCR is able to participate in the removal of both lesions.
Fig. 1. DNA Repair Pathways Involved in Removal of Non-Bulky Lesions from DNA in vivo.
WT and DNA repair mutant strains (Table 2) were transformed with damage-containing construct, and the level of TM (measured via luciferase activity and expressed as relative light units per 106 cells) was measured at 0, 45, and 120 minutes following IPTG induction using TM-LAS methodology (Supplementary Fig. 1). Each data point represents the mean of at least three replicates ± SEM. Statistical significance was calculated using a Student’s t-test. Distributions were considered to be significantly different when p < 0.05.
(A) U/Stop construct. * Denotes points at which the single mutant, ung, is significantly different from the double mutant ung mfd. All points for ung and ung mfd are significantly different from the wild type and mfd strains.
(B) U/Stop construct. All points for ung and ung uvrA(kan) are significantly different from the wild type strain. At t=45 and 120 minutes uvrA(kan) is significantly different from the wild type strain.
(C) U/Stop construct. * Denotes points at which the single mutant, mfd, is significantly different from the double mutant uvrA(Tn10) mfd. All points for uvrA(Tn10) and uvrA(Tn10) mfd are significantly different from the wild type strain, but do not significantly differ from each other at any point.
(D) 8OG/Stop construct. *Denotes points at which the single mutant, mutM is significantly different from the double mutant mutM uvrA(kan). All points after t=0 for mutM and mutM uvrA(kan) are significantly different from the wild type strain. At t=45 the uvrA(kan) strain is significantly different from the wild type strain.
(E) 8OG/Stop construct. * Denotes points at which the single mutant, uvrA(Tn10), is significantly different from the double mutant uvrA(Tn10) mfd. All points after t=0 minutes for uvrA(Tn10), mfd, and uvrA(Tn10) mfd are significantly different from the wild type strain.
By eliminating the major uracil DNA glycosylase, Ung, U remains in the DNA for a prolonged period of time and abasic sites normally resulting from Ung- or Mug-mediated removal of U will be produced more slowly [22]. If Mfd recognizes abasic sites instead of U directly, no increase in TM would be observed in ung mfd double mutants compared to the ung mutant alone because there would be fewer abasic sites to direct recruitment of Mfd and TM would be unchanged or reduced. However, when loss of Ung is combined with elimination of Mfd, there is a significant increase in TM caused by U, revealing the role of Mfd-mediated TCR of U (Fig. 1A and Table 2).
3.2 NER directs repair of U in vivo
Because of the observations in yeast documenting the overlap of DNA damage repair by BER and NER [6, 7], we wanted to examine the role of NER in the repair of U. NER could be involved in the repair of U through direct recognition of the lesion, or via initiation by TCR (Mfd-mediated), as discussed above. We observed that in strains lacking UvrA, the initiating protein of the NER pathway, following transcription induction, there was a 5- to 10-fold increase in TM at 120 minutes compared to repair-proficient (WT) cells (Fig. 1B, 1C, and Table 2). When BER (Ung) and NER (UvrA) are removed simultaneously, considerable variability in the TM levels results (Fig. 1B), although the observed trend supports roles for both BER and NER in the repair of U under non-growth conditions. It is important to note that these data do not represent NER of repair intermediates, such as abasic sites, based on previous reports from our group and others that have indicated that NER is unlikely to be involved in the repair of abasic sites [23, 24].
Because the above results do not allow us to distinguish between a direct role for NER in the repair of U versus TCR-mediated NER, we utilized a uvrA mfd double mutant. This mutant displayed a five-fold increase in TM that was consistently elevated compared to that of the mfd strain but not the uvrA strain (Fig. 1C, Table 2). These data indicate that both Mfd and UvrA can prevent U-driven TM in vivo reflecting flexibility of repair of U in vivo. Additionally, these data indicate that a subset of UvrA-mediated NER is not associated with Mfd-mediated TCR, and that there is direct repair of U by NER. These results were surprising since NER is suspected to recognize helix distortion caused by bulky DNA damage [25], and U in DNA does not cause significant helix distortion. However, the fact that NER does not have the lesion recognition specificity characteristic of the N-glycosylases of the BER pathway could confer flexibility on NER for the repair of many types of lesions, even those that are considered to be helix non-distorting.
3.3 NER is involved in the repair of 8OG in vivo
The finding that NER was capable of U repair prompted us to examine whether NER could also repair 8OG. Although it was previously reported that Mfd is important for the repair of 8OG in vivo [3], no studies have addressed the potential role of NER directly. We observed that removal of NER results in a variable increase in TM up to 5-fold, depending on the time following initiation of transcription and the specific uvrA allele used, indicating that NER is involved in the repair of 8OG in vivo (Fig. 1D, 1E, and Table 2). To further confirm the role of NER, we utilized a uvrA mutM double mutant. When UvrA is eliminated together with the 8OG DNA glycosylase for BER (MutM), there is a synergistic increase in TM of three to five-fold compared to the TM caused by removing BER alone (Fig. 1D, Table 2), indicating that BER and NER may compete for this lesion in vivo, and confirming a role for NER in the repair of 8OG. These results suggest that sharing repair of a lesion among different excision repair pathways is a cellular strategy for eliminating several non-bulky base damages.
3.4 TCR of 8OG in vivo can utilize both BER and NER
Similar to the situation with U, it was important to consider the relative roles of NER in both direct recognition of 8OG, as well as its role in TCR-mediated repair of this lesion. At various times following induction of luciferase transcription, an increase in TM up to 12-fold over WT was observed in the individual uvrA and mfd mutant strains (Fig. 1D, 1E and Table 2). The mfd mutant demonstrated consistently higher TM than the uvrA mutants. Additionally, the uvrA mfd double mutant displayed an almost three-fold increase in TM compared to the isogenic uvrA single mutant at 120 minutes (Fig. 1E, Table 2). However, there was no significant difference in TM when the double mutant uvrA mfd was compared to the mfd single mutant (Fig. 1E, Table 2).
If repair of 8OG initiated by Mfd completed via the NER pathway, mfd would be epistatic to uvrA, and there would be no elevation of TM in the mfd uvrA double mutant compared to the uvrA mutant. However, there is a significant, three-fold increase in TM in mfd uvrA compared to uvrA, demonstrating that not all repair initiated by Mfd requires UvrA (NER pathway) involvement, thus substantiating a role for BER components in TCR. This result was completely unexpected as TCR proceeding through a BER mechanism has not been demonstrated previously in any organism.
BER may be the preferred pathway coupled to TCR initiated by Mfd due to its opposite base specificity. Because 8OG can pair with adenine as well as with cytosine, many organisms have evolved two separate N-glycosylases for the processing of 8OG. MutM, discussed above, removes the 8OG lesion from the DNA when opposite a cytosine residue, but has little activity when 8OG is opposite an adenine residue [26]. If MutM were to remove the 8OG when it was paired with adenine during repair synthesis, DNA polymerase would insert a mutagenic base (T). Therefore, when 8OG is paired with adenine, a separate N-glycosylase, MutY, will remove the adenine, providing the DNA polymerase with an opportunity to insert a non-mutagenic cytosine opposite the 8OG [26]. NER lacks such opposite base specificity, so that if NER is utilized to remove 8OG from the DNA instead of BER, it could result in increased TM. This may be the reason that cells prefer to utilize BER components during TCR over NER components in the repair of 8OG. Interestingly, there is a significant increase in TM for both U and 8OG when TCR and BER are eliminated simultaneously (Fig. 1A, Table 2 and [3]), demonstrating that these pathways may compete under certain conditions. Based on these observations, we propose that TCR can utilize either NER or BER components for repair of non-bulky base damage, with BER as the predominant repair pathway (Fig. 2).
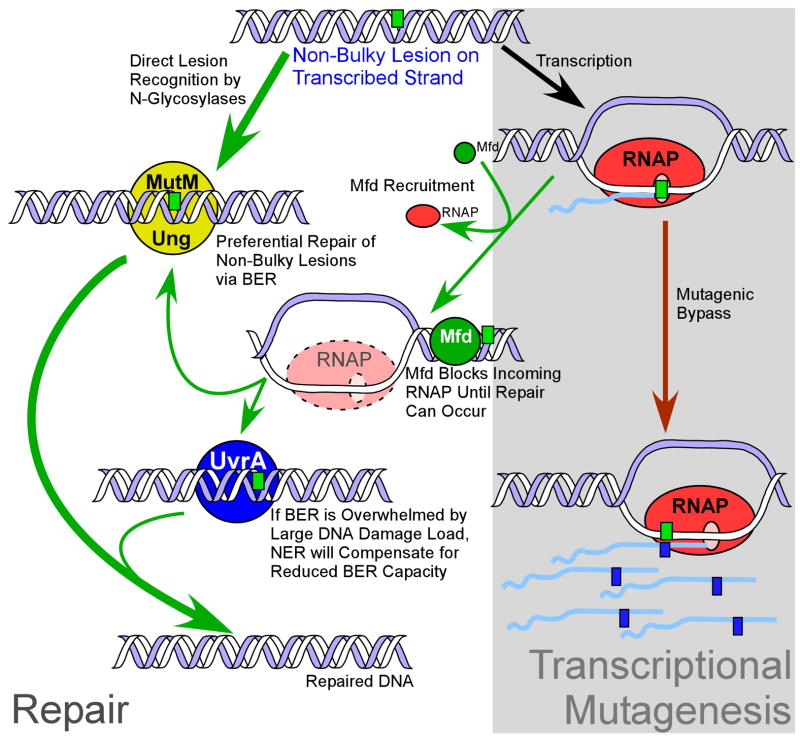
Fig. 2. Dynamic Flexibility of Interacting DNA Repair Pathways in E. coli.
Non-bulky lesions in DNA (small green box) are primarily repaired by the BER pathway. When these lesions are present on the template strand of a gene and are encountered by RNAP, at least two possible outcomes can result: 1. TM via insertion of an incorrect ribonucleotide opposite the lesion on the nascent mRNA (red arrow), or 2. Mfd-mediated TCR of the lesion (green arrows). Mfd removes RNAP from the lesion and subsequently acts to block incoming RNAP from bypassing the lesion in a mutagenic manner. Mfd leaves the damaged base accessible to BER. However, if the capacity of BER is overwhelmed by excessive DNA damage elsewhere, Mfd can recruit NER to repair the lesion, thus complementing the reduced BER capacity for the repair of small lesions.
3.3. A New Model for DNA Repair in E. coli
The dynamic flexibility of DNA repair of non-bulky lesions would provide cells with a greater capacity for repair, especially under conditions where one repair pathway is compromised or the capacity of a single pathway has been exceeded by the level of DNA damage. Such a situation could occur when a cell is exposed to a DNA damaging agent resulting in a variety of different DNA lesions introduced into the genome at different levels simultaneously. Thus a subset of such damages may only be primarily repaired by a single pathway (e.g. BER). In this study, through the use of E. coli DNA repair mutants it is possible to mimic a situation where one pathway is overwhelmed by damage, and unavailable for repair of other types of lesions. By employing TM-LAS to monitor repair, we revealed a role for TCR and NER in the repair of multiple, frequently occurring, non-bulky lesions. These results were unexpected since these pathways were suspected to only be used for bulky, helix-distorting lesions. In particular, TCR appears to be able to utilize components of either BER or NER, making the repair of damage in actively transcribed genes particularly flexible. As the majority of cells in nature are not actively dividing or replicating their DNA, dynamic flexibility of repair of active regions of the genome becomes particularly important for maintaining cell viability. Though in vitro studies of RNAP encountering many non-bulky lesions demonstrates no stalling during bypass [27–29], it is possible that in vivo the RNAP stalls a proportion of the time, recruiting Mfd to the damage and preventing further bypass by RNAP until repair can be completed. Therefore, the magnitude of increase in TM observed in the absence of Mfd is relatively modest due to the low frequency of RNAP stalling, and not because non-bulky lesions are poor substrates for TCR (Fig. 2).
Our results reveal a novel model of transcriptional encounters with non-bulky DNA damage (Fig. 2). Unrepaired lesions can be bypassed by RNAP, leading to TM by insertion of an incorrect ribonucleotide opposite the lesion into the mRNA [3]. Importantly, some proportion of the time, RNAP stalls at the damage, Mfd is recruited, and TCR is initiated. Mfd removes RNAP from the lesion and then acts as a placeholder, blocking incoming RNAP from bypassing the damage but leaving the lesion accessible to repair by BER. If BER capacity is exceeded, Mfd can recruit NER to complete repair of the lesion, compensating for the reduced capacity of BER. The discovery that NER components can be used for some level of U and 8OG repair is unexpected and reveals a plasticity of DNA repair pathway interrelationships in vivo.
Supplementary Material
Acknowledgments
We would like to thank current and past members of the Doetsch laboratory for critical reading of the manuscript and for helpful discussions. We would also like to thank Bernie Weiss for generously providing several bacterial strains used in this study and for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA120288 and National Institutes of Health Predoctoral Training Grant 5T32GM008490.
Abbreviations
- RNAP
RNA polymerase
- TM
transcriptional TM
- 8OG
8-oxoguanine
- U
uracil
- TM-LAS
transcriptional TM luciferase assay system
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair. 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 3.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 4.Grossman L, Lin C, Ahn Y. Nucleotide excision repair. In: Nickoloff J, Hoekstra M, editors. DNA Damage and Repair. Humana Press; Totowa, NJ: 1998. pp. 11–27. [Google Scholar]
- 5.Friedberg EC, Walker G, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D. C.: 2006. DNA damage; pp. 9–70. [Google Scholar]
- 6.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping Specificities of Base Excision Repair, Nucleotide Excision Repair, Recombination, and Translesion Synthesis Pathways for DNA Base Damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morey NJ, Doetsch PW, Jinks-Robertson S. Delineating the Requirements for Spontaneous DNA Damage Resistance Pathways in Genome Maintenance and Viability in Saccharomyces cerevisiae. Genetics. 2003;164:443–455. doi: 10.1093/genetics/164.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: Transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 9.Sternberg NL, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 10.Warner HR, Thompson RB, Mozer TJ, Duncan BK. The properties of a bacteriophage T5 mutant unable to induce deoxyuridine 5′-triphosphate nucleotidohydrolase. Synthesis of uracil-containing T5 deoxyribonucleic acid. J Biol Chem. 1979;254:7534–7539. [PubMed] [Google Scholar]
- 11.Hays JB, Duncan BK, Boehmer S. Recombination of uracil-containing lambda bacteriophages. J Bacteriol. 1981;145:306–320. doi: 10.1128/jb.145.1.306-320.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bregeon D, Doetsch PW. Reliable method for generating double-stranded DNA vectors containing site-specific base modifications. BioTechniques. 2004;37:760–766. doi: 10.2144/04375ST01. [DOI] [PubMed] [Google Scholar]
- 13.Seidman CE, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. Curr Protoc Mol Biol. 1997;1:1.8.1–1.8.10. doi: 10.1002/0471142727.mb0108s37. [DOI] [PubMed] [Google Scholar]
- 14.Bregeon D, Doetsch PW, Campbell JL, Modrich P. Methods Enzymol. Academic Press; New York: 2006. Assays for transcriptional mutagenesis in active genes; pp. 345–357. [DOI] [PubMed] [Google Scholar]
- 15.Smith AJ, Savery NJ. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair. 2008;7:1670–1679. doi: 10.1016/j.dnarep.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Dianov G, Bischoff C, Sunesen M, Bohr VA. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999;27:1365–1368. doi: 10.1093/nar/27.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastoriza-Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb/Ogg1 DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 20.Larsen E, Kwon K, Coin F, Egly JM, Klungland A. Transcription activities at 8-oxoG lesions in DNA. DNA Repair. 2004;3:1457–1468. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Sangurdekar D, Srienc F, Khodursky A. A classification based framework for quantitative description of large-scale microarray data. Genome Biol. 2006;R32 doi: 10.1186/gb-2006-7-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan BK, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982;151:750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clauson CL, Oestreich KJ, Austin JW, Doetsch PW. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proceedings of the National Academy of Sciences. 107:3657–3662. doi: 10.1073/pnas.0913191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedberg EC, Walker G, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D.C.: 2006. Nucleotide excision repair: General features and the process in prokaryotes; pp. 227–266. [Google Scholar]
- 26.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YH, Bogenhagen DF. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J Biol Chem. 1993;268:5849–5855. [PubMed] [Google Scholar]
- 28.Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC. Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2001;276:45367–45371. doi: 10.1074/jbc.M105282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan A, Doetsch PW. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J Biol Chem. 1998;273:21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 30.DeWitt SK, Adelberg EA. The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics. 1962;47:577–585. doi: 10.1093/genetics/47.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan BK. Isolation of insertion, deletion, and nonsense mutations of the uracil-DNA glycosylase (ung) gene of Escherichia coli K-12. J Bacteriol. 1985;164:689–695. doi: 10.1128/jb.164.2.689-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.