Abstract
We previously reported that the tumor suppressor protein p53 participates in a negative feedback loop to fine-tune PKD1 gene expression. This physiological pathway is believed to prevent polycystin-1 overexpression and thus renal cysts. The present study examined the mechanisms of p53-mediated repression of PKD1. The 5′-upstream region of the human PKD1 gene is TATA-less, GC rich and contains four consensus p53 binding sites at positions −2.7kb (BS4), −1.2kb (BS3), −0.8kb (BS2) and −0.2kb (BS1), respectively. PKD1BS1-4 are bound to endogenous p53 in vivo and in vitro. Transient transfection assays in inner medullary collecting duct cells revealed that disruption of PKD1BS1 enhances baseline PKD1 promoter activity; in contrast, disruption of PKD1BS4 suppressed PKD1 transcription. PKD1BS1 confers p53-mediated repression when substituted for the p53 enhancer element in the bradykinin B2 receptor gene, indicating that PKD1BS1 is a bona fide p53 repressor element. Moreover, PKD1BS1 requires intact BS2-4 and cellular histone deacetylase activity for full functional activity. Indeed, the PKD1BS1/4 regions are occupied by a complex containing HDAC1/2 and mSin3. These findings suggest a model whereby p53 exerts a biphasic control on PKD1 gene transcription, depending on cellular context and the cognate cis-acting element.
Keywords: Polycystic kidney disease gene, p53, Gene transcription, promoter
INTRODUCTION
Autosomal dominant polycystic kidney disease (ADPKD) is a genetically inherited disorder with an incidence of 1 in 500 to 1 in 1000 [1–4]. ADPKD is characterized by formation of renal cysts in all parts of the nephron and the collecting duct, leading to end-stage renal failure in 50% of ADPKD cases by the fifth decade of life. The polycystic disease-1 gene (PKD1) located on human chromosome 16p13.3 codes for the protein polycystin-1 (PC1) [3]. Mutations in PKD1, resulting in a dysfunctional PC1, are the major cause (85%) of all ADPKD cases, whereas the remaining cases are caused by mutations in PKD2 (15%) [4, 5]. The PC1/2 complex regulates intracellular Ca2+ entry and release, and signaling pathways such as Wnt-β-catenin, c-Jun/AP-1, Ets, Calcineurin/NFAT and JAK2/STAT1-p21 [6–11]. These signaling pathways play an important role in differentiation, proliferation, apoptosis and cell polarity and are deregulated in PC1-deficient and/or mutant cells leading to formation and expansion of tubular cysts [1–4].
Appropriate control of the PKD1 gene is crucial since over- or under expression of PKD1 in mice results in deregulated terminal differentiation and cystogenesis [5, 12–16]. Recent studies have provided insights into the transcriptional regulation of the PKD1 gene. Rodova et al. [17] have shown that β-catenin is a transcriptional activator of PKD1 through binding to a TCF/LEF element present in the PKD1 promoter. Interestingly, mice harboring a kidney specific β-catenin transgene exhibit a PKD-like phenotype [10]. Using computational gene analysis, Lantinga-van Leeuwen et al. [16] identified binding sites for transcription factors AP2, E2f, E-BOX, EGRF, ETS, MINI, MZP1, SP1 and ZBP89 in the PKD1 5′-upstream region, which are conserved in mouse, canine and human genomes. More Recently, it has been demonstrated that the transcription factor SP1 positively regulates murine pkd1 promoter activity through at least two of three SP1-response elements at position −65 (SP1A) and −46 (SP1B) with SP1A having a more prominent role than SP1B [18]. Subsequent studies demonstrated that retinoic acid and retinoic acid receptors RAR and RXR induce PKD1 promoter activity in a manner that is dependent on the interaction of SP1 motifs and protein in the PKD1 proximal promoter region [19].
Our laboratory has previously identified the tumor suppressor protein/transcription factor, p53, as a negative regulator of PKD1 gene transcription [20]. PKD1 mRNA levels are higher in kidneys of newborn p53−/− mice than wild type littermates. In transient transfection assays, p53 represses PKD1 promoter-driven transcription. Moreover, irradiation of HCT116 human colon carcinoma cells resulted in reduced levels of PKD1 mRNA in p53+/+ but not p53−/− cells, suggesting that p53 is a physiological negative regulator of PKD1 gene transcription [20]. In support of this conclusion, chromatin immunoprecipitation (ChIP) assays revealed in vivo binding of p53 to the promoter region of the PKD1 gene [20]. However, the functional relevance of these cis-acting elements has not been defined.
The present study was designed to further delineate the mechanisms underlying p53-mediated repression of PKD1 gene transcription. Our results indicate that p53 exerts dual roles in PKD1 gene regulation. Furthermore, PKD1 harbors a p53-response element which dictates active repression in association with an HDAC co-repressor complex.
METHODS
Cell Culture and Transient Transfections
Mouse Inner Medullary Collecting Duct (IMCD3) cells were obtained from the American Type Culture Collection. IMCD3 cells were maintained in Dulbecco’s Modified Eagle Medium/F12 (DMEM/F12) containing 10% Fetal Bovine Serum at 37°C in a humified incubator with 5% CO2. Transient transfections were performed utilizing Lipofectamine and PLUS reagent (Invitrogen) according to the protocols supplied by the manufacturer. Cells were co-transfected with 1μg promoter-reporter construct, 0.4μg β-galactosidase expression plasmid (to monitor transfection efficiency) with or without pCMV-p53 expression plasmid. In experiments utilizing Trichostatin A (TSA), cells were treated with TSA (50 ng/ml) or vehicle (Me2SO) two hours before transfection and again four hours after transfection. Twenty four hours post-transfection, the cells were lysed, and activities were determined utilizing enzyme assay kits for luciferase and β-galactosidase (Promega) and chloramphenicol acetyltransferase (CAT) activity as described [21]. All experiments were performed in duplicate, and results represent means ± S.E. of at least three independent experiments.
Plasmids and reporter constructs
The human −3.3kb hPKD1/+33-bp luciferase promoter construct in a pGL2-basic vector and the p53 expression plasmid pCMV-p53 were kindly supplied by J. Calvet (University of Kansas Medical Center) and G. Morris (Tulane University Cancer Center), respectively. The promoter-reporter construct pBdkrB2 −1184/+55-CAT and its derivatives lacking the P1 (P2/ΔP1) and P2 (ΔP2/P1) sites were generated in our laboratory [22]. Utilizing these pBdkrB2 constructs, a series of insertions were performed using reagents and protocols from the Quick-Change site-directed mutagenesis kit (Stratagene). The mutagenesis primers used to generate the various promoter-CAT constructs are depicted in Table 1.
Table 1.
Sequence of mutagenesis primers used to generate pBdkrB2 P2/BS1 and pBdkrB2 - ΔP2/BS1 constructs.
hPKD1 p53BS1 sequence: 5′-GGTCGCGCTGTGGCGAAGGGGGCGGAGCCTGCAC-3′
Forward mutagenesis primers
| 5′-CTGGAAGTGGAGGGGGGTCGCTGACATCACCGGCCAG |
| 5′-GTGGAGGGGGGTCGCGCTGTGTGACATCACCGGCCAG |
| 5′-GGGGGGTCGCGCTGTGGCGAAGTGACATCACCGGCCAG |
| 5′-CGCGCTGTGGCGAAGGGGGCGTGACATCACCGGCCAG |
| 5′-GTGGCGAAGGGGGCGGAGCCTTGACATCACCGGCCAG |
| 5′-CGAAGGGGGCGGAGCCTGCACTGACATCACCGGCCAG |
The mutant −3.3kb PKD1/+33-bp luciferase promoter constructs carrying deletions in the p53-binding sites (ΔBS4, ΔBS3, ΔBS2 and ΔBS1) were created as follows: mutant constructs ΔBS4 and ΔBS3 were generated through sequential mutagenesis reactions; each reaction resulted in a deletion of 5bp. A single-step mutagenesis reaction resulted in 10bp deletion in ΔBS2 and ΔBS1 constructs. The double, triple and quadruple mutant promoter-reporter constructs were engineered similarly by means of additional mutagenesis reactions. The mutagenesis primers and the promoter-luciferase constructs used are depicted in Table 2.
Table 2.
Sequence of mutagenesis primers used to generate mutant PKD1-luciferase constructs. The 10bp deletions are indicated in bold.
PKD1 p53BS1 sequence: 5′-GGTCGCGCTGTGGCGAAGGGGGCGGAGCCTGCAC-3′
PKD1 p53BS2 sequence: 5′-CTACAAGCGTGAGCCAGTTT-3′
PKD1 p53BS3 sequence: 5′-GAACTTCTGATCTTGTGATGTGCCC-3′
PKD1 p53BS4 sequence: 5′-AGTCTCACTCTGTCACCCAGGCTGGAGT-3′
Forward Mutagenesis primers
| 5′-CAGTCCCTCATCGCTGGCCCTGGCGAAGGGGGCGGAGCC | 10bp deletion p53BS1 |
| 5′-GCCTCTCGAGTACCTGGGAGAGCCAGTTTGGCTATTTTGG | 10bp deletion p53BS2 |
| 5′-GGCTAGGCTGGTCTCTCTGATCTTGTGATC | 1st 5bp deletion p53BS3 |
| 5′-CACGTTGGCTAGGCTGGTCTCTCTTGTGATCTGCCCG | 2nd 5bp deletion p53BS3 |
| 5′-CAGAGTCTCACTCTGTCACCCGAAGTGGCGGGATCTCGGC | 1st 5bp deletion p53BS4 |
| 5′-CTCACTCTGTCACCCGGAGTGAAGTGGCGG | 2nd 5bp deletion p53BS4 |
Electrophoretic Mobility Shift Assays (EMSA)
[32P]-labeled duplex oligonucleotides (~50,000 cpm) were incubated for 20 minutes at room temperature with 3.5 μg of nuclear extract from IMCD3 cells and the binding buffer (20 mM Hepes, pH 7.9, 12% glycerol, 50 mM KCl, 0.2 mM EDTA, 1 mM Spermidine, 0.5 mM Dithiothreitol and 2 μg of Poly (dI-dC)). Reactions containing an antibody against acetylated-p53 (K373, K382) (Upstate Biotechnologies) were pre-incubated with nuclear extract on ice for 30 minutes before adding the [32P] Labeled duplex oligonucleotides. The binding reaction was loaded onto a 6% acrylamide gel, and subjected to electrophoresis at 200 volts for approximately 2 hrs in 0.25 X Tris Borate EDTA solution. Following electrophoresis, the gel was soaked in a 10% glycerol solution for 10 minutes and placed onto Whatman paper, covered with plastic foil and dried for 1.5 hours at 80°C. The dried gel was then exposed to a phosphor-imager plate overnight.
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
For RT-PCR the Superscript™ First-Strand Synthesis System for RT-PCR (Invitrogen) was used. The primer sequences for the GAPDH gene were as follows: forward, 5′-AAT GCA TCC TGC ACC ACC AA-3′; reverse, 5′-GTA GCC ATA TTC ATT GTC ATA-3′, giving a 515-bp product. The PCR reaction conditions were: 94°C × 50s, 55°C × 50 s, and 72°C × 1 min and 30 s, 1.5 mM MgCl2, 25 cycles. The primer sequences for the human PKD1 gene were as follows: forward, 5′-CGC CGC TTC ACT AGC TTC GAC-3′; reverse: 5′-ACG CTC CAG AGG GAG TCC AC-3′, giving a 260-bp product. The PCR conditions were: 94°C × 50s, 62°C × 50 s, and 72°C × 1 min and 30 s, 1.5 mM MgCl2, 27 cycles. The primer sequences for the human p21waf1/cip1 gene were as follows: forward, 5′-AGC TGA GCC GCG ACT GTGAT-3′; reverse, 5′-CTG AGC GAG GCA CAA GGGTA-3′, giving a 285-bp product. The PCR conditions were: 94°C × 50s, 60°C × 50s, and 72°C × 1 min and 30s, 1.5 mM MgCl2, 25 cycles.
Western Blot Analysis
Immunoblotting was performed as described [23]. Primary antibodies included p53, FL-393 (Santa Cruz Biotechnologies, 1:500), mSin3a (Abcam, 1:4000), HDAC1 (Abcam, 1:2000), and HDAC2 (Abcam, 1:2000). The immuno-reactive bands were visualized using the enhanced chemiluminescence detection system (Amersham Biotechnologies) and captured digitally using the Alpha Innotech ChemiImager. In order to verify the correct molecular weight of the target proteins, 3μl of MagicMark™ XP Western Standard (Invitrogen) was included.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using reagents and protocols from the EZ ChIP™ Chromatin Immunoprecipitation Kit (Upstate Biotechnology), as described [24], with minor modifications. IMCD3 cells were plated in 100mm plates at a density of 2×105 cells/well and cultured in Dulbecco’s Modified Eagle medium F12 (DMEM/F12, Gibco®Cell culture systems) supplemented with 10% FBS. The following day, cells were transfected with 1μg of hPKD1 promoter construct. Twenty four hours post-transfection, cells were cross-linked using a 1% formaldehyde solution in DMEM/F12 medium for 10 min at room temperature. Glycine was added to each well to quench unreacted formaldehyde for 5 min at room temperature. Cells were washed twice in ice-cold PBS followed by lysis in SDS lysis buffer. DNA was sheared by sonication yielding DNA-fragment between 500 and 1000 bp and diluted 10-fold in ChIP/dilution buffer. After dilution, 60 μl of Protein G agarose beads were added and incubated for 1 hour at 4°C with rotation in order to remove nonspecific DNA/protein-protein G agarose complexes. After pre-clearing of chromatin, the solution was centrifuged at 4000g for 1 min, 10μl (1%) was removed from the supernatant as input, and the remaining supernatant was divided into equal amounts and used in immunoprecipitation. Immunoprecipitation was performed with antibodies to p53, acetylated histone 3 (Upstate), HDAC1 and HDAC2, and mSin3A (Abcam) overnight at 4°C with rotation. The DNA-protein-antibody complexes were recovered on protein G agarose beads. The DNA-protein complexes were eluted from the agarose beads and the DNA-protein cross-links were reversed at 65°C overnight. Next, the immunoprecipitated DNA was subjected to RNase A and Proteinase K treatment (30 minutes at 37°C and 1.5 hours at 45°C respectively), followed by purification. PCR was performed in the regions flanking the p53-binding sites BS1 or BS4.
RESULTS
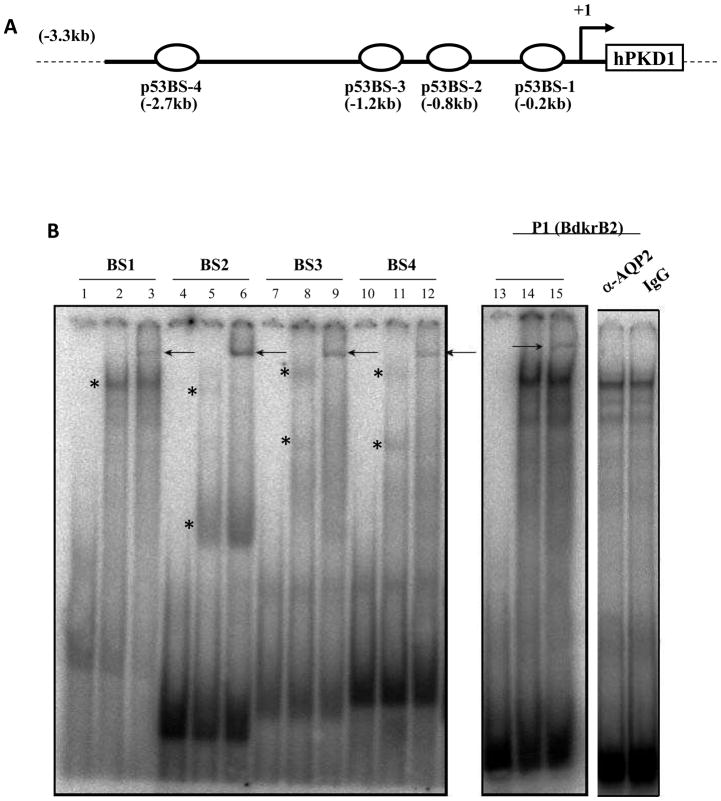
p53 binds PKD1 p53BS1-4 in Vitro
Previously, we demonstrated using ChIP-PCR that p53 binds chromatin in four regions containing putative p53 consensus sequences (BS1-4) in the PKD1 promoter [20] (Fig. 1A). Here, we used EMSA to determine the direct binding of p53 to each of these four DNA elements. Nuclear proteins extracted from IMCD3 cells were incubated with 32P-labeled oligoduplexes representing BS1-4. The results revealed the formation of one or more DNA-protein complexes (asterisk in Fig. 1B, lanes 2, 5, 8, 11 and 14). Interestingly, the binding pattern and number of complexes were somewhat different depending on the DNA sequence, presumably related to differences in p53 oligomerization and/or presence of partners in the complex. The high affinity p53 binding site (P1) in the rat Bradykinin Type 2 receptor (BdkrB2) promoter was used as a positive control [21, 22]. Addition of antibody against acetylated p53 resulted in a specific super shift (Fig. 1B, lanes 3, 6, 9, and 15). In this regards, previous studies have shown that p53 acetylation on K373 and K382 enhances p53 DNA binding affinity and transcriptional activity [25]. In contrast, addition of irrelevant antibodies against the water channel AQP2 or mouse IgG did not result in a supershift (Fig. 1B), demonstrating the specificity of the DNA binding activity. Of note, labeled BS1-4 oligoduplexes bind recombinant p53 and this can be enhanced in the presence of a p53 antibody (data not shown). These findings extend our previous data by showing that p53 binds directly and specifically to the BS1-4 DNA elements.
Figure 1. In vitro binding of p53 to the PKD1 promoter.
A. Schematic diagram showing the location of p53 consensus sequences, BS1-4. B. Electrophoretic mobility Shift Assays (EMSA). 32P-labeled oligoduplexes representing PKD1 BS1-4 were incubated with 3.5 μg of nuclear extract from IMCD3 cells. Lanes 1, 4, 7, 10 and 13: free probes; Lanes 2, 5, 8, 11 and 14: p53 BS1-4 and nuclear extract; Lanes 3, 6, 9, 12 and 15; BS 1-4 and nuclear extract in the presence of antibody against p53. Arrow indicates p53 specific super shift. P1 is an oligoduplex of the highly conserved p53 consensus sequence in the rat bradykinin B2 receptor promoter (positive control). Aquaporin 2 antibody and rabbit IgG served as negative controls.
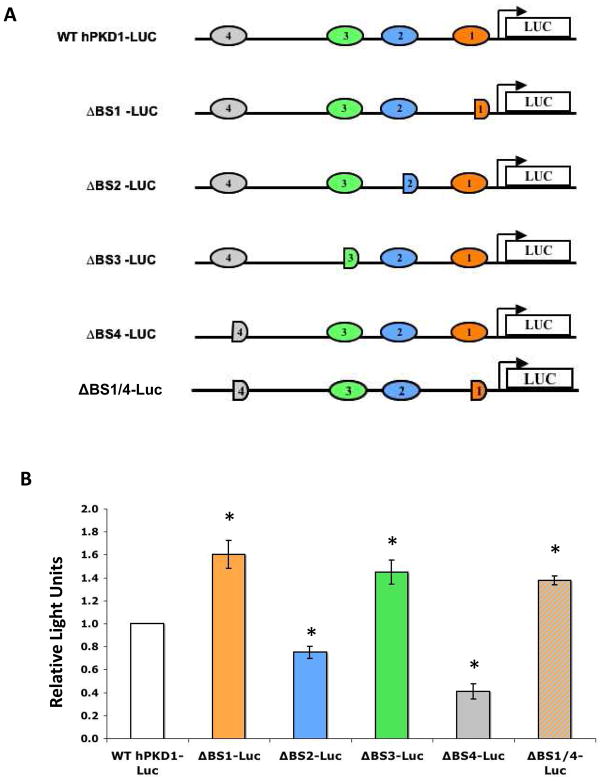
Mutagenesis of BS1-4 reveals differential effects of p53 on PKD1 promoter activity
We have shown so far that p53 binds the PKD1 promoter at four different regions: −2.7kb, −1.2kb, −0.8kb and −0.2kb, corresponding to BS4-1, respectively. To investigate the functional relevance of each of these p53-binding DNA elements, we used site-directed mutagenesis to generate mutant constructs of the parental −3.3 kb PKD1-luc construct. A schematic representation of the wild-type and mutant constructs is depicted in Fig. 2A. Mutagenesis of p53 BS1, referred to as ΔBS1-luc heretofore, resulted in 1.6-fold increase of baseline promoter activity as compared to the wild type PKD1-luc construct (Fig. 2B). This effect was also observed in HCT116 p53+/+ but not in isogenic HCT116 p53−/− cells (data not shown). These results are consistent with our previous report that p53 represses a −0.2kb PKD1-luc construct [20], and suggest that BS1 mediates transcriptional repression of PKD1.
Figure 2. Mutagenesis of BS1-4 reveals differential functions in regulation of PKD1 promoter activity.
A. Schematic representation of the parental and mutant PKD1-luc promoter constructs. B. IMCD3 cells were transiently transfected with 1 μg of PKD1-luciferase promoter constructs. pSV-LacZ (0.4 μg) was co-transfected to monitor transfection efficiency. Cell lysates were harvested 24 h post-transfection and assayed for luciferase activity. Data represent mean ± S.E. of at least three experiments, * p<0.05.
We next examined the role of BS2 located at −0.8kb of PKD1 promoter. Transient transfection assays revealed that ΔBS2-luc exhibits a modest but statistically significant decrease (−25%) in baseline activity as compared to wild type PKD1-luc construct (Figure 2B) (p<0.05). This effect was not observed in HCT116 cells (data not shown), suggesting a cell-type specific role.
Mutagenesis of BS3 at position −1.2kb of the PKD1 promoter (ΔBS3-luc) resulted in 1.4-fold increase of baseline promoter activity as compared to the wild type PKD1-luc construct in IMCD3 cells (Fig. 2B). Thus, similar to BS1, BS3 mediates transcriptional repression of hPKD1.
Mutagenesis of BS4 at position −2.7kb of the PKD1 promoter resulted in a significant 60% decrease in baseline promoter activity in both IMCD3 (Fig. 2B) and HCT116 cells (not shown) (p<0.05). Therefore, BS1 and BS3 act as repressor elements, whereas BS4 and BS2 function as enhancers. To test the latter hypothesis more rigorously, we examined the effect of ΔBS1/ΔBS4 on PKD1 promoter activity. Transfection of this double mutant construct in IMCD3 cells resulted in a significant increase of basal promoter activity as compared to the wild type construct (1.38 ± 0.04, p<0.05) (Fig. 2B), suggesting that BS4 may play a modulating role to prevent excessive repression of the PKD1 transcription by p53.
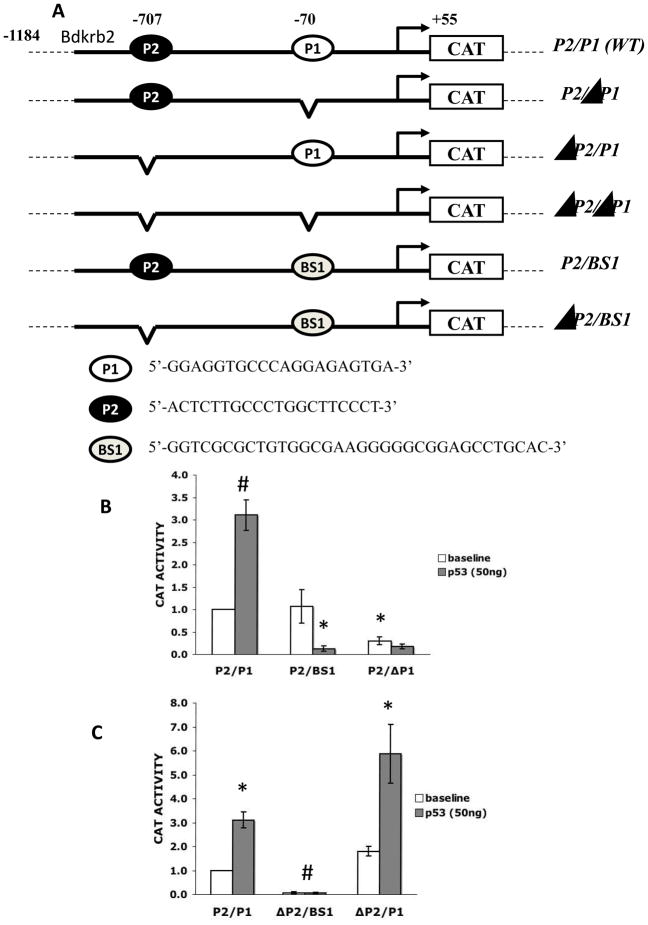
PKD1 p53BS1 mediates heterologous repression of a TATA-less gene
We previously demonstrated that BS1 is functional and responsive to p53 in the context of a synthetic TATA-Luc promoter construct. In this setting, p53 activated, rather than repressed, transcription in an orientation-independent manner [20]. However, since the PKD1 promoter lacks a TATA box, we tested whether BS1 retains its repressive function within the context of a native TATA-less promoter, in this case the rat bradykinin receptor B2 promoter (BdkrB2). The BdkrB2 promoter has two functional p53 binding sites, P1 and P2, at positions − 70 and −707 bp, relative to the transcription start site [22, 23]. BdkrB2-P1 is a bona fide positive p53-response element, and deletion of BdkrB2-P1 abrogates p53-mediated activation. In contrast, BdkrB2-P2 acts as a negative regulatory element [22, 23]. Using site-directed mutagenesis, we generated hybrid BdkrB2-(BS1)-CAT constructs, where the BS1 sequence of PKD1 was inserted in place of P1, either in the presence of P2 (P2/BS1) or absence of P2 (ΔP2/BS1). A schematic representation of the parental (P2/P1) and mutant constructs is depicted in Fig. 3A.
Figure 3. PKD1-BS1 mediates heterologous transcriptional repression.
A. A schematic representation of control and mutant rat bradykinin B2 receptor (Bdkrb2) promoter reporter constructs. The Bdkrb2 promoter has two p53-consensus sequences, P1 (enhancer) and P2 (repressor). B. Bdkrb2-CAT constructs P2/P1, P2/ΔP1 or P2/BS1 were transfected into IMCD3 cells along with the indicated amount of pCMV-p53. C. BdkrB2-CAT constructs P2/P1, ΔP2/P1 or ΔP2/BS1 were transfected into IMCD3 cells along with the indicated amount of pCMV-p53. Cell lysates were harvested 24 h post-transfection and assayed for chloramphenicol acetyltransferase activity. Data represent mean ± S.E. of at least three experiments, * p<0.05; # p<0.001.
Transient transfection assays in IMCD3 cells demonstrated that P2/BS1 has a baseline activity similar to that of the wild type P2/P1 construct. However, whereas exogenous p53 activated the P2/P1 promoter construct, p53 repressed P2/BS1 (Fig. 3B). To determine if this repression is mediated by BS1 or P2, we compared the activities of P2/BS1 and ΔP2/BS1 constructs. The results demonstrated that baseline activity of ΔP2/BS1 was significantly lower than ΔP2/P1 (Fig. 3C), consistent with the known function of P1 as a proximal enhancer element. In addition, basal ΔP2/BS1 promoter activity was higher than P2/BS1, suggesting that the P2 binding site may oppose BS1-mediated repression. Furthermore, exogenous p53 failed to activate ΔP2/BS1, whereas it activated ΔP2/P1 (Fig. 3C). These results suggest that PKD1-p53BS1 maintains its repressive function in the context of a heterologous TATA-less promoter, but acts as an enhancer element upstream of a TATA-containing promoter [20].
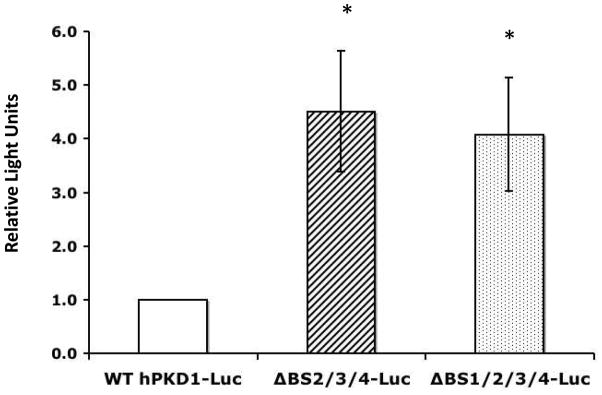
BS2-4 mutagenesis abolishes BS1-mediated repression
In order to delineate further the functionality of the p53 binding sites present in the PKD1 promoter, we generated a triple mutant construct: PKD1-ΔBS2/3/4-luc, where the p53 BS2, 3 and 4 are mutated but BS1 is left intact, and a quadruple mutant ΔBS1/2/3/4-luc, having all four sites mutated. Transient transfection of the ΔBS2/3/4-luc in IMCD3 cells resulted in a 4.5-fold increase of baseline promoter activity compared to the wild type construct (Fig. 4). Similarly, the quadruple mutant ΔBS1/2/3/4 had a 4-fold higher baseline activity than wild type construct (Fig. 4). These results, together with those in Fig. 2B, suggest that p53-mediated repression via BS1 is dependent on intact function of BS2-4, since removal of the three upstream p53 binding sites negated BS1-mediated repression.
Figure 4. PKD1-BS1 function depends on intact BS2-4.
IMCD3 cells were transiently transfected with PKD1-luciferase promoter constructs. pSV-LacZ was co-transfected to monitor transfection efficiency. Cell lysates were harvested 24 h post-transfection and assayed for luciferase activity. Data represent mean ± S.E. of at least three experiments, * p<0.05.
HDACs contribute to BS1-mediated repression
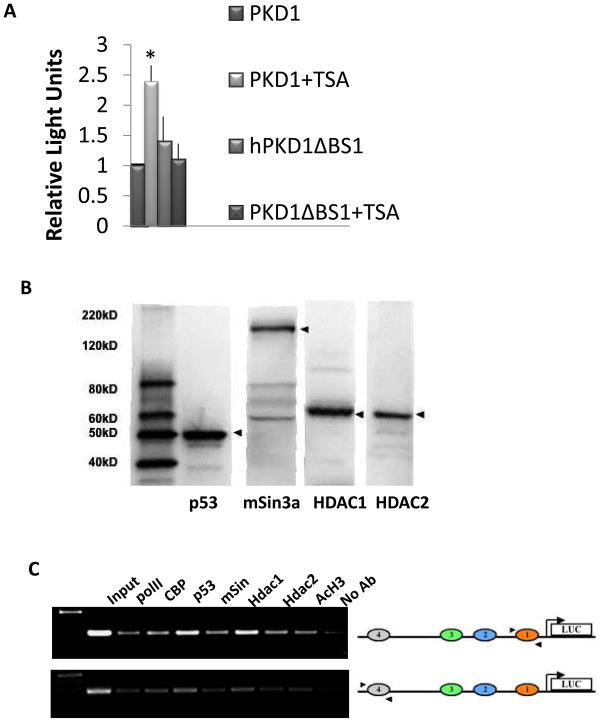
We previously demonstrated that p53-mediated repression of PKD1 is blunted by Trichostatin A (TSA), a general inhibitor of class I and II histone deacetylases (HDACs) [20]. In order to investigate the contribution of HDACs to BS1-mediated repression, IMCD3 cells were transfected with the −3.3PKD1-luc construct or its mutant derivative PKD1ΔBS1-luc and were treated with TSA or vehicle. Baseline activity of −3.3hPKD1-luc increased more than 2-fold upon TSA treatment as compared to vehicle-treated cells (Fig. 5A). In contrast, TSA failed to increase transcriptional activity of the mutant construct ΔBS1-luc, suggesting that p53 recruits a repressor complex containing HDACs to BS1.
Figure 5. Effect of Trichostatin A (TSA), an HDAC inhibitor, on PKD1 promoter activity.
A. IMCD-3 cells were treated with TSA (50 ng/ml) or vehicle 2hrs before transfection with PKD1 luciferase constructs. pSV-LacZ was co-transfected to monitor transfection efficiency. Cell lysates were harvested 24 h post-transfection and assayed for luciferase activity. Data represent mean ± S.E. of at least three experiments, *p<0.05. B. Western blot analysis demonstrating the presence of p53, mSin3A, and HDAC1/2 in IMCD3 cells. C. Chromatin immunoprecipitation analysis. PKD1-Luc promoter construct was transfected into IMCD3 cells. Twenty hour later, transfected cells were subjected to ChIP-PCR analysis as detailed in the Methods section using human PKD1-specific primers (arrowheads).
One of the basic mechanisms by which p53 exerts transcriptional repression is through interaction with and recruitment of a co-repressor complex composed of mSin3A and HDACs [26, 27]. In this regard, a p53 construct carrying mutations in a C-terminal domain required for proper interaction with mSin3a and HDAC1 [28] failed to repress PKD1 promoter [20]. Accordingly, we performed ChIP assays to determine whether a p53-mSin3A-HDAC complex assembles on the PKD1 promoter utilizing primer which can amplify human, not mouse, PKD1 promoter sequences in regions flanking BS1/4. The human PKD1-Luc construct was transiently transfected into IMCD3; twenty-four hrs later, cells were subjected to immunoprecipitation using antibodies against p53, mSin3A, HDAC1 and HDAC2, and acetylated histone H3. Western blot analysis confirmed the specificity of the antibodies used, as well as expression of p53, mSin3A, HDAC1 and HDAC2 in IMCD3 cells (Fig. 5B). The results revealed that BS1/4 are bound by p53, mSin3, and HDAC1/2 (Fig. 5C). These regions are also bound with CBP (a histone acetyltransferase), consistent with the presence of baseline acetylated Histone H3.
DISCUSSION
The present study demonstrates the presence of four p53-response elements in the human PKD1 promoter; two of these sites, BS1 (proximal) and BS4 (distal) are functional antagonists. Further, PKD1 harbors a p53-response element (BS1) which dictates active repression in association with an HDAC co-repressor complex.
The upregulation of PKD1 promoter activity as a result of BS1 mutagenesis is consistent with the fact that p53 represses a −200bp PKD1 promoter-reporter construct, which contains the BS1 motif [20]. Besides p53BS1, the −200bp proximal PKD1 promoter region contains Ets binding sites, which respond to the Ets factors Ets-1 and Fli-1, as well as multiple Sp1 binding sites [7, 19]. Deletion of the 5′ decamer of BS1 did not disrupt any of the above-mentioned binding sites; therefore, the increase in baseline activity of the ΔBS1 mutant construct is likely due to disruption of p53-BS1 interactions.
We previously reported that BS1 is functional when placed upstream of a TATA-box in the context of a synthetic reporter construct; in this setting, interaction of p53 with BS1 mediates transcriptional activation rather than repression [20]. However, since the PKD1 promoter is TATA-less, we examined BS1 function in the context of a native TATA-less promoter. To this end, we took advantage of a well characterized p53-target gene, the bradykinin B2 receptor (BdkrB2). The BdkrB2 promoter contains two p53 binding sites, P1 and P2, at positions −70 and −707 bp, relative to transcription start site, respectively. P1 mediates transcriptional activation, whereas P2 mediates repression [22]. We swapped P1 with BS1 and demonstrated that although baseline activity was similar as compared to the wild type construct (P2/P1), exogenous p53 repressed the P2/BS1-reporter construct but activated the P2/P1-reporter construct. Moreover, removal of the P2 site (ΔP2/BS1) reduced baseline promoter activity to a level lower than ΔP2/P1; and addition of p53 failed to induce activation. We therefore conclude that BS1 functions as a repressor within the context of a TATA-less promoter. In this regard, there is evidence that p53 associates with the transcriptional machinery to modulate gene transcription [29, 30].
p53 is a transcription factor which can activate or repress gene transcription depending on the target promoter and the nature of co-recruited cofactors (e.g., HDAC/mSin3 vs. CBP/p300) [26]. We reasoned that p53 represses PKD1 via recruitment of an HDAC/mSin3a complex. Since TSA can no longer induce activation of PKD1 transcription in the absence of BS1, it is reasonable to conclude that HDAC recruitment to the PKD1 promoter depends on intact BS1-p53 interactions. On the other hand, the decrease of PKD1 promoter activity when both BS1 and BS4 were mutated suggests that BS4 is involved in “linking” of distal and proximal DNA response elements by p53. Although we did not study the precise mechanism by which BS1 and BS4 interact functionally, it is conceivable that it involves stabilization of the basal transcription machinery or blockade of access of negatively acting factors. Our ChIP analysis showing the presence of a complex composed of HDAC1/2, mSin3A and p53 bound to BS1 and BS4 supports a looping model whereby distal and proximal p53-response elements are juxtaposed.
Although this and a previous study demonstrate that PKD1 is a target of p53 [20], there is evidence that p53 is downstream of PC1 signaling. For example, PC1 knockdown in HEK293 cells compromises the ability to increase p53 levels following DNA damage. Therefore, PC1 regulates a G1 checkpoint via p53 activation [31]. Further, mouse pkd1−/− cells have a tendency to undergo immortalized proliferation, associated with downregulation of the PC1-JNK-p53 pathway [32]. Thus, p53 and PC1 are components of an autofeedback pathway which functions to tightly regulate the expression of p53 and PC1, as aberrant expression of either protein lead to impaired nephron development [12, 15, 33].
Acknowledgments
This work was supported by NIH grants RO1-DK62250 and DK-56264. DVB was pre-doctoral student and partially supported by a grant from the Institutional Award program of the National Center for research Resources (P20RR017659) and the Tulane Renal and Hypertension Center of excellence. We thank Drs. Zubaida Saifudeen and Oliver Wessely for their inputs and insightful discussions about the project.
Abbreviations
- ADPKD
Adult Polycystic Kidney Disease
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sutters M, Germino GG. Autosomal dominant polycystic kidney disease: molecular genetics and pathophysiology. J Lab Clin Med. 2003;141:91–101. doi: 10.1067/mlc.2003.13. [DOI] [PubMed] [Google Scholar]
- 2.Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 3.Wilson PD. Polycystic kidney disease. N Engl J Med. 2004;350:151–64. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Taulman PD, Yoder BK. Cystic kidney disease: all roads lead to the cilium. Physiology (Bethesda) 2004;19:225–30. doi: 10.1152/physiol.00003.2004. [DOI] [PubMed] [Google Scholar]
- 5.Ong AC, Harris PC. Molecular pathogenesis of ADPKD: the polycystin complex gets complex. Kidney Int. 2005;67:1234–47. doi: 10.1111/j.1523-1755.2005.00201.x. [DOI] [PubMed] [Google Scholar]
- 6.Puri S, Magenheimer BS, Maser RL, Ryan EM, Zien CA, Walker DD, Wallace DP, Hempson SJ, Calvet JP. Polycystin-1 activates the calcineurin/NFAT (nuclear factor of activated T-cells) signaling pathway. J Biol Chem. 2004;279:55455–64. doi: 10.1074/jbc.M402905200. [DOI] [PubMed] [Google Scholar]
- 7.Puri S, Rodova M, Islam MR, Magenheimer BS, Maser RL, Calvet JP. Ets factors regulate the polycystic kidney disease-1 promoter. Biochem Biophys Res Commun. 2006;342:1005–13. doi: 10.1016/j.bbrc.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 8.Parnell SC, Magenheimer BS, Maser RL, Zien CA, Frischauf AM, Calvet JP. Polycystin-1 activation of c-Jun N-terminal kinase and AP-1 is mediated by heterotrimeric G proteins. J Biol Chem. 2002;277:19566–72. doi: 10.1074/jbc.M201875200. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, Arnould T, Sellin LK, Benzing T, Fan MJ, Gruning W, Sokol SY, Drummond I, Walz G. The polycystic kidney 1 gene product modulates Wnt signaling. J Biol Chem. 1999;274:4947–53. doi: 10.1074/jbc.274.8.4947. [DOI] [PubMed] [Google Scholar]
- 10.Saadi-Kheddouci S, Berrebi D, Romagnolo B, Cluzeaud F, Peuchmaur M, Kahn A, Vandewalle A, Perret C. Early development of pkd in transgenic mice expressing an activated mutant of the beta-catenin gene. Oncogene. 2001;20:5972–81. doi: 10.1038/sj.onc.1204825. [DOI] [PubMed] [Google Scholar]
- 11.Manzati E, Aguiari G, Banzi M, Manzati M, Selvatici R, Falzarano S, Maestri I, Pinton P, Rizzuto R, del Senno L. The cytoplasmic C-terminus of polycystin-1 increases cell proliferation in kidney epithelial cells through serum-activated and Ca2+-dependent pathways. Exp Cell Res. 2005;304:391–406. doi: 10.1016/j.yexcr.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Thivierge C, Kurbegovic A, Couillard M, Guillaume R, Cote O, Trudel M. Overexpression of PKD1 causes polycystic kidney disease. Mol Cell Biol. 2006;26:1538–48. doi: 10.1128/MCB.26.4.1538-1548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillaume R, D’Agati V, Daoust M, Trudel M. Murine Pkd1 is a developmentally regulated gene from morula to adulthood: role in tissue condensation and patterning. Dev Dyn. 1999;214:337–48. doi: 10.1002/(SICI)1097-0177(199904)214:4<337::AID-AJA6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Ward CJ, Turley H, Ong AC, Comley M, Biddolph S, Chetty R, Ratcliffe PJ, Gattner K, Harris PC. Polycystin, the polycystic kidney disease 1 protein, is expressed by epithelial cells in fetal, adult, and polycystic kidney. Proc Natl Acad Sci U S A. 1996;93:1524–8. doi: 10.1073/pnas.93.4.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard L, Sloane-Stanley JA, Sharpe JA, Aspinwall R, Lu W, Buckle V, Strmecki L, Walker D, Ward CJ, Alpers CE, Zhou J, Wood WG, Harris PC. A human PKD1 transgene generates functional polycystin-1 in mice and is associated with a cystic phenotype. Hum Mol Genet. 2000;9:2617–27. doi: 10.1093/hmg/9.18.2617. [DOI] [PubMed] [Google Scholar]
- 16.Lantinga-van Leeuwen IS, Leonhard WN, Dauwerse H, Baelde HJ, van Oost BA, Breuning MH, Peters DJ. Common regulatory elements in the polycystic kidney disease. Eur J Hum Genet. 2005;13:649–59. doi: 10.1038/sj.ejhg.5201392. [DOI] [PubMed] [Google Scholar]
- 17.Rodova M, Islam MR, Maser RL, Calvet JP. The polycystic kidney disease-1 promoter is a target of the bta catenin/T-cell factor pathway. J Biol Chem. 2002;277:29577–83. doi: 10.1074/jbc.M203570200. [DOI] [PubMed] [Google Scholar]
- 18.Jeon JO, Yoo KH, Park JH. Expression of the Pkd1 gene is momentously regulated by Sp1. Nephron Exp Nephrol. 2007;107:e57–64. doi: 10.1159/000108643. [DOI] [PubMed] [Google Scholar]
- 19.Islam MR, Puri S, Rodova M, Magenheimer BS, Maser RL, Calvet JP. Retinoic acid-dependent activation of the polycystic kidney disease-1 (PKD1) promoter. Am J Physiol Renal Physiol. 2008;295:F1845–54. doi: 10.1152/ajprenal.90355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Bodegom D, Saifudeen Z, Dipp S, Puri S, Magenheimer BS, Calvet JP, El-Dahr SS. The polycystic kidney disease-1 gene is a target for p53-mediated transcriptional repression. J Biol Chem. 2006;281:31234–44. doi: 10.1074/jbc.M606510200. [DOI] [PubMed] [Google Scholar]
- 21.Saifudeen Z, Du H, Dipp S, El-Dahr SS. The bradykinin type 2 receptor is a target for p53-mediated transcriptional activation. J Biol Chem. 2000;275:15557–62. doi: 10.1074/jbc.M909810199. [DOI] [PubMed] [Google Scholar]
- 22.Marks J, Saifudeen Z, Dipp S, El-Dahr SS. Two functionally divergent p53-response elements in the rat bradykinin B2 receptor promoter. J Biol Chem. 2003;278:34158–66. doi: 10.1074/jbc.M304543200. [DOI] [PubMed] [Google Scholar]
- 23.Shen B, Harrison-Bernard LM, Fuller AJ, Vanderpool V, Saifudeen Z, El-Dahr SS. The bradykinin B2 receptor gene is a target of angiotensin II type 1 receptor signaling. J Am Soc Nephrol. 2007;18:1140–9. doi: 10.1681/ASN.2006101127. [DOI] [PubMed] [Google Scholar]
- 24.Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4 and CBP: implications for terminal nephron differentiation. Am J Physiol Renal Physiol. 2005;288:F899–909. doi: 10.1152/ajprenal.00370.2004. [DOI] [PubMed] [Google Scholar]
- 25.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–54. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 26.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–8. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 28.Hong TM, Chen JJ, Peck K, Yang PC, Wu CW. p53 amino acids 339-346 represent the minimal p53 repression domain. J Biol Chem. 2001;276:1510–5. doi: 10.1074/jbc.M008231200. [DOI] [PubMed] [Google Scholar]
- 29.Farmer G, Friedlander P, Colgan J, Manley JL, Prives C. Transcriptional repression by p53 involves molecular interactions distinct from those with the TATA box binding protein. Nucleic Acids Res. 1996;24:4281–8. doi: 10.1093/nar/24.21.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Bae Y, Jeong W, Ahn C, Kang S. Depletion of PKD1 by antisense oligonucleotide induces premature G1/S-phase transition. Eur J Hum Genet. 2004;12:433–40. doi: 10.1038/sj.ejhg.5201136. [DOI] [PubMed] [Google Scholar]
- 32.Nishio S, Hatano M, Nagata M, Horie S, Koike T, Tokuhisa T, Mochizuki T. Pkd1 regulates immortalized proliferation of renal tubular epithelial cells through p53 induction and JNK activation. J Clin Invest. 2005;115:910–8. doi: 10.1172/JCI22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godley LA, Kopp JB, Eckhaus M, Paglino JJ, Owens J, Varmus HE. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev. 1996;10:836–50. doi: 10.1101/gad.10.7.836. [DOI] [PubMed] [Google Scholar]