Abstract
The repair of DNA double-strand breaks (DSB) by homologous recombinational repair (HRR) underlies the high radioresistance and low mutability observed in S-phase mammalian cells. To evaluate the contributions of HRR and nonhomologous end-joining (NHEJ) to overall DSB repair capacity throughout the cell cycle after γ-irradiation, we compared HRR-deficient RAD51D-knockout 51D1 to CgRAD51D-complemented 51D1 (51D1.3) CHO cells for survival and chromosomal aberrations (CAs). Asynchronous cultures were irradiated with 150 or 300 cGy and separated by cell size using centrifugal elutriation. Cell survival of each synchronous fraction (~20 fractions total from early G1 to late G2/M) was measured by colony formation. 51D1.3 cells were most resistant in S, while 51D1 cells were most resistant in early G1 (with survival and chromosome-type CA levels similar to 51D1.3) and became progressively more sensitive throughout S and G2. Both cell lines experienced significantly reduced survival from late S into G2. Metaphases were collected from every third elutriation fraction at the first post-irradiation mitosis and scored for CAs. 51D1 cells irradiated in S and G2 had ~2-fold higher chromatid-type CAs and a remarkable ~25-fold higher level of complex chromatid-type exchanges compared to 51D1.3 cells. Complex exchanges in 51D1.3 cells were only observed in G2. These results show an essential role for HRR in preventing gross chromosomal rearrangements in proliferating cells and, with our previous report of reduced survival of G2-phase NHEJ-deficient prkdc CHO cells [Hinz et al. DNA Repair 4, 782–792, 2005], imply reduced activity/efficiency of both HRR and NHEJ as cells transition from S to G2.
Keywords: radiosensitivity, cell cycle, homologous recombinational repair, non-homologous end-joining, chromosomal aberrations, complex exchange
1. Introduction
DNA double strand breaks (DSBs) are considered the most critical lesion induced by ionizing radiation (IR), many chemotherapeutic agents and endogenous oxidative metabolism. The misrepair or lack of repair of DSBs underlies the generation of chromosomal aberrations (CA), which dictate to a great extent the proliferative and carcinogenic potential of cells exposed to these genotoxic agents [1]. Mammalian cells employ two major pathways to repair DSBs: nonhomologous end joining (NHEJ) and homologous recombinational repair (HRR). NHEJ operates throughout the cell cycle to rejoin the majority of IR-induced DSBs [2–5]. NHEJ is mediated by the DNA-dependent protein kinase, consisting of the DNA end-binding Ku70–Ku80 heterodimer and the catalytic subunit DNA-PKcs, the XRCC4–LigIV–XLF scaffold/ligation complex, and several accessory proteins (reviewed in [4,5]). More recently-identified sub-pathways of NHEJ include microhomology-mediated end-joining (MMEJ), a Ku-independent process that requires 5–25 nucleotides of microhomology at DSB ends (NHEJ typically requires 0–4 nucleotides) [6], and an alternative/backup DNA-PK-independent pathway (B-NHEJ) that employs base excision repair and single-strand break repair proteins (PARP1, XRCC1, LIGI/LIGIIIα) to ligate DSB ends [7].
In mammalian cells, HRR repairs a subset of IR-induced DSBs in the S and G2 phases of the cell cycle using the sister chromatid as a repair template and mediates the recovery of broken replication forks (so-called one-sided DSBs) during DNA replication in S phase [2–4,8–10]. Repair of frank DSBs through HRR in replicated regions of the genome is thought to occur predominantly through synthesis-dependent strand annealing resulting in gene conversion, as opposed to the reciprocal exchange of sequences following Holliday junction resolution [4,10]. HRR is mediated by the RAD51 recombinase along with numerous accessory proteins including BRCA1, BRCA2, and the weakly conserved RAD51 paralogs [4,8]. Rad51-independent single-strand annealing (SSA), an error-prone HRR pathway that employs the ERCC1–XPF endonuclease and results in deletion or exchange of sequences between homologous repeats, is suggested to play only a minor role in IR-induced DSB repair in mammalian cells [4,11–13].
The five RAD51 paralogs RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 share ~20–30% protein sequence identity to Rad51 [8] and form distinct subcomplexes (RAD51B–C– D–XRCC2 and RAD51C–XRCC3) that are required for RAD51 localization to DSBs and efficient HRR [14–16]. RAD51 paralog null mutants in vertebrate cells are defective in RAD51 nuclear focus formation and show increased spontaneous CAs and sensitivity to DNA-damaging agents including IR [8,13,17–27]. It is important to consider that null mutations in core HRR genes such as RAD51, BRCA1, and BRCA2 are lethal in dividing cells, whereas null mutations in the Rad51 paralogs are embryonic lethal but not cell lethal (reviewed in [28]). Thus, HRR-defective mutant cell lines retain residual HRR activity that is sufficient for (reduced) proliferation and may contribute to overall DSB repair capacity.
The impact of NHEJ or HRR deficiency on cell killing and CA induction in Chinese hamster cells following irradiation during different phases of the cell cycle is described in several reports [2,9,13,19–21,27,29]. Compared with wild-type cells, NHEJ-deficient cells show greatly increased radiosensitivity in G0/G1 when HRR does not operate due to the lack of a sister chromatid. HRR-deficient cells are generally less radiosensitive than NHEJ-deficient cells and show increased cellular and chromosomal radiosensitivity in S and G2 [2,9,13,27]. Since the Chinese hamster mutant cell lines studied to date are not isogenic and presumably have adventitious mutations, it is unclear whether HRR capacity influences the recovery of G1-irradiated cells once they progress into S.
This study employs a well-characterized RAD51D-null mutant (51D1) and its isogenic gene-complemented derivative (51D1.3) generated in our laboratory by Cre-Lox gene targeting in CHO AA8 cells [17]. Our goals were to determine how RAD51D influences the survival of synchronous cells irradiated throughout the cell cycle and provide further understanding of how HRR prevents lethal CA induction. We have previously shown that 51D1 rad51d cells are ~1.5-fold more sensitive to γ-rays and have 3-fold higher levels of spontaneous CAs (primarily chromatid-type), a 12-fold increased rate of hprt gene mutation, and 4–10-fold higher rates of gene amplification at the DHFR and CAD loci respectively, relative to isogenic controls [17]. Since CHO cells are Tp53-defective due to a single amino acid change at position 211 and do not undergo IR-induced G1-phase delay or apoptosis [30,31], lethal CAs are the primary cause of cell killing in these cell lines [32]. We show that, compared with control cells, HRR-deficient rad51d cells have significantly higher chromatid-type CA induction in S and G2, which is inversely correlated with their survival measured by colony-forming ability after γ-irradiation.
2. Materials and Methods
2.1. Cell culture and irradiations
Derivation of the isogenic 51D1 rad51d and CgRAD51D-complemented 51D1 (51D1.3) CHO cell lines is described in [17]. Cultures of 51D1 and 51D1.3 cells were grown in suspension at 37°C in αMEM medium (GIBCO/Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 100 U/ml penicillin G (Sigma), and 100 µg/ml streptomycin sulfate (Sigma) [33]. Cultures of ~2 × 108 exponentially growing cells were centrifuged at 300 × g, resuspended in fresh ice-cold culture medium in 50-ml conical tubes, and irradiated with 150 or 300 cGy of 662 keV γ-rays with a J. L. Shepard Mark-I cesium-137 beam irradiator at a dose-rate of ~2.5 Gy/min. Irradiated cells were then synchronized at 4°C by centrifugal elutriation using a Beckman Coulter J6-MI centrifuge at 2800 rpm and an initial flow rate of 19–20 ml/min. Successive fractions were collected by increasing the flow rate in increments of 1 ml/min to obtain ~20 fractions total.
To determine the cell cycle distribution of each fraction, 5 × 105 cells were centrifuged at 300 × g, resuspended in fresh 37°C growth medium supplemented with 10 µg/ml 5-bromo-2′-deoxyuridine (BrdU), and cultured at 37°C for 30 min. Cells were then centrifuged at 300 × g, rinsed with PBS and fixed with 70% ethanol, treated with 5 µg/ml RNase (Qiagen), and denatured with 2M HCl. Cell suspensions were rinsed with PBS and incubated at 37°C with 1:100 Alexa-488-conjugated anti-BrdU mouse monoclonal antibody (PRB-1, Molecular Probes/Invitrogen) in PBS for 30 min, rinsed with PBS, and stained with 50 µg/ml propidium iodide. The proportion of BrdU-positive (S phase) cells and total DNA content was measured by multi-parameter flow cytometry using a FACscan (Becton Dickinson) and data were analyzed using BD CellQuest™ software. The survival of the asynchronous cultures before and after irradiation and of each elutriated fraction was determined by single-cell colony formation ability by plating 300–600 cells in triplicate in 10-cm culture dishes with complete medium. After 8–10 days of growth at 37°C, the dishes were rinsed with PBS, fixed with 95% ethanol, and stained with Gram Crystal Violet (Becton Dickinson). Colonies of ≥50 viable cells were scored as survivors.
2.2. Chromosome aberration (CA) analysis
For the first, every subsequent third, and final elutriation fractions spanning the cell cycle (7 fractions total), ~2 × 105 cells per fraction were centrifuged at 300 × g, resuspended into fresh 37°C culture medium, aliquoted into two 15 ml conical tubes, and incubated at 37°C. Mitotic collection intervals were based upon the population doubling time of these cell lines (~12 h for 51D1.3 cells and ~16 h for 51D1 cells, with a 1–2h allowance for IR-induced cell cycle delay) and the cell cycle position of the particular elutriation fraction. For example, the first elutriation fractions containing early G1-phase 51D1 cells were cultured for ~16–18 h before metaphase collection, while the final elutriation fractions containing late G2/M-phase cells were cultured for 2 h prior to collection. Cultures were treated at 37°C with 0.1 µg/ml KaryoMAX® colcemid solution (GIBCO/Invitrogen) for 2–2.5 h, collected and centrifuged at 300 × g for 3 min. The cell pellet was resuspended in 10 ml of 37°C 75 mM KCl hypotonic buffer, and incubated for 7 min at 37°C. Two ml of fresh 3:1 methanol:acetic acid (Carnoy’s) fixative was added to the cell suspension in hypotonic buffer and gently mixed. After 2 min, the suspensions were centrifuged at 200 × g for 3 min and 4 ml of fresh fixative was added drop-wise. This procedure was repeated twice and cell suspensions were dropped onto cold, wet slides, air-dried and desiccated for 24 h at 37°C. Slides were stained in a 10% Giemsa solution (Gurr® R66, BDH Chemicals, Ltd.) prepared in a pH 6.8-buffered solution (Sorenson’s buffer), washed vigorously in McIlvaine’s rinsing buffer (approximately 18 mM citric acid/16 mM disodium phosphate prepared in distilled water) and distilled water, and dried with an air jet. Slides were mounted with CytoSeal™ 60 mounting medium (Microm International) and a coverslip.
Approximately 50–75 diploid metaphase spreads per elutriation fraction in four independent experiments per cell line were scored using a 100× oil-immersion objective and 2× optivar on a Zeiss Axiophot microscope. Only metaphase spreads with minimal chromosome overlap and cytoplasmic interference were examined. The modal chromosome number for both cell lines was 21 and ranged from 20–22. Mitotic indices of the elutriation fractions were consistently high, ranging from 9–36% for the 51D1 samples and 11–40% for the 51D1.3 samples. The Savage classification scheme for scoring chromosomal aberrations was used [34]. Chromosome-type aberrations include dicentrics, centric and acentric rings, and terminal deletions; chromatid-type aberrations include chromatid gaps and breaks, isobreaks, and symmetrical and asymmetrical chromatid-type exchanges. For this study, chromatid gaps are defined as fully discontinuous chromatid fragments detached at a distance less than the width of the chromatid arm [35]. Chromatid breaks are defined as fully discontinuous chromatid fragments either displaced at a distance greater than the width of the chromatid from the original chromosome or no longer aligned with the original chromatid axis. Chromatid fragments that did not appear to be completely severed from the chromatid were considered achromatic lesions and were not scored. Complex chromatid-type exchanges (requiring ≥3 breaks on ≥2 chromosome arms [36,37]) were also recorded along with the number of chromosomes involved in these exchanges, but no attempt was made to estimate the number of actual exchange events involved in these grossly complex aberrations. Statistical significance for differences between 51D1 and 51D1.3 cell survival values and CA frequencies were tested using Student’s t-tests for two independent sample distributions (SigmaPlot, Systat Software, Inc.).
3. Results
3.1. Increased radiosensitivity of rad51d cells in S and G2, but not G1
Our experimental approach involved irradiating exponentially growing populations of 51D1 rad51D cells and CgRad51D-complemented 51D1 (51D1.3) cells and then separating these populations into highly synchronous fractions by centrifugal elutriation, as previously reported [2]. The relative survival of 19–20 elutriation fractions of the HRR-deficient 51D1 cells and HRR-proficient 51D1.3 cells irradiated with 150 or 300 cGy is shown in Fig. 1. Survival is plotted as a function of the fraction of cells in S phase (as determined by BrdU labeling and flow cytometry), with the early G1-phase fractions on the left and late G2/M-phase fractions on the right (see also SI Figs. 1 and 2 for representative FACS profiles and survival plotted against elutriation fraction number). The relative survival of irradiated asynchronous cultures before elutriation is shown in Fig. 1 on the far right and is in close agreement with our previous data [17]. Plating efficiencies before elutriation for the four independent experiments conducted per cell line ranged from 0.6–0.75 for the 51D1 cells and 0.85–0.95 for the 51D1.3 cells, also in close agreement with our previous data [17]. The elutriation protocol did not significantly reduce the plating efficiency of the elutriated fractions (data not shown).
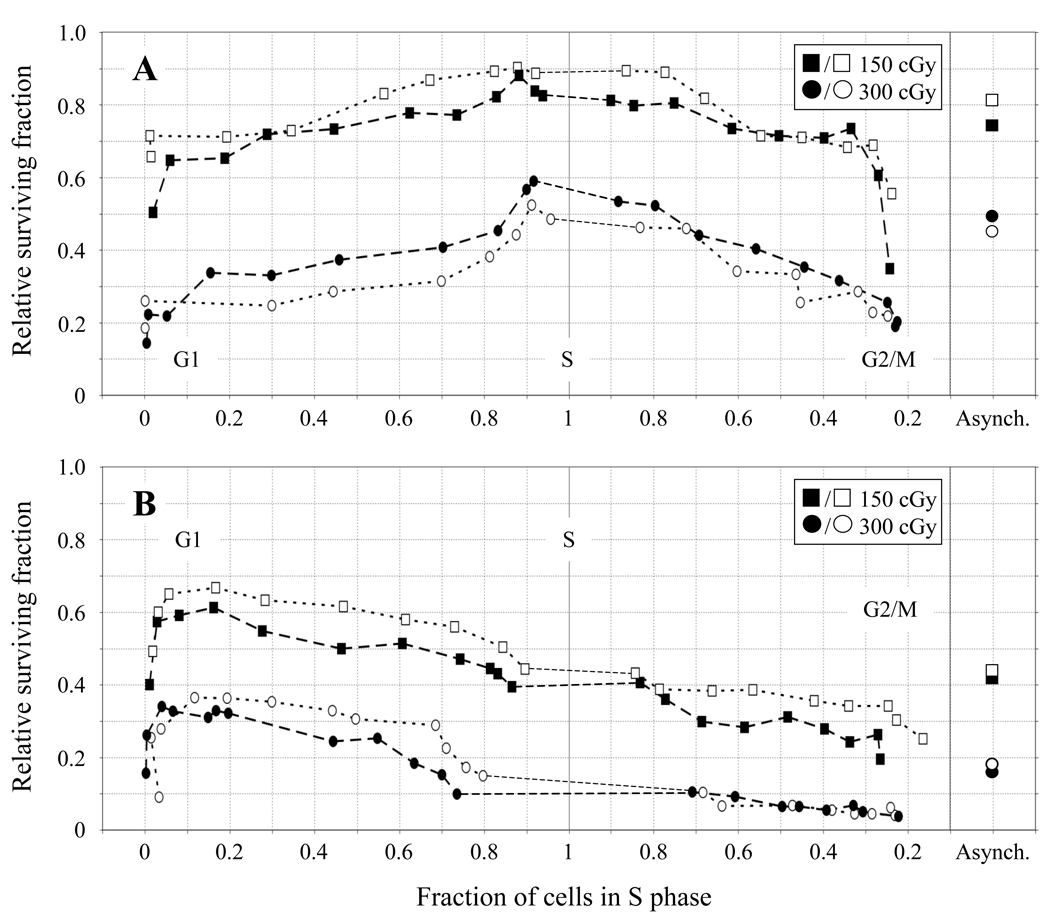
Fig. 1.
Relative survival of 51D1.3 (A) and 51D1 (B) cells after 150 cGy (squares) and 300 cGy (circles) γ-irradiation plotted against fraction of cells in S phase. Survival values are corrected for plating efficiency of unirradiated cells. Open and filled symbols represent survival data for two independent experiments conducted at each dose. The relative survival of the asynchronous cell cultures is shown on the far right (“Asynch.”).
The survival of 51D1.3 cells in Fig. 1A shows the classic pattern of S-phase radioresistance previously observed by us and other groups using wild-type Chinese hamster cell lines [2,9,38]. At 300 cGy, 51D1.3 survival increases ~2.5-fold (from ~0.2 to ~0.5) as cells exit G1 and enter S, remains high throughout S, and subsequently declines by ~2.5-fold as the cells enter G2/M. At 150 cGy, the pattern of 51D1.3 survival is qualitatively similar to the pattern at 300 cGy, although less pronounced. In contrast, the survival of 51D1 rad51d cells in Fig. 1B is maximal in G1, and cells become increasingly more sensitive as they progress through S and into G2/M. This reduction from G1 to G2/M is ~3-fold (from ~0.6 to 0.2) at 150 cGy and ~7-fold (from 0.35 to 0.05) at 300 cGy. For both cell lines, the survival of the pre-elutriated asynchronous cultures most closely matches the survival of the S-phase fractions, which would be expected since the majority of cells in the asynchronous cultures are in S phase. At 150 cGy, asynchronous 51D1 survival is ~1.8-fold lower than 51D1.3 survival; this difference increases to ~2.8- fold for 300 cGy.
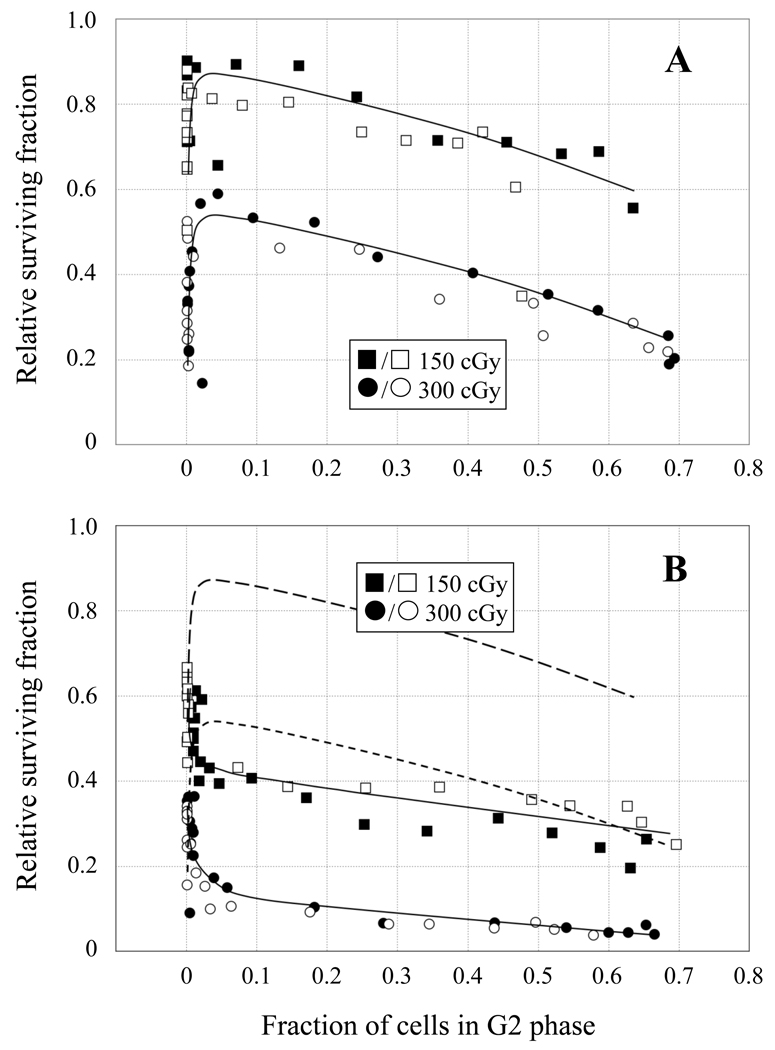
Importantly, the two IR doses result in nearly equivalent cell killing of both cell lines in the G1-phase fractions (~60–70% survival for 150 cGy; ~25–35% survival for 300 cGy), supporting the notion that HRR does not contribute significantly to the survival of cells irradiated in G1. In S phase, 51D1 cell survival is ~2-fold lower than the 51D1.3 cells at 150 cGy and ~4-fold lower at 300 cGy. Both cell lines experience a significant decline in survival (p < 0.05) as cells exit S and progress through G2. For both cell lines, the lowest survival is observed for the late G2/M fractions and the early G1 fractions, consistent with the report that mitosis is the most radiosensitive portion of the cell cycle [38]. In Fig. 2, survival is plotted as a function of cells in G2 to more clearly illustrate the reduced survival of both cell lines in G2 compared to S phase. 51D1.3 survival declines ~1.5–2-fold as cells move from S into G2 after both 150 and 300 cGy (Fig. 2A), and 51D1 survival declines similarly (Fig. 2B). Compared to G2-phase 51D1.3 cells, G2-phase 51D1 cells have ~2.5-fold lower survival at 150 cGy and ~5-fold lower survival at 300 cGy, as seen in Fig. 2B. Flow cytometry analyses indicated that ~60–70% of cells in the final elutriation fractions were G2/M-phase cells with 4N DNA content (SI Figs. 1 and 2) while the mitotic indices for these fractions were 10–15% following the 2 h colcemid treatment (i.e., 5– 7.5% of cells per h entered mitosis). Therefore, the majority of cells in these final fractions were in G2 at the time of irradiation, and mitotic cells are unlikely to contribute significantly to the pattern of decreased G2-phase survival in Figs. 1 and 2.
Fig. 2.
Relative survival of 51D1.3 (A) and 51D1 (B) cells after 150 cGy (squares) and 300 cGy (circles) γ-irradiation plotted against fraction of cells in G2 phase. Open and filled symbols represent survival data for two independent experiments conducted at each dose. Solid lines in both panels are best fit to the survival data; dashed lines in (B) are the best-fit lines of 51D1.3 survival data from (A) for comparison.
3.2. High levels of IR-induced chromatid-type aberrations in rad51d cells
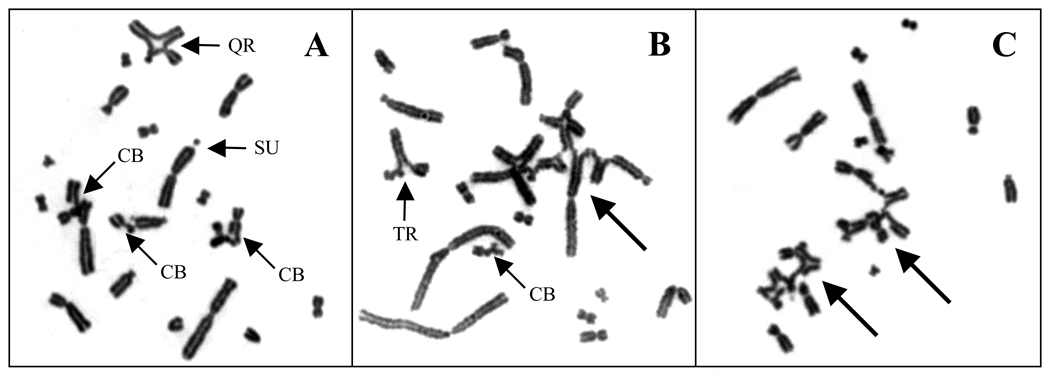
To determine the cause of these cell cycle-associated radiosensitivity patterns, we examined CA induction in approximately one-third of the elutriation fractions spanning the cell cycle. The majority of 51D1 metaphases contained both chromosome-type aberrations (i.e., those induced in non-replicated chromosomal regions in G1/S-phase cells) and chromatid-type aberrations (i.e., those induced in replicated chromosomal regions in S/G2-phase cells). Occasionally both classes of aberrations affected the same chromosome(s) (e.g., a dicentric with a chromatid-type intrachange). Examples of IR-induced CAs observed in 51D1 metaphases are shown in Fig. 3 and come from elutriation fractions 7–13, which contain predominantly S-phase cells (~50–90%) at the time of irradiation. Fig. 3A shows a 51D1 metaphase after 150 cGy with several chromatid breaks and inter- and intrachromosomal chromatid-type exchanges (quadriradial, sister union). Fig. 3B shows a 51D1 metaphase after 300 cGy with inter- and intrachromosomal chromatid-type exchanges and a complex chromatid-type exchange involving 7 chromosomes (marked by arrow). Similarly, Fig. 3C shows a 51D1 metaphase after 300 cGy with two complex exchanges (marked by arrows), one involving 4 chromosomes and the other involving 5 chromosomes.
Fig. 3.
Examples of IR-induced CAs in S-phase 51D1 cells. (A) 150 cGy, elutriation fraction 13 (~75% S); metaphase with chromatid breaks (CB) and inter- and intra-chromosomal chromatid-type exchanges, including a quadriradial (QR) and sister union (SU), marked by smaller arrows. (B) 300 cGy, elutriation fraction 7 (~60% S); metaphase with an inter-chromosomal chromatid-type exchange [triradial (TR), marked by smaller arrow] and complex exchange involving seven chromosomes (marked by larger arrow). (C) 300 cGy, elutriation fraction 10 (~75% S); metaphase with two complex exchanges involving four and five chromosomes (marked by larger arrows).
The frequencies of chromosome-type and chromatid-type CAs induced by 150 and 300 cGy plotted in Figs. 4 and 5 and reported in SI Tables 1–4 are total observed frequencies and have not been corrected for the spontaneous CA levels in unirradiated cultures, on which we reported previously [17]. 51D1 cells have 3-fold higher levels of spontaneous CAs (primarily chromatid-type gaps and breaks; ~3/cell) compared to 51D1.3 cells (~1/cell). Simple chromatid-type exchanges also occur spontaneously in 51D1 cells at low frequency (~1 in 25 cells), but not in 51D1.3 cells. Figs. 4 and 5 include data for 4 independent experiments conducted with each cell line (two at 150 cGy, two at 300 cGy) normalized per unit dose for comparison purposes.
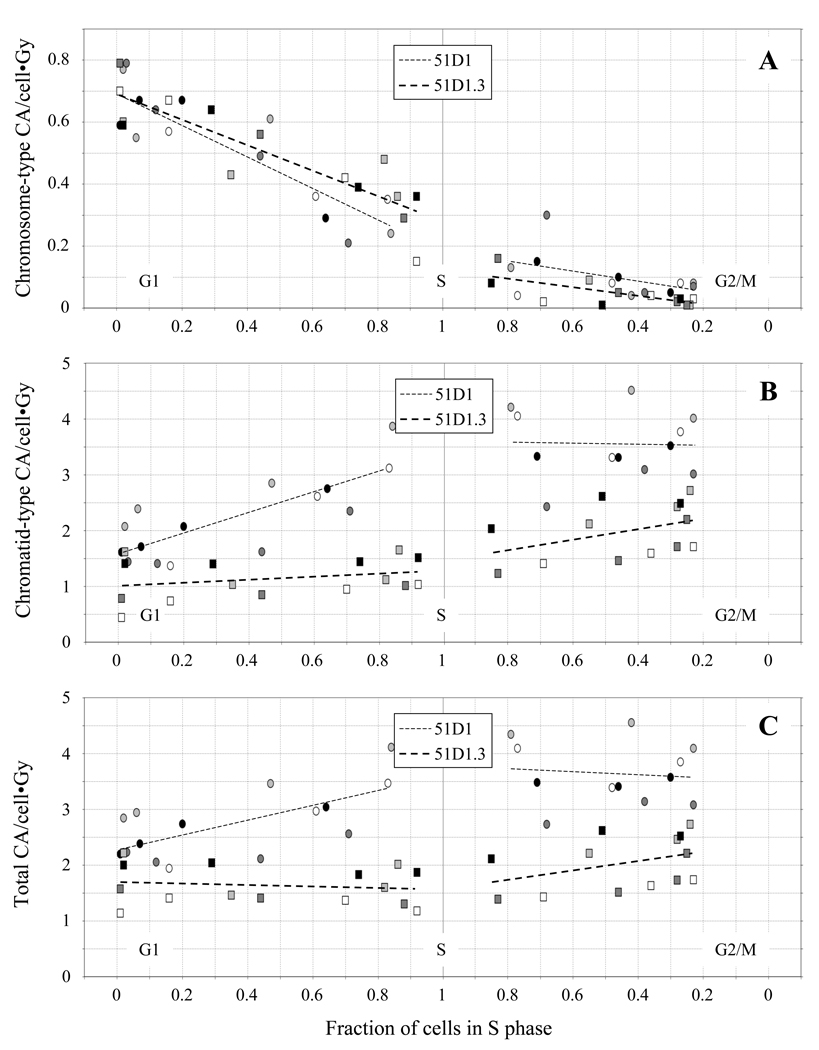
Fig. 4.
Frequencies of chromosome-type CAs (A), chromatid-type CAs (B), and total CAs (C) in 51D1 cells (circles) and 51D1.3 cells (squares) after 150 and 300 cGy γ-irradiation plotted against fraction of cells in S phase. CA frequencies are normalized per unit dose; different symbol fill colors represent four independent experiments performed with each cell line.
Fig. 5.
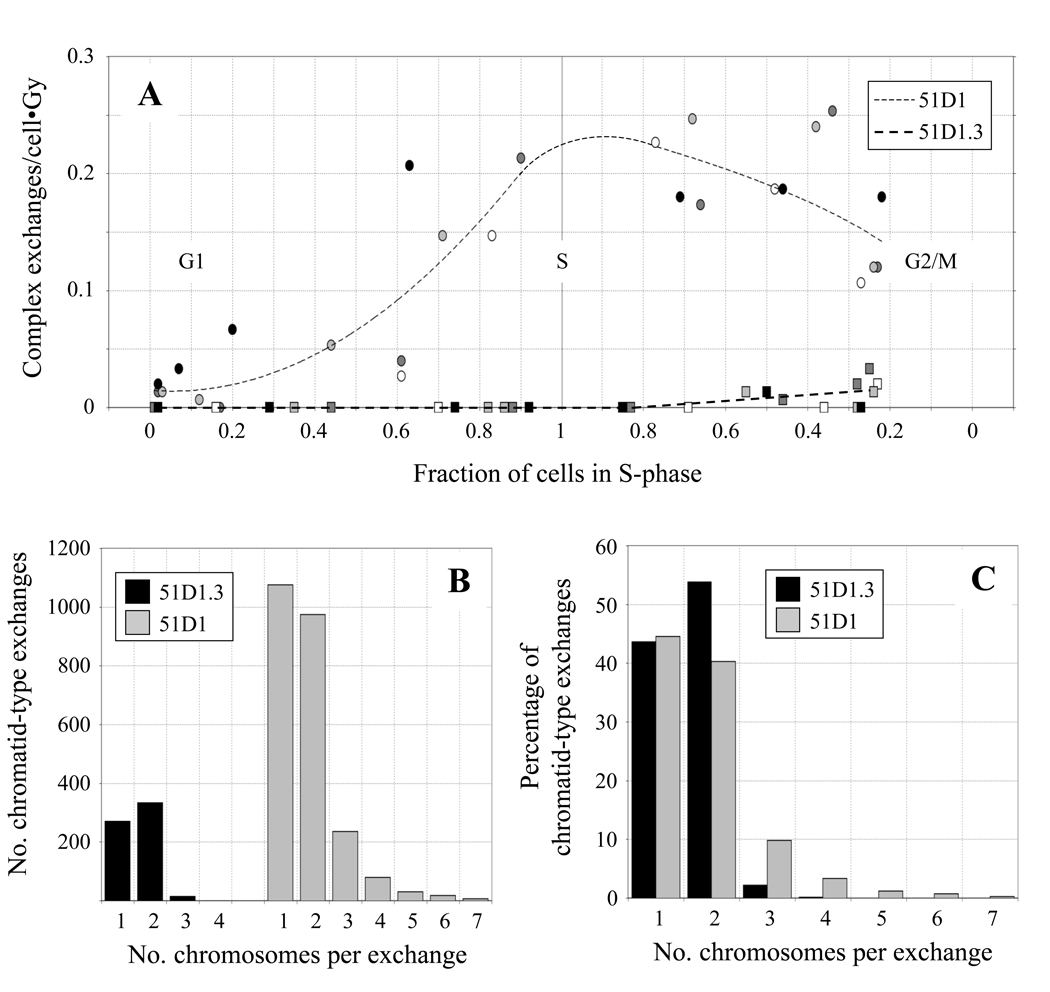
(A) Frequencies of complex exchange aberrations in 51D1 and 51D1.3 cells after 150 and 300 cGy γ-irradiation plotted against fraction of cells in S phase. CA frequencies are normalized per unit dose; symbols and fill colors are defined in Fig. 4. (B) Distribution of the number of chromosomes per chromatid-type exchange in 51D1 and 51D1.3 cells. (C) Distribution of the number of chromosomes per chromatid-type exchange relative to the percentage of chromatid-type exchanges in 51D1 and 51D1.3 cells.
In Fig. 4, the frequencies of chromosome-type CAs, chromatid-type CAs, and total CAs (excluding the complex aberrations discussed below) are plotted in panels A–C, respectively, as a function of the fraction of cells in S phase. In Fig. 4A, chromosome-type CAs (dicentrics, rings, deletions) in both cell lines are highest in G1, decrease during S, and return to background levels in G2 (~0.01/cell [17]). For the G1 fractions, approximately 0.22 dicentrics, 0.07 centric rings, and 0.35 chromosome-type deletions were induced per cell/Gy (see SI Tables 1–4). Chromosome-type CA frequencies do not differ significantly between cell lines (p = 0.83). This result implies that HRR does not participate in DSB repair events that result in chromosome-type exchanges in G1-irradiated cells.
In Fig. 4B, 51D1 cells show significantly higher (~1.5–2-fold, p < 0.01) levels of chromatid-type CAs (gaps, breaks, and exchanges) throughout the cell cycle compared with 51D1.3 cells (see also SI Tables 1–4). Levels of chromatid-type CAs increase as 51D1 cells progress from G1 to S and remain high through G2. In contrast, 51D1.3 cells show relatively little increase in these types of aberrations from G1 to early S, followed by an increase from late S through G2. Approximately 0.7 chromatid breaks and 1 simple chromatid exchange are induced per cell/Gy in late S/G2-phase 51D1 cells compared to ~0.3 breaks and 0.35 exchanges induced per cell/Gy in late S/G2-phase 51D1.3 cells. In contrast to the ~2–4-fold increase in breaks and ~5–10-fold increase in exchanges as cells progress from G1 to late S/G2, only a modest increase in gaps (~20–40%) above background levels was observed after irradiation in the late S/G2 fractions of both cell lines. In Fig. 4C, the pattern of total CA induction follows the chromatid-type CA pattern of Fig. 4B, with 51D1 cells having significantly higher total CAs than 51D1.3 cells (p < 0.01). These results suggest that higher levels of IR-induced chromatid-type CAs are primarily responsible for the decreased survival observed in both HRR-deficient 51D1 cells and HRR-proficient 51D1.3 cells as they progress from S to G2.
3.3 High levels of IR-induced complex chromosomal aberrations in rad51d cells
By definition, the generation of complex CAs requires ≥3 breaks on ≥2 chromosomes [36], or more technically, on ≥1 chromosome in the case of complex intrachromosomal exchanges [37]. Examples of complex CAs in S-phase 51D1 cells can be seen in Fig. 3B and 3C. These complex CAs appear to be concatamers of symmetrical and asymmetrical inter- and intrachanges with their accompanying acentric fragments. These exchange figures suggest they were generated in S or G2 by the NHEJ-mediated misrepair of DSBs distributed across several non-homologous chromosomes. Given that the final elutriation fractions contain 20–30% S-phase cells, we cannot specify if these events occurred during NHEJ-mediated repair of one-sided DSBs at broken replication forks in S phase or during NHEJ-mediated repair of two-sided DSBs in post-replicative chromosomal regions. However, the complex CAs observed in the final fractions of both cell lines must have been generated in G2 since they were collected 2 h after irradiation (similar to the protocol of the G2 chromosomal radiosensitivity assay [39]). Complex exchange formation was not an artifact of the elutriation process, as identical levels of complex exchanges were observed in cultures grown as adherent monolayers in tissue culture flasks, irradiated with the same doses, and collected 3 h and 8 h post-irradiation to examine G2 phase and S phase-irradiated cells, respectively, according to the protocol of Natarajan et al. [21] (see SI Table 5). Spontaneous complex exchanges were not detected in either cell line.
Frequencies of IR-induced complex chromatid-type CAs in 51D1 and 51D1.3 cells are normalized per unit dose and plotted in Fig. 5A as a function of the fraction of cells in S phase (see also SI Tables 1–4). Compared to 51D1.3 cells, the induction of complex chromatid-type exchanges in 51D1 cells is much higher (p < 10−7). Similar to the pattern of simple chromatid-type CAs in Fig. 4B, complex chromatid-type exchanges in 51D1 cells increase as cells enter S. However, distinct from the Fig. 4B pattern, levels of complex chromatid-type CAs decrease somewhat as 51D1 cells enter G2/M. In contrast, 51D1.3 cells show no complex exchanges in early S-phase followed by a low level in late S/G2. Overall, complex chromatid-type CAs occurred in 51D1.3 cells in 1 in ~100 cells after 150 cGy and 1 in ~25 cells after 300 cGy compared to 1 in ~3–4 cells after 150 cGy and 1 in ~1.5–2 cells after 300 cGy in 51D1 cells.
Histograms of the number of chromosomes involved in chromatid-type exchanges in the 51D1 and 51D1.3 cells are shown in Fig. 5B and 5C, which include data for both simple and complex exchanges. On average, 1.6 chromosomes are involved in 51D1.3 chromatid-type exchanges and 1.8 chromosomes are involved in 51D1 chromatid-type exchanges. The large majority of chromatid-type exchanges in 51D1.3 cells are simple, involving only 1 chromosome (intrachange; 44% of total) or 2 chromosomes (interchange; 54% of total). Very few complex exchanges are observed in these HRR-proficient cells (~2% of total chromatid-type exchanges) and were limited to 3–4 chromosomes; in fact, only one 4-chromosome complex exchange was observed in ~2000 51D1.3 cells scored.
Interestingly, the overall complexity of chromatid-type interchanges is higher in 51D1 cells than in 51D1.3 cells (Figs. 5B and 5C). While interchanges constitute 56% of total 51D1 chromatid-type exchanges, the fraction of simple interchanges is 40% (versus 54% in 51D1.3 cells), and the fraction of complex exchanges is 15% (versus ~2% in 51D1.3 cells). While the maximum number of chromosomes per 51D1 complex exchange was 7, metaphases containing 2–4 independent complex exchanges involving 3–7 chromosomes each were observed (see example in Fig. 3C). In these cases, a maximum of 13 chromosomes per cell (~60% of the total CHO genome) were involved in complex exchange events. Cells with multiple complex exchanges accounted for ~16% of 51D1 cells with complex exchanges and ~2% of 51D1 cells with chromatid-type exchanges. Only single complex exchange events were observed in 51D1.3 cells. Overall, IR-induced complex exchanges in 51D1 cells occurred at ~25-fold higher frequency per unit dose compared to 51D1.3 cells.
4. Discussion
Our results support the hypothesis that HRR-deficient mammalian cells repair IR-induced DSBs (including one-sided DSBs) predominantly by error-prone NHEJ in the S and G2 phases of the cell cycle. 51D1.3 cells display the classical wild-type cell cycle response to IR [27,38], being most resistant in S and most sensitive in mitosis, whereas 51D1 rad51d cells show a pattern analogous to other HRR-deficient hamster cell lines [2,9,27], being most resistant in G1 and becoming progressively more sensitive through S and G2/M. Both 51D1 and 51D1.3 cells have nearly equivalent survival in the earliest G1-phase fractions. However, compared to 51D1.3 cells, 51D1 cells have lower survival in the late G2/M-phase fractions. These results imply that HRR contributes significantly to IR-induced DSB repair and cell survival in G2 but not in G1. A general pattern of declining survival is clearly evident as both wild-type and HRR-deficient cells progress from S to G2/M post-irradiation (Figs. 1 and 2 and [2]). We have reported a similar pattern of reduced survival through the cell cycle in NHEJ-deficient prkdc CHO cells (V3) [2], which are HRR-proficient. Thus, the capacity of both pathways to repair IR-induced DSBs appears to decline as cells prepare for mitosis, an issue that could not be resolved in an earlier study [40].
Chromatid-type (as well as chromosome-type) aberrations were observed in G1-irradiated 51D1 cells, suggesting unrepaired single-strand DNA damage was converted to onesided DSBs during replication. Along with other HRR-deficient CHO/V79-derived cell lines [18–22], this phenotype of chromatid-type CA induction in G0/G1-irradiated cells is also shared with human ataxia-telangiectasia cells [41] and NHEJ-defective rodent cells [21,42], suggesting possible roles for ATM and NHEJ in the repair of broken replication forks (see also [43,44]). However, we observed only limited induction of chromatid gaps compared to the prominent induction of chromatid breaks and exchanges in late S/G2-phase 51D1 and 51D1.3 cells, suggesting this class of chromatid-type CAs does not significantly influence cell survival. Controversies regarding the origin and nature of chromatid gaps (including discrepancies in the definitions of chromatid “gaps” versus “breaks” based on the degree of fragment separation) and their role in IR-induced cell killing are further discussed in [45,46].
We have shown that chromatid-type exchanges are a major class of spontaneous and IR-induced CAs in 51D1 rad51d cells compared to control cells. Studies examining rodent cells deficient in BRCA1, BRCA2, and the other RAD51 paralogs [18–25,47–49] likewise report excess levels of chromatid-type exchanges. A recent cytogenetic study [21] examining several DSB repair-deficient Chinese hamster cell lines (including 51D1) irradiated in G1, S or G2 documented CA induction quantitatively similar to our data but did not report complex CA induction in 51D1 cells, a key finding of our study. The induction of simple and complex chromatid-type exchanges in G2-phase 51D1 and 51D1.3 cells is consistent with their reduced survival in this phase and is indicative of declining DSB repair efficiency. Complex CA induction also occurs in S-phase 51D1 cells, suggesting that chromosome mis-rejoining occurs as broken replication forks are inappropriately resolved by NHEJ rather than HRR. The appearance of complex CAs only in G2-phase 51D1.3 cells suggests that there is likely an increased probability of mis-rejoining IR-induced DSBs in this phase cells regardless of which repair pathway initiates repair. The non-conservative HRR sub-pathway SSA could potentially be responsible for generating complex CAs since levels of chromatid exchanges following IR or UV-irradiation are significantly reduced in ercc1 and xpf mouse and Chinese hamster cells compared to wild-type cells [12,50,51].
In conclusion, our results show that HRR capacity is absolutely required for the proper repair of a subset of IR-induced DSBs in S/G2-phase cells and also suggest the efficiency of DSB repair declines from S to G2. HRR is an important defense against tumorigenesis as evidenced by the presence of gross chromosomal rearrangements in spontaneous tumors derived from BRCA1 and BRCA2-mutant mice [47–49,52]. Likewise, cytogenetic studies of BRCA1 and BRCA2-deficient breast cancers suggest that chromosomal rearrangements figure prominently in the etiology of these tumors in humans [53–55]. Several reports suggest that somatic (non-tumor) cells derived from heterozygous BRCA1/2 mutation carriers have impaired DNA repair capacity judged by higher rates of IR-induced chromatid breakage, implying higher susceptibility for spontaneous chromosomal rearrangements in these individuals [56–58]. High levels of chromatid-type exchanges (triradials and quadriradials) are also seen in Fanconi anemia patients, of which the clinical subgroups D1, J, and N result from biallelic mutations in the HRR genes BRCA2, BRIP1/BACH1, and PALB2, respectively (reviewed in [59]). Taken together, this evidence clearly indicates that HRR plays an essential role in maintaining genomic integrity in replicating cell populations both spontaneously and after treatment with DNA damaging agents. In addition, the strong correlation between IR-induced CAs and cellular lethality in Tp53-defective CHO cells in this study is particularly informative for understanding the role of CAs in the killing of Tp53-defective human tumor cells during radiotherapy.
Supplementary Material
Acknowledgements
We would like to thank Dr. Joel Bedford and Dr. Hatsumi Nagasawa for their insightful comments. This research was supported by NIH National Cancer Institute grant CA112566 from the U.S. Department of Health and Human Services. This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contracts W-7405-Eng-48 and DE-AC52-07NA27344.
Abbreviations
- CA
chromosomal aberration
- DSB
DNA double-strand break
- HRR
homologous recombinational repair
- IR
ionizing radiation
- NHEJ
non-homologous end joining
- SSA
single-strand annealing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Pfeiffer P, Goedecke W, Kuhfittig-Kulle S, Obe G. Pathways of DNA double-strand break repair and their impact on the prevention and formation of chromosomal aberrations, Cytogenet. Genome Res. 2004;104:7–13. doi: 10.1159/000077460. [DOI] [PubMed] [Google Scholar]
- 2.Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair. 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 4.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 8.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 9.Tamulevicius P, Wang M, Iliakis G. Homology-directed repair is required for the development of radioresistance during S phase: interplay between double-strand break repair and checkpoint response. Radiat. Res. 2007;167:1–11. doi: 10.1667/RR0751.1. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankenberg-Schwager M, Gebauer A, Koppe C, Wolf H, Pralle E, Frankenberg D. Single-strand annealing, conservative homologous recombination, nonhomologous DNA end joining, and the cell cycle-dependent repair of DNA double-strand breaks induced by sparsely or densely ionizing radiation. Radiat. Res. 2009;171:265–273. doi: 10.1667/RR0784.1. [DOI] [PubMed] [Google Scholar]
- 14.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Schild D, Thelen MP, Thompson LH. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D–mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 19.Fuller LF, Painter RB. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 1988;193:109–121. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 20.Tucker JD, Jones NJ, Allen NA, Minkler JL, Thompson LH, Carrano AV. Cytogenetic characterization of the ionizing radiation-sensitive Chinese hamster mutant irs1. Mutat. Res. 1991;254:143–152. doi: 10.1016/0921-8777(91)90005-a. [DOI] [PubMed] [Google Scholar]
- 21.Natarajan AT, Berni A, Marimuthu KM, Palitti F. The type and yield of ionising radiation induced chromosomal aberrations depend on the efficiency of different DSB repair pathways in mammalian cells. Mutat. Res. 2008;642:80–85. doi: 10.1016/j.mrfmmm.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deans B, Griffin CS, O'Regan P, Jasin M, Thacker J. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003;63:8181–8187. [PubMed] [Google Scholar]
- 24.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 25.Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009;69:863–872. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheong N, Wang X, Wang Y, Iliakis G. Loss of S-phase-dependent radioresistance in irs-1 cells exposed to X-rays. Mutat. Res. 1994;314:77–85. doi: 10.1016/0921-8777(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 28.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 29.Iliakis GE, Okayasu R. Radiosensitivity throughout the cell cycle and repair of potentially lethal damage and DNA double-strand breaks in an X-ray-sensitive CHO mutant. Int. J. Radiat. Biol. 1990;57:1195–1211. doi: 10.1080/09553009014551291. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q, Hill RP. Radiosensitivity, apoptosis and repair of DNA double-strand breaks in radiation-sensitive Chinese hamster ovary cell mutants treated at different dose rates. Radiat. Res. 1996;146:636–645. [PubMed] [Google Scholar]
- 31.Lee H, Larner JM, Hamlin JL. Cloning and characterization of Chinese hamster p53 cDNA. Gene. 1997;184:177–183. doi: 10.1016/s0378-1119(96)00592-6. [DOI] [PubMed] [Google Scholar]
- 32.Borgmann K, Dede M, Wrona A, Brammer I, Overgaard J, Dikomey E. For X-irradiated normal human fibroblasts, only half of cell inactivation results from chromosomal damage. Int. J. Radiat. Oncol. Biol. Phys. 2004;58:445–452. doi: 10.1016/j.ijrobp.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 33.Thompson LH, Fong S, Brookman K. Validation of conditions for efficient detection of HPRT and APRT mutations in suspension-cultured Chinese hamster ovary cells. Mutat. Res. 1980;74:21–36. doi: 10.1016/0165-1161(80)90188-0. [DOI] [PubMed] [Google Scholar]
- 34.Savage JR. Classification and relationships of induced chromosomal structual changes. J. Med. Genet. 1976;13:103–122. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu TC, Wu X, Trizna Z. Mutagen sensitivity in humans. A comparison between two nomenclature systems for recording chromatid breaks, Cancer Genet. Cytogenet. 1996;87:127–132. doi: 10.1016/0165-4608(95)00301-0. [DOI] [PubMed] [Google Scholar]
- 36.Cornforth MN. Analyzing radiation-induced complex chromosome rearrangements by combinatorial painting. Radiat. Res. 2001;155:643–659. doi: 10.1667/0033-7587(2001)155[0643:ariccr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Boei JJ, Vermeulen S, Moser J, Mullenders LH, Natarajan AT. Intrachanges as part of complex chromosome-type exchange aberrations. Mutat. Res. 2002;504:47–55. doi: 10.1016/s0027-5107(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 38.Sinclair WK, Morton RA. Variations in X-ray response during the division cycle of partially synchronized Chinese hamster cells in culture. Nature. 1963;199:1158–1160. doi: 10.1038/1991158a0. [DOI] [PubMed] [Google Scholar]
- 39.Wilson PF, Nagasawa H, Fitzek MM, Little JB, Bedford JS. G2-phase chromosomal radiosensitivity of primary fibroblasts from hereditary retinoblastoma family members and some apparently normal controls. Radiat. Res. 2010;173:62–70. doi: 10.1667/RR1943.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagasawa H, Latt SA, Lalande ME, Little JB. Effects of X-irradiation on cell-cycle progression, induction of chromosomal aberrations and cell killing in ataxia telangiectasia (AT) fibroblasts. Mutat. Res. 1985;148:71–82. doi: 10.1016/0027-5107(85)90209-x. [DOI] [PubMed] [Google Scholar]
- 42.Nagasawa H, Little JB, Inkret WC, Carpenter S, Raju MR, Chen DJ, Strniste GF. Response of X-ray-sensitive CHO mutant cells (xrs-6c) to radiation. II. Relationship between cell survival and the induction of chromosomal damage with low doses of alpha particles. Radiat. Res. 1991;126:280–288. [PubMed] [Google Scholar]
- 43.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RT, Gotoh E, Mullinger AM, Ryan AJ, Shiloh Y, Ziv Y, Squires S. Targeting double-strand breaks to replicating DNA identifies a subpathway of DSB repair that is defective in ataxia-telangiectasia cells. Biochem. Biophys. Res. Comm. 1999;261:317–325. doi: 10.1006/bbrc.1999.1024. [DOI] [PubMed] [Google Scholar]
- 45.Bryant PE. Repair and chromosomal damage. Radiother. Oncol. 2004;72:251–256. doi: 10.1016/j.radonc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Savage JR. On the nature of visible chromosomal gaps and breaks, Cytogenet. Genome Res. 2004;104:46–55. doi: 10.1159/000077465. [DOI] [PubMed] [Google Scholar]
- 47.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 49.Donoho G, Brenneman MA, Cui TX, Donoviel D, Vogel H, Goodwin EH, Chen DJ, Hasty P. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36:317–331. doi: 10.1002/gcc.10148. [DOI] [PubMed] [Google Scholar]
- 50.Melton DW, Ketchen AM, Nunez F, Bonatti-Abbondandolo S, Abbondandolo A, Squires S, Johnson RT. Cells from ERCC1-deficient mice show increased genome instability and a reduced frequency of S-phase-dependent illegitimate chromosome exchange but a normal frequency of homologous recombination. J. Cell Sci. 1998;111(Pt 3):395–404. doi: 10.1242/jcs.111.3.395. [DOI] [PubMed] [Google Scholar]
- 51.Chipchase MD, Melton DW. The formation of UV-induced chromosome aberrations involves ERCC1 and XPF but not other nucleotide excision repair genes. DNA Repair. 2002;1:335–340. doi: 10.1016/s1568-7864(02)00010-1. [DOI] [PubMed] [Google Scholar]
- 52.Weaver Z, Montagna C, Xu X, Howard T, Gadina M, Brodie SG, Deng CX, Ried T. Mammary tumors in mice conditionally mutant for Brca1 exhibit gross genomic instability and centrosome amplification yet display a recurring distribution of genomic imbalances that is similar to human breast. cancer, Oncogene. 2002;21:5097–5107. doi: 10.1038/sj.onc.1205636. [DOI] [PubMed] [Google Scholar]
- 53.Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir RB, Borg A, Kallioniemi OP. Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res. 1997;57:1222–1227. [PubMed] [Google Scholar]
- 54.Gretarsdottir S, Thorlacius S, Valgardsdottir R, Gudlaugsdottir S, Sigurdsson S, Steinarsdottir M, Jonasson JG, Anamthawat-Jonsson K, Eyfjord JE. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 1998;58:859–862. [PubMed] [Google Scholar]
- 55.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Ann. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 56.Buchholz TA, Wu X, Hussain A, Tucker SL, Mills GB, Haffty B, Bergh S, Story M, Geara FB, Brock WA. Evidence of haplotype insufficiency in human cells containing a germline mutation in BRCA1 or BRCA2. Int. J. Cancer. 2002;97:557–561. doi: 10.1002/ijc.10109. [DOI] [PubMed] [Google Scholar]
- 57.Barwell J, Pangon L, Georgiou A, Kesterton I, Langman C, Arden-Jones A, Bancroft E, Salmon A, Locke I, Kote-Jarai Z, Morris JR, Solomon E, Berg J, Docherty Z, Camplejohn R, Eeles R, Hodgson SV. Lymphocyte radiosensitivity in BRCA1 and BRCA2 mutation carriers and implications for breast cancer susceptibility. Int. J. Cancer. 2007;121:1631–1636. doi: 10.1002/ijc.22915. [DOI] [PubMed] [Google Scholar]
- 58.Docherty Z, Georgiou A, Langman C, Kesterton I, Rose S, Camplejohn R, Ball J, Barwell J, Gilchrist R, Pangon L, Berg J, Hodgson S. Is chromosome radiosensitivity and apoptotic response to irradiation correlated with cancer susceptibility? Int. J. Radiat. Biol. 2007;83:1–12. doi: 10.1080/09553000600932968. [DOI] [PubMed] [Google Scholar]
- 59.Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins: mechanistic insights. Mutat. Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.