Abstract
Regulators of G protein signalling (RGS) proteins are united into a family by the presence of the RGS domain which serves as a GTPase-activating protein (GAP) for various Galpha subunits of heterotrimeric G proteins. Through this mechanism, RGS proteins regulate signalling of numerous G protein-coupled receptors. In addition to RGS domains, RGS proteins contain diverse regions of various lengths that regulate intracellular localization, GAP activity or receptor selectivity of RGS proteins, often through interaction with other partners. However, it is becoming increasingly appreciated that through these non-RGS regions, RGS proteins can serve non-canonical functions distinct from inactivation of Galpha subunits. This review summarizes the data implicating RGS proteins in (i) regulation of G protein signalling by non-canonical mechanisms, (ii) regulation of non-G protein signalling, (iii) signal transduction from receptors not coupled to G proteins, (iv) activation of mitogen-activated protein kinases, and (v) non-canonical functions in the nucleus.
Keywords: Regulator of G protein signalling, Heterotrimeric G protein, G protein-coupled receptor, Cell migration, Gene transcription, Nucleus
1. Introduction
Seven-transmembrane G-protein-coupled receptors signal through heterotrimeric G proteins by promoting GDP-to-GTP exchange on the Gα subunit, resulting in dissociation of Gα-GTP from Gβγ subunits and activation of their specific effectors. Gα subunits hydrolyze GTP through their intrinsic GTPase activity, which leads to inactivation of G proteins. This process is promoted by the regulators of G protein signalling (RGS) that function as GTPase-activating proteins (GAP) for Gα subunits [1, 2]. RGS proteins are united into a family by the presence of the RGS domain which serves as a GAP with some degree of specificity for various Gα subunits and receptors [3]. In addition to RGS domains, RGS proteins contain diverse regions of various lengths, often containing other defined domains. These unique regions may regulate intracellular localization, GAP activity or receptor selectivity of RGS proteins, often through interaction with other proteins (reviewed in [4–6]). However, it is becoming increasingly appreciated that the non-RGS regions of RGS proteins can serve non-canonical functions distinct from inactivation of Gα subunits, or even from G protein signalling entirely. This review summarizes the examples of five such novel types of RGS functions: (i) regulation of G protein signalling by RGS proteins through non-canonical mechanisms, (ii) regulation of non-G protein signalling by RGS proteins; (iii) RGS proteins as signal transducers, (iv) scaffolding function of RGS proteins in activation of mitogen-activated protein kinases, and (v) nuclear functions of RGS proteins.
2. Regulation of G protein signalling by RGS proteins through non-canonical mechanisms
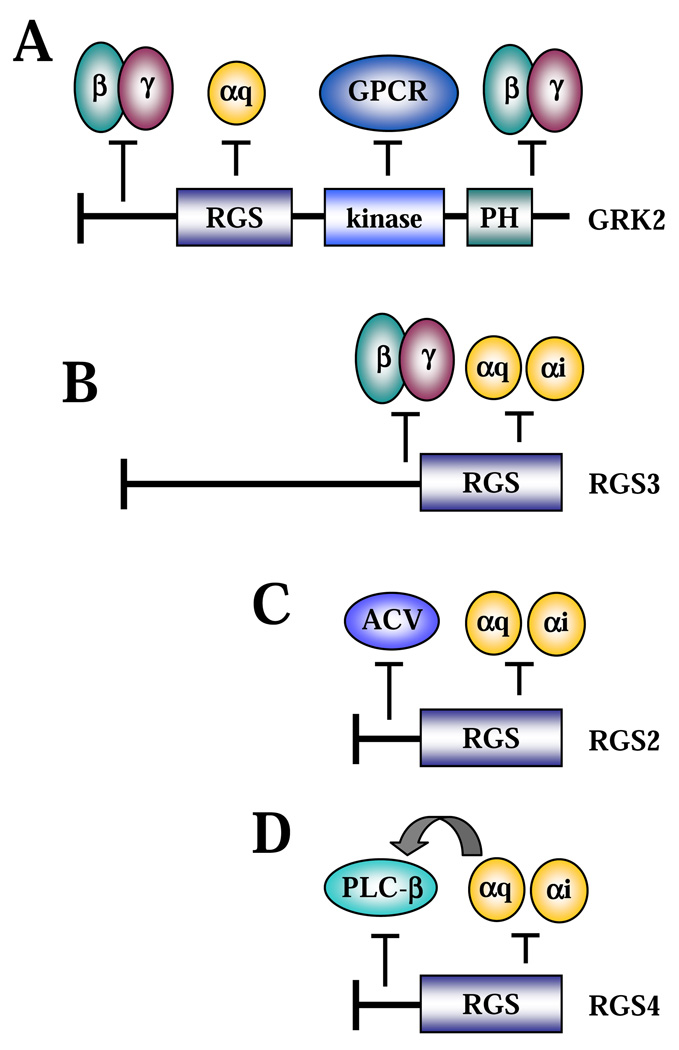
2.1. Regulation of Gβγ signalling by RGS proteins (Figs. 1A, 1B)
Figure 1. Regulation of G protein signalling by RGS proteins through non-canonical mechanisms.
A, B, regulation of Gβγ signalling by GRK2 and RGS3, respectively. C, regulation of ACV by RGS2. D, regulation of PLCβ by RGS4.
Gβγ subunits mediate activation of a number of GPCR effectors, including phospholipase C β (PLCβ) and adenylyl cyclase (AC) [7, 8]. G protein-coupled receptor kinase GRK2, also known as β-adrenergic receptor kinase, is the RGS family member that has been known for many years to interact with Gβγ subunits via its C-terminus [9]. In this manner, GRK2 acts as a Gβγ effector and functions to inhibit GPCR activity by either (i) phosphorylation of GPCRs, which leads to receptor desensitization [10], or (ii) inhibition of Gαq signalling by direct interaction via its RGS-homology domain [11, 12]. At the same time, binding of GRK2 to Gβγ prevents activation of other Gβγ effectors; and thus the C-terminus of GRK2 is widely used to dissect Gβγ signalling [13]. Recently, it was found that the N-terminus of GRK2 could also interact with Gβγ and inhibit Gβγ-induced inositol trisphosphate (IP3) production independently of its C-terminus or RGS domain [14] (Fig. 1A). In addition to GRK2, RGS3 can also bind Gβ1γ2, at least in part through a region N-terminal to the RGS domain [15] (Fig. 1B). Ectopic RGS3 inhibited Gβ1γ2-induced production of IP3,activation of extracellular signal regulated protein kinase (ERK) and Akt [15], or activation of a small GTPase Rac [16]. Considering the requirement for Gβγ signalling in chemotaxis, Gβγ binding may account for the more potent inhibition of migration of lymphoid cells to interleukin-8 or monocyte chemoattractant protein (MCP-1) by RGS3 as compared to RGS1 or RGS2 [17].
2.2. Regulation of adenylyl cyclases by RGS proteins (Fig.1C)
Adenylyl cyclases are commonly activated by Gαs [18] and inhibited by Gαi [19] subunits. However, many RGS proteins, such as RGS1, RGS2, RGS3, and RGS13, can also inhibit AC activation by Gαs [20–23]. This effect of RGS proteins is unlikely to be a result of GAP activity on Gαs, as (i) there is no evidence that these RGS proteins function as effective GAPs for Gαs, (ii) this inhibition (by RGS2) still occurs in the presence of the nonhydrolyzable analog GTPγS, and (iii) RGS proteins can inhibit forskolin stimulated AC in the absence of activated Gαs [23]. There is strong evidence that RGS proteins regulate AC activity through direct interaction with AC, at least in the case of RGS2 [22]. Deletion and alanine-scanning mutagenesis studies identified three critical amino acids in the N-terminus of RGS2 (outside of the RGS domain) that were required for its interaction with adenylyl cyclase V (ACV). Mutation of these residues or deletion of the N-terminus abrogated ACV interaction and suppression of cAMP production [24]. A recent study identified four different RGS2 protein products starting at methionines 1, 5, 16, and 33 of the full-length RGS2, which were results of alternative translation initiation sites. Consistent with the RGS2-AC interaction data, only the longer fragments containing the N-terminus inhibited cAMP production by constitutively active Gαs, whereas all RGS2 isoforms equally suppressed Gαq-induced IP3 generation and calcium flux [25]. Furthermore, the role of endogenous RGS2 in the regulation of AC was suggested by a study that showed that the increased expression of RGS2 in osteoblasts by ATP or forskolin accounted for the reduced parathyroid hormone-related peptide (PTHrP)-induced cAMP accumulation in wild type, but not in RGS2-knockout cells [26].
2.3. Regulation of phospholipase Cβ by RGS proteins (Fig. 1D)
PLCβ is an established effector of Gαq and Gβγ [27–29]. Activation of PLCβ is traditionally inhibited by various RGS proteins through accelerating GTP hydrolysis by Gαq [30–35], or non-canonically by competing with Gβγ [13, 15]. However, it was also suggested that RGS2, RGS4, and Gα-interacting protein GAIP can inhibit PLCβ in a GAP-independent manner through interfering with the Gαq-PLCβ interaction, as (i) they blocked PLCβ activation by constitutively-activated Gαq-GTPγS [34, 35], and (ii) excess amounts of PLCβ blocked the RGS4-Gαq interaction [34]. Consistent with this observation, recent studies showed that the N-terminal 33 residues of RGS4 (distinct from its RGS domain) can directly interact with the C-terminal tail of PLCβ [36]. Since the C terminus of PLCβ also binds Gαq, these data are consistent with the observation that the RGS4-PLCβ interaction is competed by activated Gαq [34]. Thus, RGS4 can inhibit PLCβ activity by disrupting its association with activated Gαq.
3. Regulation of non-G protein signalling by RGS proteins
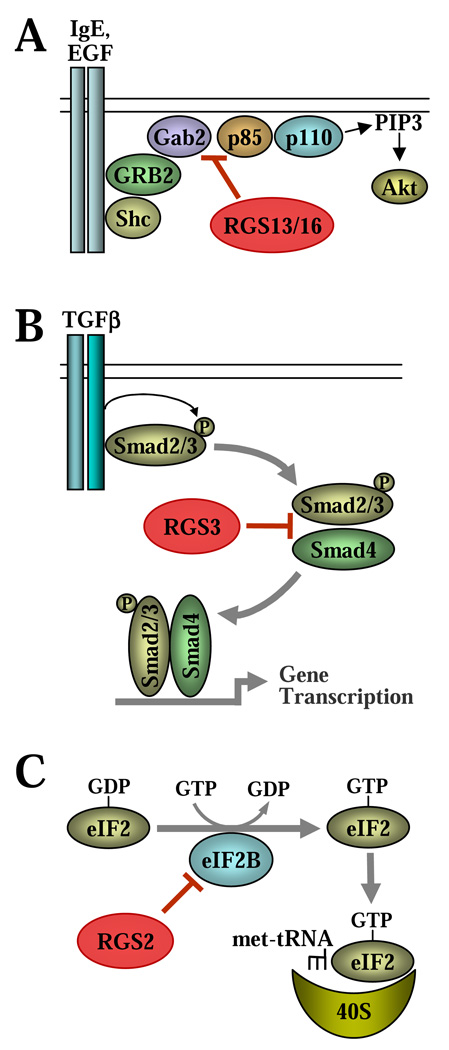
3.1. Regulation of receptor tyrosine kinase signalling by RGS proteins (Fig. 2A)
Figure 2. Regulation of non-G protein signalling by RGS proteins.
A, inhibition of PI3 kinase by RGS13 and RGS16. B, regulation of TGF-β signalling by RGS3. C, translational control by RGS2.
Recently, it was discovered that the p85α subunit of PI3 kinase interacts with several RGS proteins (RGS1, 4, 5, 13, and 16), of which RGS13 and RGS16 have been further characterized [37, 38]. In mast cells, exposure to antigen activates the receptor tyrosine kinase FcεRI and leads to activation of many signalling pathways. Notably, PI3 kinase is critical for antigen-induced degranulation of mast cells [39]. The mechanism of PI3 kinase activation involves recruitment of the regulatory p85 subunit to the IgE receptor complex by a tyrosine-phosphorylated adaptor protein called Grb2 associated binder 2 (Gab2) [40]. It was shown that RGS13 binds the p85 subunit and inhibits IgE-induced PI3K activity by interfering with the p85-Gab2 interaction [37]. The region of interaction was mapped to the N-terminal 51 residues of RGS13, which partially overlaps with the RGS domain (residues 34–149). Reconstitution of mast cells with RGS13 or the N-terminal 51-residue fragment inhibited mast cell degranulation, whereas knockout of RGS13 enhanced signalling events downstream of PI3 kinase (PIP3 production, phosphorylation of Akt and PLC-γ1, and calcium flux) and promoted degranulation of mast cells [37].
Similarly, RGS16 was shown to interact with p85α in the MCF7 breast cancer cell line, and this required residues 31–60 of RGS13 distinct from the RGS domain [38]. Binding of RGS16 to p85 also inhibited the p85-Gab1 interaction resulting in inhibition of EGF-induced AKT phosphorylation and proliferation of MCF7 cells, as shown by both RGS16-overexpression and knockdown approaches [38]. Interestingly, the p85-interacting regions of RGS13 and RGS16 have no significant homology, suggesting that the mechanisms of interaction are distinct.
3.2. Regulation of TGF-β signalling by RGS3 (Fig. 2B)
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that controls growth, survival and the phenotype of many cells. TGF-β signalling is largely mediated by phosphorylation of the “receptor-activated” R-Smads (Smad2/3/5/8) by the TGF-β receptor family, which leads to heteromerization of R-Smads with the “common-mediator” Co-Smad (Smad4). The R-Smad/Smad4 interaction is required for their accumulation in the nucleus and activation of specific gene transcription in cooperation with a variety of other co-activators [41, 42]. Regulation of TGF-β signalling is canonically mediated by the “inhibitory” I-Smads (Smad6 and Smad7) that interfere with receptor-mediated phosphorylation of R-Smads [43, 44].
A high-throughput screening of the components of TGF-β signalling for interacting partners predicted a potential interaction between several Smads and RGS3 (supplemental data in [45]). Subsequently, we demonstrated coimmunoprecipitation of ectopically expressed RGS3 with Smad2, Smad3 and Smad4, as well as showed interaction of endogenous RGS3 with Smad3 and Smad4 in EL4 thyoma cell line [46]. This interaction was mapped to the 270–379 region of RGS3 outside of the RGS domain, and to the Mad homology 2 (MH2) domain of Smad3 that mediates Smad heteromerization. Accordingly, RGS3 interfered with TGF-β - induced dimerization of Smad3 with Smad4 without affecting R-Smad phosphorylation; this translated to inhibition of Smad-mediated gene transcription by overexpression of RGS3 but not of its RGS domain [46]. We are currently investigating how endogenous RGS3 regulates TGF-β signalling and cell responses.
3.3. Regulation of protein synthesis by RGS2 (Fig. 2C)
In eukaryotes, initiation of mRNA translation during protein synthesis is controlled by a family of eukaryotic initiation factors (eIFs). The heterotrimeric GTPase eIF2 is an essential factor for protein synthesis that forms a ternary complex with GTP and the initiator Met-tRNA bringing it to the 40S ribosomal subunit. GTP binding of eIF2 is promoted by a pentameric guanine nucleotide exchange factor (GEF), eIF2B [47].
Recently, a yeast two-hybrid screen identified the epsilon subunit of eIF2B (eIF2Bε) as a binding partner of RGS2, and the interaction between eIF2Bε and RGS2 was demonstrated at the overexpressed and endogenous levels of these proteins [48]. Functionally, RGS2 inhibited the GEF activity of eIF2B, translation efficiency and protein synthesis; and these effects were independent on its GAP activity. The interaction was mapped to a 37-residue region which is homologous to the eIF2Bε-binding region of eIF2β. Mutation of conserved residues within this region abolished the eIF2Bε-RGS2 interaction and regulation of translation by RGS2 [48]. Interestingly, the eIF2Bε-binding region of RGS2 overlaps with the N-terminal part of the RGS domain, and the residues critical for eIF2Bε-binding by RGS2 are also conserved in some other RGS proteins. As such, it would be interesting to examine if other RGS proteins bind eIF2Bε and how this interaction is affected by G proteins.
4. RGS proteins as signal transducers
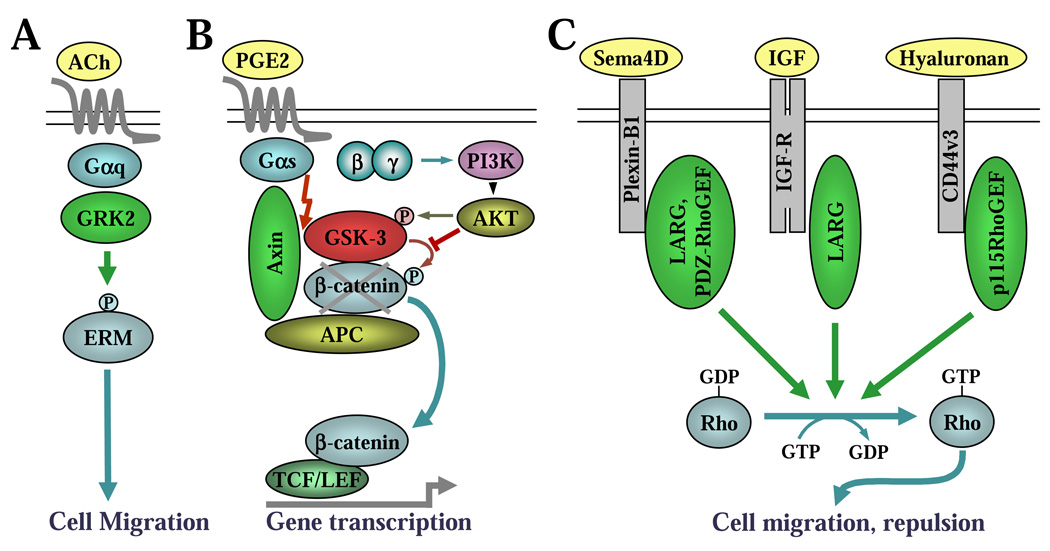
4.1. GRK2 as an effector of Gαq in control of cell migration (Fig. 3A)
Figure 3. RGS proteins as signal transducers.
A, GRK2 as an effector of Gαq in control of cell migration. B, Axin as an effector of Gαs in control of β-catenin signalling. C, Activation of RhoA by RGS-RhoGEFs induced by non-GPCR receptors.
The recent resolution of the crystal structure of a Gαq-GRK2-Gβγ complex suggested that the RGS homology domain of GRK2 binds Gαq as an effector rather than as a GAP, since the GRK2-binding residues in Gαq are analogous to the effector binding sites in Gαs and Gαt [49]. These results raise the possibility that Gαq can induce GRK2 to activate its own effector cascade [50]. Indeed, several recent studies implicate GRK2 as an effector that propagates G protein signalling.
In one such study, acetylcholine-induced membrane ruffling of hepatoma Hep2 cells expressing the muscarinic M1 receptor was abolished by expression of either a catalytically inactive GRK2 mutant or the RGS domain of GRK2. However, this response was unaffected by expression of the mutant RGS domain of GRK2 that could not bind Gαq [51]. Subsequently, the authors identified threonine-567 of ezrin as a phospho-substrate of GRK2, and phosphorylation of this residue was previously shown to stabilize the active form of ezrin [52]. Ezrin belongs to a family of ERM (ezrin, radixin, moesin) proteins that are involved in actin remodeling and cellular migration. Acetylcholine-induced phosphorylation of ezrin was reduced by expression of the RGS domain of GRK2 [51]. Another study also showed that GRK2 could phosphorylate radixin. Knockdown of GRK2 decreased the migration of Madin-Darby canine kidney (MDCK) cells, which correlated with decreased levels of phospho-ERM proteins [53]. Thus, these data suggest a model wherein Gαq-recruited GRK2 phosphorylates ERM proteins to promote cell migration.
4.2. Axin as an effector of Gαs in control of β-catenin signalling (Fig. 3B)
Beta-catenin is a key effector of Wnt signalling that promotes cell proliferation in many cell types through activation of the T cell–factor (TCF) / lymphoid enhancer factor (LEF) transcription factors [54]. The transcriptional activity of β-catenin is normally repressed by a complex consisting of adenomatous polyposis coli (APC), axin and glycogen synthase kinase 3 (GSK3), which phosphorylates β-catenin, leading to its ubiquitination and proteasomal degradation. Activation of Frizzled receptors by Wnt ligand disrupts this complex and inhibits phosphorylation of β-catenin, leading to its stabilization, accumulation in the nucleus and activation of TCF/LEF-dependent gene transcription [55, 56].
A member of the β-catenin-disruption complex, axin, has an atypical RGS domain that directly interacts with activated Gαs, but has no GAP activity [57]. Instead, this interaction appears to mediate a cross-talk between Gαs and β-catenin signalling in colon cancer cells. Prostaglandin E2 (PGE2) is known to act through a Gαs-coupled receptor and stimulate proliferation of colon cancer cells. Recently, it was discovered that PGE2 promoted β-catenin protein accumulation and activation, which was required for PGE2-induced proliferation [57]. Importantly, this effect of PGE2 was almost completely abolished by overexpression of the RGS domain of axin. Activation of β-catenin by PGE2 is not mediated by a canonical cAMP/ PKA pathway, but requires a different dual mechanism that involves: (i) binding of activated Gαs to axin (through the RGS domain) leading to a disruption of the axin-GSK3 complex, and (ii) simultaneous activation of Gβγ (presumably released from GTP-Gαs) resulting in a canonical Gβγ-mediated activation of PI3 kinase and AKT-dependent phosphorylation and inactivation of GSK-3 [57]. The first mechanism represents the function of axin as an effector of Gαs.
4.3. Non-canonical mechanisms of RhoA activation by RGS-RhoGEFs (Fig. 3C)
The RhoGEF family of RGS proteins, which consists of p115RhoGEF, leukemia-associated RhoGEF (LARG) and PDZ-RhoGEF, is a unique family that functions as both a GTPase activating protein (GAP) and an effector for Gα12/13. These RGS-RhoGEFs contain an RGS homology domain that binds to and accelerates the inactivation of Gα12 and Gα13 [58–60]. On the other hand, binding to Gα12/13 stimulates the guanine nucleotide exchange factor (GEF) activity of these RhoGEFs through the dbl-homology (DH) domain which promotes the exchange of GDP for GTP on monomeric Rho GTPases, leading to their activation. The adjacent pleckstrin homology (PH) domain is critical for full GEF activity and aids in anchoring the RhoGEFs to other signalling proteins and subcellular localization [61, 62]. Thus, RGS-RhoGEFs provide a direct link between Gα12/13- coupled GPCRs and Rho activation.
In addition to having the RGS and DH/PH domains, PDZ-RhoGEF and LARG also contain a PSD-95/Dlg/ZO-1 (PDZ) domain that has been implicated in binding to the C-termini of various transmembrane proteins [62, 63]. It was originally proposed that through binding to the C-terminus of some GPCRs (LPA1 and LPA2, previously referred to as Edg2 and Edg4), the PDZ domain of LARG and of PDZ-RhoGEF may provide receptor selectivity of these proteins in terms of (i) transducing the signal to RhoA and (ii) regulating the activity of Gα12/13 induced by a given GPCR [64]. However, the PDZ domain of RGS-RhoGEFs can also interact with receptors not coupled to G proteins. The PDZ domains of LARG and of PDZ-RhoGEF bind to the cytoplasmic domain of plexin-B1 [65–68] that activates RhoA upon stimulation with its ligand semaphorin 4D [68]. Rho activation through these two RGS-RhoGEFs is required for semaphorin-induced growth cone collapse of neuronal cells and migration of endothelial and breast carcinoma cells [69, 70]. The PDZ domain of LARG can also bind the C-terminus of the receptor tyrosine kinase insulin-like growth factor receptor (IGF-R1) β-subunit. Stimulation of MDCK cells with IGF-1 activated Rho and its effector Rho-associated kinase, and enhanced the formation of actin stress fibers in a LARG-dependent manner [71].
It was also reported that CD44v3, a hyaluronan receptor found in human metastatic breast tumor cells, functionally couples to p115RhoGEF. Binding of hyaluronan to CD44v3 promotes p115RhoGEF-mediated activation of RhoA [72]. Additional studies have found that hyaluronan-bound CD44 also recruits LARG to stimulate RhoA activation in human head and neck squamous carcinoma cells [73]. Interestingly, the CD44-LARG complex has been also reported to interact with the EGF receptor and to promote Ras-mediated stimulation of Raf-1 and ERK pathway and cell proliferation [73]. However, the mechanism of RGS-RhoGEF recruitment by CD44 and EGF receptor is not clear, as the functional domains that mediate this interaction are not yet defined.
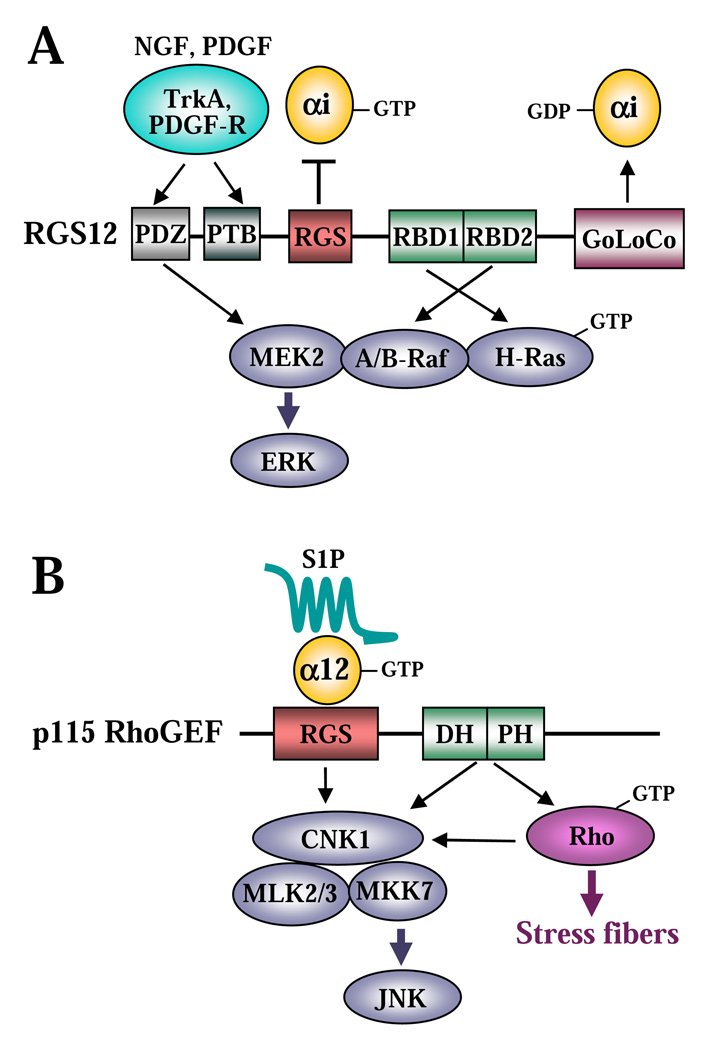
4.4. Activation of MAP kinase signalling through RGS proteins (Fig. 4)
Figure 4. Activation of MAP kinase signalling through RGS proteins.
A, activation of ERK through RGS12. B, activation of JNK through p115-RhoGEF.
RGS12 and RGS14 are unique within the RGS family as they contain a tandem repeat of two regions homologous to the Ras-binding domain (RBD) of Raf, which was originally predicted through a bioinformatics analysis [74]. Subsequently, it was biochemically demonstrated that RGS12 interacts with activated H-Ras through its RBD domains [75]. Interestingly, the same RBD repeat region provides the interaction of RGS12 with A-Raf and B-Raf (effectors of Ras), whereas the PDZ domain of RGS12 mediates its binding to MEK2 (an effector of Raf) [75]. Furthermore, RGS12 can also interact with some receptor tyrosine kinases (RTK), such as the platelet derived growth factor (PDGF) receptor [76] and the nerve growth factor (NGF) receptor tyrosine kinase TrkA[75], possibly through its PDZ domain and/or phosphotyrosine binding domain (PTB) [76]. Thus, RGS12 may serve as a scaffold for the RTK/Ras/Raf/MEK module, suggesting its role in controlling ERK signalling (Fig. 4A). Indeed, RGS12 facilitated ERK activation by PDGF in CHO-K1 cells and by NGF in PC12 cells, and RGS12 was required for NGF-mediated neurite outgrowth of PC12 cells [75]. A similar Ras/Raf/MEK scaffolding role was recently demonstrated for RBD-containing RGS14 [77, 78].
RGS proteins may also mediate Jun kinase activation. It has been reported that p115-RhoGEF interacts with the scaffolding protein connector enhancer of KSR1 (CNK1), surprisingly through its DH/PH and RGS domains, suggesting multiple interacting interfaces [79]. CNK1 appears to serve as a bridge in the complex formation between p115RhoGEF and the components of the Jun kinase pathway (MLK2, MLK3 and MKK7), providing a mechanism for Jun kinase activation by RhoA (Fig. 4B). Endogenous CNK1 mediates Jun kinase activation induced by serum or sphingosine 1-phosphate (S1P), or by overexpression of constitutively active Gα12, p115RhoGEF, or constitutively active RhoA. Interestingly, endogenous CNK1 is dispensable for RhoA activation and stress fiber formation by p115RhoGEF [79].
5. Nuclear localization and function of RGS proteins
We originally discovered that a truncated variant of RGS3 (RGS3T), which lacks a large N-terminal portion of RGS3 but contains a short N-terminus and the C-terminal RGS domain, is localized to the nucleus when transfected in CHO cells [80]. Nearly at the same time, the nuclear localization was reported for ectopic RGS2 and RGS10 in COS-7 cells, as well as for endogenous RGS10 in neuroglioma (H4) cells [81]. Throughout the past decade, it became clear that nuclear localization is a common feature of several RGS proteins, as demonstrated by many groups in various cell types at both ectopic and endogenous levels of RGS expression (summarized in table 1). It is noteworthy that localization of RGS proteins to the nucleus may depend on cell type and the level of expression. Thus, overexpressed RGS13 is largely nuclear in COS-7 and HEK293 cells, but is cytoplasmic in HeLa cells and B lymphocytes [82]. Therefore, intracellular localization of RGS proteins, especially at the endogenous level of their expression, could vary and should be examined in a cell-specific context.
Table 1.
Reported nuclear localization of RGS proteins.
| RGS protein | Cells | Endogenous or Ectopic |
Reference |
|---|---|---|---|
| RGS2 | COS-7 HEK293 Neuroblastoma SH-SY5Y Astrocytoma 1321N1 PC3 |
Ectopic Ectopic Ectopic Endogenous Ectopic |
[81, 93, 97] [25, 87, 90] [99] [100, 101] [92] |
| RGS3T | CHO COS-7 |
Ectopic Ectopic |
[80] [97] |
| RGS6L | COS-7 | Ectopic | [94] |
| RGS6S | COS-7 | Ectopic | [88, 97] |
| RGS8 | DDT1MF2 | Ectopic | [83] |
| RGS9-2 | Forebrain neurons COS-7 |
Endogenous Ectopic |
[85] [85] |
| RGS10 | COS-7 Neuroglioma (H4) HEK293 PC3 |
Ectopic Endogenous Ectopic Ectopic |
[81] [81] [95] [92] |
| RGS12TS-S | HEK293 COS-7 |
Endogenous Ectopic |
[84, 102] [84, 102] |
| RGS13 | COS-7 HEK293 NIH3T3 |
Ectopic Ectopic Ectopic |
[82, 97] [82, 96] [96] |
| RGS14 | HeLa HeLa NIH3T3 |
Endogenous Ectopic Endogenous |
[98] [86, 89, 98] [86] |
| RGS20 (RGSZ) | COS-7 | Ectopic | [97] |
5.1. Mechanisms of nuclear localization of RGS proteins
Several mechanisms have been proposed for the nuclear targeting of RGS proteins. Our data suggests that accumulation of RGS3T in the nucleus is mediated by a unique N-terminal region outside of the RGS domain of RGS3T. The deletion of this region (leaving only the RGS domain) results in equal distribution of the protein between the nucleus and cytoplasm [80]. This distribution could possibly be because of the small size of the RGS domain (14 kDa), which allows for its passive movement through the nuclear pore. The concept that nuclear targeting of RGS proteins is driven by regions outside of the RGS domain has been supported by others, as exemplified by RGS8 [83], RGS12TS-S [84], RGS9-2 [85]and RGS14 [86].
On the other hand, it is also proposed that the RGS domains of some RGS proteins (RGS2, RGS4, RGS6, RGS10, RGS14, RGS16) have an intrinsic ability to localize to the nucleus through nuclear targeting motifs or through passive diffusion, unless other regions outside of the RGS domain drive their nuclear exclusion [81, 86–88]. For example, the RGS domains of RGS4 and RGS16 are localized to the nucleus, while full length RGS4 and RGS16 are cytoplasmic [81]. The cytoplasmic localization of RGS4 and RGS16 is mediated by nuclear extraction signals (NES) outside of their RGS domains, and mutation of the NES leads to nuclear accumulation of full length RGS4 and RGS16 [81]. Likewise, nuclear localization of RGS14 is partially mediated by its RGS domain [86], while the cytoplasmic targeting of RGS14 is mediated by its NES [86] and/or GoLoco domain [89]. Thus, nuclear targeting of RGS proteins can be controlled by both the RGS domain and the regions outside of it.
A number of studies suggest that the nuclear/cytoplasmic distribution of RGS proteins is a regulated process. Nuclear-localized RGS2, RGS4 and RGS8 migrate to the plasma membrane when co-transfected with GTPase-deficient mutants of Gα subunits that permanently associate with RGS proteins [83, 90, 91]. The nuclear-to-plasma membrane translocation of RGS2 and RGS4 was also observed upon overexpression of G protein coupled receptors (β2-adrenergic, AT1 angiotensin II, or M2 muscarinic receptors) [90]. Furthermore, agonist-induced translocation of RGS2 and RGS10 from the nucleus to the cytoplasm by melatonin [92] or translocation of RGS2 by prostaglandin F2α [93] was shown, although this could be mediated by G protein signalling (protein kinase C activation) rather than by a direct recruitment of RGS proteins by activated Gα subunits [92]. Thus, extraction of RGS proteins from the nucleus to the cytoplasm and plasma membrane by activated G proteins could be one mechanism of regulated intracellular localization of RGS proteins.
Nuclear targeting of RGS proteins could be also mediated by interaction with other proteins, or by protein phosphorylation. The long isoform of RGS6 (RGS6L) binds a transcriptional repressor, Dnmt1-associated protein (DMAP1) through a unique region outside of its RGS domain and is recruited to the nucleus by DMAP1 [94]. Phosphorylation of RGS10 by protein kinase A (PKA) promotes its nuclear localization, but currently the mechanism is unknown [95]. RGS13 accumulates in the nucleus through the binding of phosphorylated (by PKA) cAMP-response element binding protein CREB [82, 96]. Finally, RGS2, RGS3T, RGS6, RGS13, and RGS20 (RGSZ) translocate to the nucleoli upon mild or proteotoxic stress conditions [97], whereas RGS14 translocates from the cytoplasm to promyelocytic leukemia nuclear bodies upon mild heat stress, but not proteotoxic or transcription-linked stresses [86].
5.2. Nuclear functions of RGS proteins
The nuclear function of RGS proteins is currently under intense investigation, but two major possibilities can already be postulated. As described in section 5.1, activation of G proteins leads to translocation of several RGS proteins from the nucleus to the plasma membrane [83, 90, 91, 93]. Thus, the nucleus may serve to sequester RGS proteins from plasma membrane-localized G proteins, thus controlling the availability of RGS proteins for the regulation of G protein signalling. This concept is consistent with the “fine-tuning” model in which initial G protein signalling is allowed while the simultaneous recruitment of RGS proteins to the plasma membrane ensures a transient response.
More intriguingly, arising evidence suggests that nuclear RGS proteins may serve functions potentially unrelated to the canonical regulation of G protein signalling at the plasma membrane. We showed that overexpressed nuclear RGS3T induces apoptosis of CHO cells, whereas the cytoplasmic full length RGS3 does not [80]. The apoptotic effect of RGS3T is not mediated by the RGS domain, which on its own is diffusely localized between the nucleus and the cytoplasm and does not induce apoptosis [80]. Overexpression of the nuclear RGS12TS-S variant in COS-7 cells inhibited DNA synthesis and cell cycle progression, possibly through direct interference with the basal gene transcription machinery. This effect of RGS12TS-S was mediated by its unique N-terminal domain, and not by the RGS domain [84]. In contrast, RGS6 is recruited to the nucleus through interaction with DMAP1 and inhibits its repressor activity, thus promoting gene transcription [94]. RGS13 binds phosphorylated CREB and inhibits its transcriptional activity in the nucleus by interfering with binding of CREB to DNA and to CREB-binding protein CBP. Again this regulation occurs in a GAP-independent manner [96]. Finally, RGS14 segregates to the centrosomes and astral mictotubules, regulates microtubule dynamics, and thus controls chromosomal segregation during mitosis of HeLa cells. Knockdown of RGS14 in HeLa cells results in an increased number of multinucleated cells and a decrease in cell proliferation [98]. Interestingly, this function of RGS14 could be mediated by its RGS domain and may involve regulation of nucleotide cycling of G proteins, although this activity may be distinctly localized away from its traditional association with seven-transmembrane receptors [98].
6. Conclusions
It has become appreciated that the RGS family encompasses a set of diverse proteins with several roles extending beyond GAP activity. The multitude of RGS functions may partially account for the existence of many RGS proteins, far surpassing what is necessary to regulate the few G protein subtypes. However, several non-canonical functions of RGS proteins summarized here were shown by overexpression approach and should be validated at the endogenous expression levels of RGS proteins. In most cases, the GAP-independent functions of RGS proteins are mediated by the interaction with other partners through regions outside of the RGS domain. Therefore, RGS proteins may simultaneously interact with activated Gα subunits and other signalling molecules, suggesting that RGS proteins may mediate crosstalk between G protein signalling and previously unrelated cellular processes. We anticipate that future studies in the RGS field will expand our understanding of the multifunctional roles of RGS proteins.
Acknowledgments
This study was supported by National Institutes of Health Awards R01 HL071755 (N. O. D.), R01 GM85058 (N.O.D), T32 HD 07009 (N.S.), T32 HL 07605 (D.M.Y.), and American Heart Association Fellowship Awards 10PRE2630163 (N.S.), 0825868G (D.M.Y).
Abbreviations
- AC
adenylyl cyclase
- CHO
chinese hamster ovary
- CNK1
connector enhancer of KSR1
- CREB
cAMP-response element binding protein
- DH
dbl-homology
- DMAP1
Dnmt1-associated protein 1
- EGF
epidermal growth factor
- eIF
eukaryotic initiation factor
- ERK
extracellular signal regulated protein kinase
- ERM
ezrin radixin moesin
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GPCR
G protein-coupled receptor
- Grb2
growth factor receptor-bound protein
- GRK
G protein receptor kinase
- GSK
glycogen synthase kinase
- HEK
human embryonic kidney
- IP3
inositol trisphosphate
- LARG
leukemia-associated RhoGEF
- LPA
lysophosphatidic acid
- LEF
lymphoid enhancer factor
- MDCK
Madin-Darby canine kidney
- MEK2
MAPK/ERK kinase
- MH2
Mad homology 2
- MKK
mitogen-activated kinase kinase
- MLK
mixed-lineage kinase
- NES
nuclear extraction signal
- NGF
nerve growth factor
- PDGF
platelet-derived growth factor
- PDZ
PSD-95/Dlg/ZO-1 domain
- PH
pleckstrin homology
- PI3K
phosphoinositide (3) kinase
- PIP3
phosphatidylinositol (3,4,5) trisphosphate
- PKA
protein kinase A
- PLC
phospholipase C
- PDGF
platelet derived growth factor
- PGE2
Prostaglandin E2
- PTB
phosphotyrosine binding domain
- RBD
Ras-binding domain
- RGS
regulator of G protein signalling
- RTK
receptor tyrosine kinase
- S1P
sphingosine 1-phosphate
- TCF
T-cell factor
- TGF-β
transforming growth factor-β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Druey KM, Blumer KJ, Kang VH, Kehrl JH. Nature. 1996;379(6567):742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 2.Hunt TW, Fields TA, Casey PJ, Peralta EG. Nature. 1996;383(6596):175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- 3.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. Cell Signal. 2006;18(5):579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Hollinger S, Hepler JR. Pharmacol Rev. 2002;54(3):527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 6.Willars GB. Semin Cell Dev Biol. 2006;17(3):363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Federman AD, Conklin BR, Schrader KA, Reed RR, Bourne HR. Nature. 1992;356(6365):159–161. doi: 10.1038/356159a0. [DOI] [PubMed] [Google Scholar]
- 8.Katz A, Wu D, Simon MI. Nature. 1992;360(6405):686–689. doi: 10.1038/360686a0. [DOI] [PubMed] [Google Scholar]
- 9.Pitcher JA, Inglese J, Higgins JB, Arriza JL, Casey PJ, Kim C, Benovic JL, Kwatra MM, Caron MG, Lefkowitz RJ. Science. 1992;257(5074):1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 10.Strasser RH, Benovic JL, Caron MG, Lefkowitz RJ. Proc Natl Acad Sci U S A. 1986;83(17):6362–6366. doi: 10.1073/pnas.83.17.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. J Biol Chem. 1999;274(48):34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- 12.Day PW, Carman CV, Sterne-Marr R, Benovic JL, Wedegaertner PB. Biochemistry. 2003;42(30):9176–9184. doi: 10.1021/bi034442+. [DOI] [PubMed] [Google Scholar]
- 13.Koch WJ, Hawes BE, Inglese J, Luttrell LM, Lefkowitz RJ. J Biol Chem. 1994;269(8):6193–6197. [PubMed] [Google Scholar]
- 14.Eichmann T, Lorenz K, Hoffmann M, Brockmann J, Krasel C, Lohse MJ, Quitterer U. J Biol Chem. 2003;278(10):8052–8057. doi: 10.1074/jbc.M204795200. [DOI] [PubMed] [Google Scholar]
- 15.Shi CS, Lee SB, Sinnarajah S, Dessauer CW, Rhee SG, Kehrl JH. J Biol Chem. 2001;276(26):24293–24300. doi: 10.1074/jbc.M100089200. [DOI] [PubMed] [Google Scholar]
- 16.Vogt A, Lutz S, Rumenapp U, Han L, Jakobs KH, Schmidt M, Wieland T. Cell Signal. 2007;19(6):1229–1237. doi: 10.1016/j.cellsig.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Bowman EP, Campbell JJ, Druey KM, Scheschonka A, Kehrl JH, Butcher EC. J. Biol. Chem. 1998;273(43):28040–28048. doi: 10.1074/jbc.273.43.28040. [DOI] [PubMed] [Google Scholar]
- 18.Harris BA, Robishaw JD, Mumby SM, Gilman AG. Science. 1985;229(4719):1274–1277. doi: 10.1126/science.3839937. [DOI] [PubMed] [Google Scholar]
- 19.Taussig R, Iniguez-Lluhi JA, Gilman AG. Science. 1993;261(5118):218–221. doi: 10.1126/science.8327893. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee TK, Eapen AK, Fisher RA. J Biol Chem. 1997;272(24):15481–15487. doi: 10.1074/jbc.272.24.15481. [DOI] [PubMed] [Google Scholar]
- 21.Johnson EN, Druey KM. J Biol Chem. 2002;277(19):16768–16774. doi: 10.1074/jbc.M200751200. [DOI] [PubMed] [Google Scholar]
- 22.Roy AA, Baragli A, Bernstein LS, Hepler JR, Hebert TE, Chidiac P. Cell Signal. 2006;18(3):336–348. doi: 10.1016/j.cellsig.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, Dennis JC, Morrison EE, Vodyanoy V, Kehrl JH. Nature. 2001;409(6823):1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 24.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. J Biol Chem. 2003;278(18):15842–15849. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 25.Gu S, Anton A, Salim S, Blumer KJ, Dessauer CW, Heximer SP. Mol Pharmacol. 2008;73(1):1–11. doi: 10.1124/mol.107.036285. [DOI] [PubMed] [Google Scholar]
- 26.Roy AA, Nunn C, Ming H, Zou MX, Penninger J, Kirshenbaum LA, Dixon SJ, Chidiac P. J Biol Chem. 2006;281(43):32684–32693. doi: 10.1074/jbc.M604416200. [DOI] [PubMed] [Google Scholar]
- 27.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Science. 1991;251(4995):804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 28.Smrcka AV, Sternweis PC. J Biol Chem. 1993;268(13):9667–9674. [PubMed] [Google Scholar]
- 29.Taylor SJ, Chae HZ, Rhee SG, Exton JH. Nature. 1991;350(6318):516–518. doi: 10.1038/350516a0. [DOI] [PubMed] [Google Scholar]
- 30.Dulin NO, Sorokin A, Reed E, Elliott S, Kehrl JH, Dunn MJ. Mol Cell Biol. 1999;19(1):714–723. doi: 10.1128/mcb.19.1.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C, Hepler JR, Gilman AG, Mumby SM. Proc Natl Acad Sci U S A. 1997;94(12):6159–6163. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neill JD, Duck LW, Sellers JC, Musgrove LC, Scheschonka A, Druey KM, Kehrl JH. Endocrinology. 1997;138(2):843–846. doi: 10.1210/endo.138.2.5034. [DOI] [PubMed] [Google Scholar]
- 33.Yan Y, Chi PP, Bourne HR. J Biol Chem. 1997;272(18):11924–11927. doi: 10.1074/jbc.272.18.11924. [DOI] [PubMed] [Google Scholar]
- 34.Hepler JR, Berman DM, Gilman AG, Kozasa T. Proc Natl Acad Sci U S A. 1997;94(2):428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heximer SP, Watson N, Linder ME, Blumer KJ, Blumer KJ, Hepler JR. Proc Natl Acad Sci U S A. 1997;94(26):14389–14393. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dowal L, Elliott J, Popov S, Wilkie TM, Scarlata S. Biochemistry. 2001;40(2):414–421. doi: 10.1021/bi001923+. [DOI] [PubMed] [Google Scholar]
- 37.Bansal G, Xie Z, Rao S, Nocka KH, Druey KM. Nat Immunol. 2008;9(1):73–80. doi: 10.1038/ni1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang G, Bansal G, Xie Z, Druey KM. J Biol Chem. 2009;284(32):21719–21727. doi: 10.1074/jbc.M109.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali K, Bilancio A, Thomas M, Pearce W, Gilfillan AM, Tkaczyk C, Kuehn N, Gray A, Giddings J, Peskett E, Fox R, Bruce I, Walker C, Sawyer C, Okkenhaug K, Finan P, Vanhaesebroeck B. Nature. 2004;431(7011):1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- 40.Nishida K, Yoshida Y, Itoh M, Fukada T, Ohtani T, Shirogane T, Atsumi T, Takahashi-Tezuka M, Ishihara K, Hibi M, Hirano T. Blood. 1999;93(6):1809–1816. [PubMed] [Google Scholar]
- 41.Feng XH, Derynck R. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 42.Massague J, Seoane J, Wotton D. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 43.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature. 1997;389(6651):622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 44.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 45.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. Science. 2005:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 46.Yau DM, Sethakorn N, Taurin S, Kregel S, Sandbo N, Camoretti-Mercado B, Sperling AI, Dulin NO. Mol Pharmacol. 2008;73(5):1356–1361. doi: 10.1124/mol.108.044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimball SR. Int J Biochem Cell Biol. 1999;31(1):25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR, Chidiac P. J Cell Biol. 2009;186(5):755–765. doi: 10.1083/jcb.200811058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lodowski DT, Barnhill JF, Pyskadlo RM, Ghirlando R, Sterne-Marr R, Tesmer JJ. Biochemistry. 2005;44(18):6958–6970. doi: 10.1021/bi050119q. [DOI] [PubMed] [Google Scholar]
- 50.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Science. 2005;310(5754):1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 51.Cant SH, Pitcher JA. Mol Biol Cell. 2005;16(7):3088–3099. doi: 10.1091/mbc.E04-10-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. J Cell Biol. 1998;140(3):647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kahsai AW, Zhu S, Fenteany G. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11(2):233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 55.Liu C, Li Y, Semenov M, Han C, Baeg G, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108(6):837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 56.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272(5264):1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 57.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Science. 2005;310(5753):1504–1510. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 58.Fukuhara S, Chikumi H, Gutkind JS. FEBS Lett. 2000;485(2–3):183–188. doi: 10.1016/s0014-5793(00)02224-9. [DOI] [PubMed] [Google Scholar]
- 59.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. J Biol Chem. 1999;274(9):5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 60.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. Science. 1998;280(5372):2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 61.Dutt P, Nguyen N, Toksoz D. Cell Signal. 2004;16(2):201–209. doi: 10.1016/s0898-6568(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 62.Fukuhara S, Chikumi H, Gutkind JS. Oncogene. 2001;20(13):1661–1668. doi: 10.1038/sj.onc.1204182. [DOI] [PubMed] [Google Scholar]
- 63.Kuner R, Swiercz JM, Zywietz A, Tappe A, Offermanns S. Eur J Neurosci. 2002;16(12):2333–2341. doi: 10.1046/j.1460-9568.2002.02402.x. [DOI] [PubMed] [Google Scholar]
- 64.Yamada T, Ohoka Y, Kogo M, Inagaki S. J Biol Chem. 2005;280(19):19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
- 65.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. Proc Natl Acad Sci U S A. 2002;99(19):12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirotani M, Ohoka Y, Yamamoto T, Nirasawa H, Furuyama T, Kogo M, Matsuya T, Inagaki S. Biochem Biophys Res Commun. 2002;297(1):32–37. doi: 10.1016/s0006-291x(02)02122-8. [DOI] [PubMed] [Google Scholar]
- 67.Perrot V, Vazquez-Prado J, Gutkind JS. J Biol Chem. 2002;277(45):43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 68.Swiercz JM, Kuner R, Behrens J, Offermanns S. Neuron. 2002;35(1):51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 69.Pasterkamp RJ, Kolodkin AL. Curr Opin Neurobiol. 2003;13(1):79–89. doi: 10.1016/s0959-4388(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 70.Puschel AW. Adv Exp Med Biol. 2007;600:12–23. doi: 10.1007/978-0-387-70956-7_2. [DOI] [PubMed] [Google Scholar]
- 71.Taya S, Inagaki N, Sengiku H, Makino H, Iwamatsu A, Urakawa I, Nagao K, Kataoka S, Kaibuchi K. J Cell Biol. 2001;155(5):809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bourguignon LYW, Singleton PA, Zhu H, Diedrich F. Hyaluronan-mediated CD44 Interaction with RhoGEF and Rho Kinase Promotes Grb2-associated Binder-1 Phosphorylation and Phosphatidylinositol 3-Kinase Signaling Leading to Cytokine (Macrophage-Colony Stimulating Factor) Production and Breast Tumor Progression. 2003:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 73.Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. J Biol Chem. 2006;281(20):14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- 74.Ponting CP. J Mol Med. 1999;77(10):695–698. doi: 10.1007/s001099900054. [DOI] [PubMed] [Google Scholar]
- 75.Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP. Embo J. 2007;26(8):2029–2040. doi: 10.1038/sj.emboj.7601659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sambi BS, Hains MD, Waters CM, Connell MC, Willard FS, Kimple AJ, Pyne S, Siderovski DP, Pyne NJ. Cell Signal. 2006;18(7):971–981. doi: 10.1016/j.cellsig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Shu FJ, Ramineni S, Hepler JR. Cell Signal. 2010;22(3):366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. PLoS One. 2009;4(3):e4884. doi: 10.1371/journal.pone.0004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaffe AB, Hall A, Schmidt A. Curr Biol. 2005;15(5):405–412. doi: 10.1016/j.cub.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 80.Dulin NO, Pratt P, Tiruppathi C, Niu J, Voyno-Yasenetskaya T, Dunn MJ. J Biol Chem. 2000;275(28):21317–21323. doi: 10.1074/jbc.M910079199. [DOI] [PubMed] [Google Scholar]
- 81.Chatterjee TK, Fisher RA. J Biol Chem. 2000;275(31):24013–24021. doi: 10.1074/jbc.M002082200. [DOI] [PubMed] [Google Scholar]
- 82.Shi GX, Harrison K, Wilson GL, Moratz C, Kehrl JH. J Immunol. 2002;169(5):2507–2515. doi: 10.4049/jimmunol.169.5.2507. [DOI] [PubMed] [Google Scholar]
- 83.Saitoh O, Masuho I, Terakawa I, Nomoto S, Asano T, Kubo Y. J Biol Chem. 2001;276(7):5052–5058. doi: 10.1074/jbc.M006917200. [DOI] [PubMed] [Google Scholar]
- 84.Chatterjee TK, Fisher RA. Mol Cell Biol. 2002;22(12):4334–4345. doi: 10.1128/MCB.22.12.4334-4345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bouhamdan M, Michelhaugh SK, Calin-Jageman I, Ahern-Djamali S, Bannon MJ. Biochim Biophys Acta. 2004;1691(2–3):141–150. doi: 10.1016/j.bbamcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 86.Cho H, Kim DU, Kehrl JH. J Biol Chem. 2005;280(1):805–814. doi: 10.1074/jbc.M408163200. [DOI] [PubMed] [Google Scholar]
- 87.Heximer SP, Lim H, Bernard JL, Blumer KJ. J Biol Chem. 2001;276(17):14195–14203. doi: 10.1074/jbc.M009942200. [DOI] [PubMed] [Google Scholar]
- 88.Chatterjee TK, Liu Z, Fisher RA. J Biol Chem. 2003;278(32):30261–30271. doi: 10.1074/jbc.M212687200. [DOI] [PubMed] [Google Scholar]
- 89.Shu FJ, Ramineni S, Amyot W, Hepler JR. Cell Signal. 2007;19(1):163–176. doi: 10.1016/j.cellsig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 90.Roy AA, Lemberg KE, Chidiac P. Mol Pharmacol. 2003;64(3):587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- 91.Masuho I, Itoh M, Itoh H, Saitoh O. J Neurochem. 2004;88(1):161–168. doi: 10.1046/j.1471-4159.2003.02139.x. [DOI] [PubMed] [Google Scholar]
- 92.Rimler A, Jockers R, Lupowitz Z, Sampson SR, Zisapel N. J Pineal Res. 2006;40(2):144–152. doi: 10.1111/j.1600-079X.2005.00290.x. [DOI] [PubMed] [Google Scholar]
- 93.Wu YL, Chuang HH, Kou YR, Lee TS, Lu SH, Huang YC, Nishi Y, Yanase T. Chin J Physiol. 2008;51(5):282–291. [PubMed] [Google Scholar]
- 94.Liu Z, Fisher RA. J Biol Chem. 2004;279(14):14120–14128. doi: 10.1074/jbc.M309547200. [DOI] [PubMed] [Google Scholar]
- 95.Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ. J Biol Chem. 2001;276(35):32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- 96.Xie Z, Geiger TR, Johnson EN, Nyborg JK, Druey KM. Mol Cell. 2008;31(5):660–670. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chatterjee TK, Fisher RA. J Biol Chem. 2003;278(32):30272–30282. doi: 10.1074/jbc.M212688200. [DOI] [PubMed] [Google Scholar]
- 98.Martin-McCaffrey L, Willard FS, Oliveira-dos-Santos AJ, Natale DR, Snow BE, Kimple RJ, Pajak A, Watson AJ, Dagnino L, Penninger JM, Siderovski DP, D'Souza SJ. Dev Cell. 2004;7(5):763–769. doi: 10.1016/j.devcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 99.Song L, Zmijewski JW, Jope RS. Biochem Biophys Res Commun. 2001;283(1):102–106. doi: 10.1006/bbrc.2001.4742. [DOI] [PubMed] [Google Scholar]
- 100.Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS. Biochim Biophys Acta. 2001;1541(3):201–211. doi: 10.1016/s0167-4889(01)00144-6. [DOI] [PubMed] [Google Scholar]
- 101.Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS. Arch Biochem Biophys. 2001;392(2):192–196. doi: 10.1006/abbi.2001.2430. [DOI] [PubMed] [Google Scholar]
- 102.Chatterjee TK, Fisher RA. J Biol Chem. 2000;275(38):29660–29671. doi: 10.1074/jbc.M000330200. [DOI] [PubMed] [Google Scholar]