Abstract
Mammalian mitochondria synthesize a set of thirteen proteins that are essential for energy generation via oxidative phosphorylation. The genes for all of the factors required for synthesis of the mitochondrially-encoded proteins are located in the nuclear genome. A number of disease-causing mutations have been identified in these genes. In this manuscript, we have elucidated the mechanisms of translational failure for two disease states characterized by lethal mutations in mitochondrial elongation factor Ts (EF-Tsmt) and elongation factor Tu (EF-Tumt).
EF-Tumt delivers the aminoacyl-tRNA (aa-tRNA) to the ribosome during the elongation phase of protein synthesis. EF-Tsmt regenerates EF-Tumt:GTP from EF-Tumt:GDP. A mutation of EF-Tsmt (R325W) leads to a two-fold reduction in its ability to stimulate the activity of EF-Tumt in poly(U)-directed polypeptide chain elongation. This loss of activity is caused by a significant reduction in the ability of EF-Tsmt R325W to bind EF-Tumt, leading to a defect in nucleotide exchange.
A mutation of Arg336 to Gln in EF-Tumt causes infantile encephalopathy caused by defects in mitochondrial translation. EF-Tumt R336Q is as active as the wild-type protein in polymerization using E. coli 70S ribosomes and E. coli [14C]Phe-tRNA but is inactive in polymerization with mitochondrial [14C]Phe-tRNA and mitochondrial 55S ribosomes. The R336Q mutation causes a two-fold decrease in ternary complex formation with E. coli aa-tRNA but completely inactivates EF-Tumt for binding to mitochondrial aa-tRNA. Clearly the R336Q mutation in EF-Tumt has a far more drastic effect on its interaction with mitochondrial aa-tRNAs than bacterial aa-tRNAs.
Keywords: EF-Tu, EF-Ts, translation, elongation, mitochondria, disease
1. Introduction
During protein biosynthesis, elongation factor Tu (EF-Tu) forms a ternary complex with aminoacyl-tRNA (aa-tRNA) and GTP and promotes the binding of the aa-tRNA to the A-site of the ribosome [1]. Following codon:anticodon interactions, an EF-Tu:GDP complex is released from the ribosome. Elongation factor Ts (EF-Ts) binds to the EF-Tu:GDP complex and promotes the exchange of GDP for GTP, regenerating the active EF-Tu:GTP complex [2].
Mammalian mitochondria have a specific protein biosynthetic system responsible for the synthesis of thirteen polypeptides of the respiratory chain complexes. Protein synthesis in this organelle requires elongation factors that correspond to EF-Tu and EF-Ts (EF-Tumt and EF-Tsmt). Both of these proteins are encoded in the nuclear genome, synthesized in the cell cytoplasm, and subsequently imported into the mitochondrion. While mutations in mitochondrial DNA are well known to cause disease, few mutations have been identified in nuclear genes that encode mitochondrial proteins required for protein biosynthesis in this organelle. Recently, a mutation in the gene coding for EF-Tsmt (Arg312 to Trp)1 was reported leading to encephalomyopathy in one patient and hypertrophic cardiomyopathy in another [3]. Both patients died at 7 weeks of age. This mutation is predicted to disrupt important interactions between EF-Tumt and EF-Tsmt.
EF-Tsmt is a 30.7 kDa protein that consists of an N-terminal domain and a core domain divided into N- and C-terminal subdomains (Figure 1A) [4]. EF-Tumt and EF-Tsmt interact through extensive surface contacts. The N-terminal domain and subdomain N of the core of EF-Tsmt interact directly with the G-domain (domain I) of EF-Tumt, while subdomain C of the core contacts domain III of EF-Tumt.
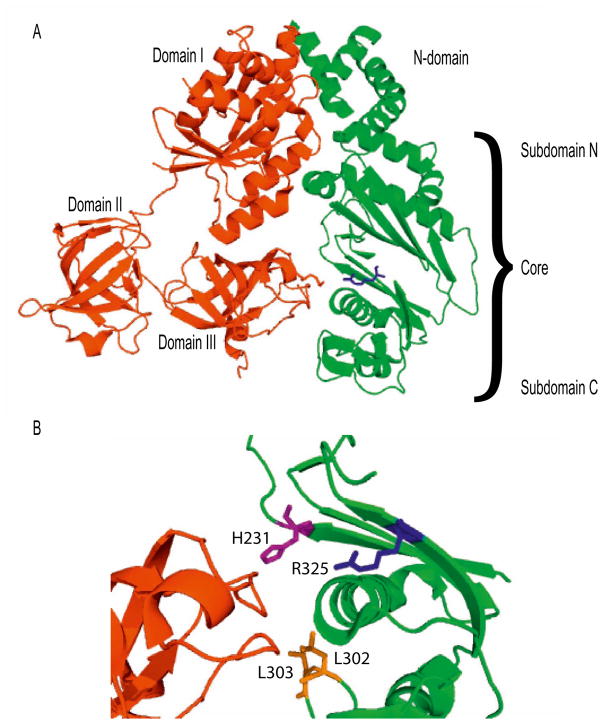
Figure 1.
Structural view of the interaction of EF-Tumt with EF-Tsmt. A. Overall structure of the EF-Tumt:EF-Tsmt complex (PDB 1XB2). EF-Tumt is shown in red, and EF-Tsmt is shown in green. EF-Tsmt R325W is highlighted in blue. B. Close-up view of the interaction of residues near the R325W mutation. Domain III of EF-Tumt is shown in red, subdomain C of EF-Tsmt is shown in green, and EF-Tsmt R325W is shown in blue. Residues known to be important for the interaction of EF-Tsmt with EF-Tumt are shown as purple (H231) and orange (L302 and L303) sticks [5].
Arg325 of EF-Tsmt is highly conserved and is found in most bacterial and mitochondrial factors [3]. This residue is located in a β-strand in subdomain C of the core (shown in blue in Figure 1A and Figure 1B). While it does not make direct contact with EF-Tumt, other nearby residues in this region do so. Molecular modeling has predicted that the R325W mutation could disrupt the tertiary structure of the subdomain C of the core, altering its interactions with domain III of EF-Tumt [3]. Mutations in this region have been shown to reduce the affinity of EF-Tsmt for EF-Tumt about six to seven-fold [5]. For example, mutation of His231 to Ala caused a 6-fold decrease in the binding constant governing the interaction of EF-Tsmt with EF-Tumt. Mutation of L302 and L303 (shown in orange in Figure 1B) caused a 7-fold decrease in the binding constant governing the interaction of EF-Tumt and EF-Tsmt. We have directly examined the effect of the R325W mutation by testing the ability of the mutated protein to bind directly to EF-Tumt and to stimulate its activity in poly(U)-directed polymerization.
EF-Tumt is a 45.1 kDa protein composed of three domains. Domain I contains the guanine nucleotide binding site. All three domains are involved in binding the aa-tRNA [6], while domains I and III interact with EF-Tsmt. The stable binding of guanine nucleotides and EF-Tsmt to EF-Tumt are mutually exclusive, allowing EF-Tsmt to serve as a nucleotide exchange factor for EF-Tumt [7,8].
A mutation in EF-Tumt, (Arg336 to Gln), was discovered in an infant with lactic acidosis and fetal encephalopathy [9]. The infant later died at an age of 14 months. Arg336 of EF-Tumt is located in domain II of the protein, in a region known to interact with the acceptor stem of the aa-tRNAs. EF-Tumt R336Q was previously found to be unable to form a ternary complex with mitochondrial Ser-tRNA [10]. The mutated factor was also deficient in poly(U)-directed poly(Phe) synthesis using mitochondrial Phe-tRNA and E. coli 70S ribosomes. However, no studies were carried out with mitochondrial ribosomes and mitochondrial elongation factor G1 (EF-G1mt) [10]. In this manuscript, the activity of the mutated EF-Tumt is examined on mitochondrial ribosomes, and the effect of the mutation on its ability to form ternary complexes with several additional mitochondrial aa-tRNAs has been examined.
2. Materials and methods
2.1 Materials
[14C]phenylalanine and [35S]methionine were purchased from Perkin Elmer Life Sciences, Inc. E. coli tRNA was obtained from Boehringer Mannheim. E. coli [14C]Phe-tRNA and the human mitochondrial [35S]Met-tRNA transcript were prepared as described [11,12]. Mitochondrial tRNA was purified from bovine mitoplasts using the Qiagen RNA/DNA maxi kit. The tRNAPhe was aminoacylated using [14C]Phe as described [13]. E. coli ribosomes were purified from E. coli W cells; mitochondrial 55S ribosomes, and EF-G1mt were purified as described [14,15].
2.2 Cloning, expression, and purification of proteins
BMtu/pET24c and BMts/pET24c, the E. coli expression vectors for C-terminal histidine-tagged bovine EF-Tumt and EF-Tsmt, are described in [16,17] and the proteins expressed from them are referred to as the wild-type (WT) proteins. Expression vectors for the mutant EF-Tumt (R336Q) and mutant EF-Tsmt (R325W) were constructed by using the Quik Change site-directed mutagenesis kit (Stratagene). EF-Tumt and EF-Tsmt were purified using described protocols [4,16,18].
2.3 Circular dichroism of EF-Tsmt and EF-Tsmt R325W
Purified EF-Tsmt and EF-Tsmt R325W were dialyzed in 1000 volumes CD buffer containing: 10 mM MgSO4, 50 mM potassium phosphate buffer (pH 7.6), and 5% glycerol for 4 h with a change of buffer after 2 h. CD spectra of EF-Tsmt and EF-Tsmt R325W were obtained using an Aviv Model 62DS Circular Dichroism Spectrometer. Data points were collected in 1 nm steps, and the data were analyzed using the CD Pro software.
2.4 Poly (Phe) polymerization assays
To test the ability of EF-Tsmt to stimulate the activity of EF-Tumt in poly(Phe) directed polymerization, reaction mixtures (25 μL) contained 50 mM Tris-HCl (pH 7.8), 1 mM dithiothreitol (DTT), 0.1 mM spermine, 6 mM MgCl2, 80 mM KCl, 0.5 mM GTP, 1.25mM phosphor(enol)pyruvate (PEP), 0.4 U pyruvate kinase, 3 μg poly(U), 8.3 pmol (0.33 μM) E. coli [14C]Phe-tRNA, 5 pmol (0.2 μM) EF-G1mt, 0.25 μM E. coli 70S ribosomes, 0.5 pmol (20 nM) EF-Tumt, and 0.025–0.075 pmol (1–3 nM) EF-Tsmt. The reaction mixtures were incubated at 37 °C for 30 min and analyzed as described [19]. A blank representing the amount of label retained on the filter in the absence of EF-Tsmt was not subtracted in order to illustrate the level of polymerization obtained in the presence of EF-Tumt alone.
To test the poly(Phe) polymerization activity of WT and mutated EF-Tumt in the E . coli system, reaction mixtures (25 μL) contained 50 mM Tris-HCl (pH 7.8), 1 mM DTT, 0.1 mM spermine, 6 mM MgCl2, 80 mM KCl, 0.5 mM GTP, 1.25 mM PEP, 0.4 U pyruvate kinase, 3 μg poly(U), 8.3 pmol (0.3 μM) E. coli [14C]Phe-tRNA, 5 pmol (0.2 μM) EF-G1mt, 16 μg (0.25 μM) E. coli 70S ribosomes, and 0.2–0.8 pmol (8–32 nM) EF-Tumt (WT or R336Q) in a 1:1 mix with EF-Tsmt. Reaction mixtures were incubated at 37 °C for 30 min, at which time they were quenched by the addition of 5% TCA and processed as described [19]. A blank representing the amount of label retained on the filter in the absence of EF-Tumt (~0.3 pmol) was subtracted from each value.
For assays performed in the mitochondrial system, the reaction mixtures (25 μL) contained 50 mM Tris-HCl (pH 7.8), 1 mM DTT, 0.1 mM spermine, 7.5 mM MgCl2, 40 mM KCl, 0.5 mM GTP, 1.25 mM PEP, 0.4 U pyruvate kinase, 3 μg poly(U), 3 pmol (0.12 μM) mitochondrial [14C]Phe-tRNA, 5 pmol (0.2 μM) EF-G1mt, 3 pmol (0.12 μM) mitochondrial 55S ribosomes, and 0.2–0.8 pmol EF-Tumt (WT or the R336Q mutated protein) in a 1:1 mixture with EF-Tsmt. Reactions were incubated at 37 °C for 30 min, que nched by the addition of 5% TCA, and processed as described [19]. A blank representing the amount of label retained on the filter in the absence of EF-Tumt (~0.08 pmol) was subtracted from each value.
2.5 Physical interaction of EF-Tumt and EF-Tsmt measured using a gel-shift assay
The binding of EF-Tsmt to EF-Tumt was studied using a gel-shift assay according to [20]. Briefly, EF-Tsmt (40 pmol) and EF-Tumt:GDP (0–40 pmol) were incubated on ice for 5 min in a reaction mixture (10 μL ) containing: 50 mM Tris-HCl (pH 7.5), 70 mM KCl, 1.5 mM EDTA, 10% glycerol, 1 mM DTT, 0.004% bromophenol blue. The samples were resolved by native polyacrylamide gel electrophoresis (PAGE). The electrophoresis was performed at 4°C (450 V, 2 h) on a 1 mm × 20 cm × 10 cm 9% PAGE gel (acrylamide:bisacrylamide 29:1) containing 8 mM Tricine-NaOH (pH 8.2), 1 mM EDTA and 5% glycerol. The running buffer contained 8 mM Tricine-NaOH (pH 8.2) and 1 mM EDTA. The proteins were visualized using SYPRO Ruby (Molecular Probes).
2.6 Ternary complex formation measured using hydrolysis protection
The ability of EF-Tumt and EF-Tumt R336Q to protect aa-tRNA from spontaneous deacylation was monitored as described [21], with slight modifications. Ternary complexes were formed in reaction mixtures (100 μL) containing 75 mM Tris-HCl (pH 7.5), 75 mM NH4Cl, 15 mM MgCl2, 7.5 mM DTT, 60 μg/ml bovine serum albumin, 1 mM GTP, 2.4 mM PEP, 0.1 U pyruvate kinase, 12.5 pmol (0.125 μM) E. coli [14C]Phe-tRNA, and 0–330 pmol (0–3.3 μM) EF-Tumt (WT or R336Q). The samples were incubated for the indicated times at 30 °C, and precipitated in cold 5% TCA. The remaining aa-tRNA was collected on filter papers (3MM, Whatman) and the amount of EF-Tu:GTP:[14C]Phe-tRNA was quantified using a liquid scintillation counter.
2.7 Ternary complex formation measured using RNase protection
Ternary complex formation assays were carried out in reaction mixtures (50 μL) containing 20 mM HEPES-KOH (pH 7.0), 1 mM DTT, 6.7 mM MgCl2, 68 mM KCl, 0.5 mM GTP, 1.25mM PEP, 0.8 U pyruvate kinase, 7.2 pmol (0.14 μM) E. coli [14C]Phe-tRNA, and 0 – 20 pmol (0.1–0.4 μM) EF-Tumt (WT or R336Q). For experiments using mitochondrial tRNAs, 5.3 pmol (0.11 μM) native bovine mitochondrial [14C]Phe-tRNA or 9.6 pmol (0.19 μM) of the human mitochondrial [35S]Met-tRNA transcript and 0–60 pmol (0–1.2 μM) EF-Tumt (WT or R336Q) were used. The reactions were incubated for 15 min at 0 °C, at which time 10 μg RNase A was allowed to digest the sample for 30 s. Ice cold 5% TCA was added, and the remaining aa-tRNA was precipitated at 0 °C for 10 min and analyzed as described [12,22]. A blank representing the amount of label retained on the filters in the absence of EF-Tumt (~0.3 pmol using E. coli [14C]Phe-tRNA, ~0.1 pmol using mitochondrial [14C]Phe-tRNA and ~0.2 pmol using [35S]Met-tRNA) was subtracted from each value.
3. Results and Discussion
3.1 Biochemical consequences of the R325W mutation in EF-Tsmt
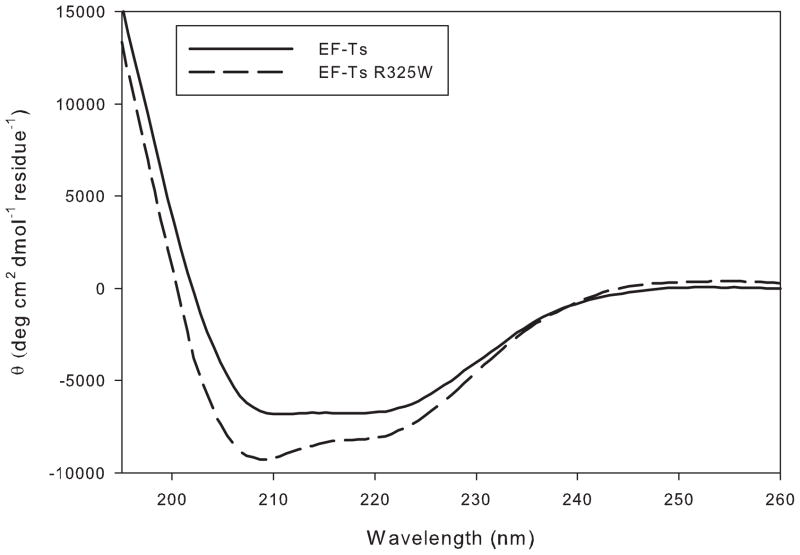
A mutated derivative of bovine EF-Tsmt (R325W) was prepared using site-directed mutagenesis corresponding to the lethal mutation in humans [3]. To determine whether the mutation adversely affected the structure of EF-Tsmt, the CD spectra of the mutated derivative was compared to that of the WT factor. As indicated in Figure 2, the α-helical content of the mutated protein did not change significantly with respect to the wild-type factor, suggesting that the protein retained its normal secondary structure. A thermal melt monitored by CD indicated that both EF-Tsmt and EF-Tsmt R325W unfold at the same temperature (data not shown), indicating that the mutation has not greatly destabilized the protein.
Figure 2.
Circular dichroism spectra of EF-Tsmt and EF-Tsmt R325W. CD spectra of EF-Tsmt (WT and R325W) obtained as described in Materials and Methods.
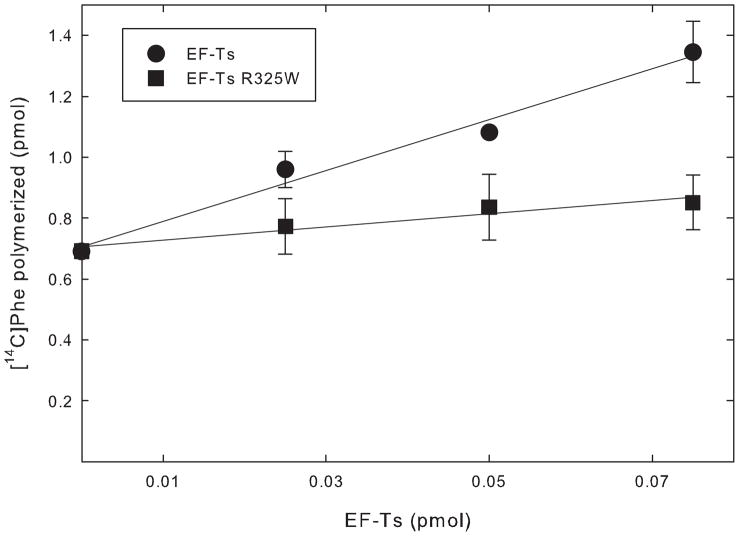
To investigate the biochemical effect of the R325W mutation, the ability of EF-Tsmt and its R325W variant to stimulate the activity of EF-Tumt in poly(Phe) polymerization was examined. EF-Tumt alone has measurable activity in polymerization, as indicated by the considerable amount of Phe polymerized in the absence of EF-Tsmt (Figure 3, y-intercept). However, its activity is enhanced by the presence of EF-Tsmt due to an increase in the guanine nucleotide exchange rate, allowing EF-Tumt to be used more catalytically. Wild-type EF-Tsmt stimulated the activity of EF-Tumt more than 2-fold under the reaction conditions used (Figure 3). This value is consistent with previous observations [23]. However, the stimulation of EF-Tumt by the R325W variant was reduced substantially, and only a 1.2-fold stimulation was observed (Figure 3). This reduction could arise from the failure of the mutated EF-Tsmt to interact with EF-Tumt:GDP effectively, reducing GDP release and formation of an EF-Tumt:EF-Tsmt complex. The reduction could also reflect a failure of the EF-Tumt:EF-Tsmt complex to bind GTP, which is necessary to release EF-Tumt for another round of polymerization.
Figure 3.
Activity of EF-Tsmt and EF-Tsmt R325W in poly(Phe) polymerization. The abilities of EF-Tsmt and EF-Tsmt R325W to function in a poly(U)-directed peptide polymerization assay were tested using EF-Tumt and E. coli [14C]Phe-tRNA. EF-Tsmt is shown in closed squares and EFTsmt R325W is shown in closed circles. The assays were performed multiple times and data were normalized with respect to the point containing 0.05 pmol of EF-Tsmt.
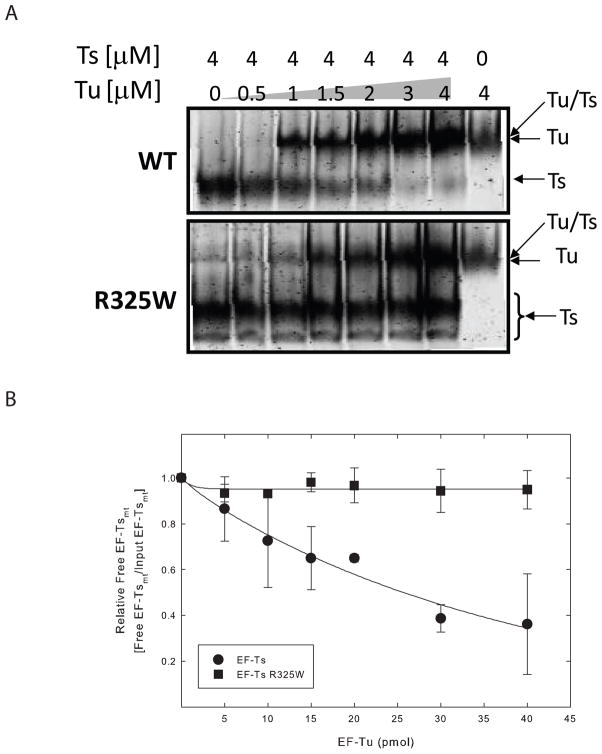
To investigate the interaction between EF-Tumt and EF-Tsmt R325W directly, the binding of the two proteins was studied using a gel-shift assay (Figure 4). EF-Tsmt or EF-Tsmt R325W was incubated with increasing amounts of EF-Tumt. After complex formation, the proteins were separated on native-PAGE. This gel system separates EF-Tsmt from the EF-Tumt:EF-Tsmt complex. However, the mobility of EF-Tumt:EF-Tsmt complex was quite similar to that for EF-Tumt, and the resolution of the complex from free EF-Tumt was difficult. Thus, the efficiency of complex formation was assessed by the quantity of free EF-Tsmt remaining. Wild-type EF-Tsmt was effective in forming a complex with EF-Tumt in a dose dependent manner. Approximately 60% of EF-Tsmt bound EF-Tumt when incubation was carried out using equal molar amounts of the two factors (Figure 4A, upper panel and Figure 4B, closed circles). In contrast, EF-Tsmt R325W was ineffective in binding EF-Tumt and remained free, even when incubated with an equal amount of EF-Tumt (Figure 4A, lower panel and Figure 4B, closed squares). These results indicate that the reduction in poly(U)-dependent poly(Phe) synthesis with the mutated EF-Tsmt arises to a large extent from a failure of the mutated EF-Tsmt to interact with EF-Tumt.
Figure 4.
Physical interaction of EF-Tsmt and EF-Tsmt R325W with EF-Tumt. The abilities of EF-Tsmt and EF-Tsmt R325W to bind to EF-Tumt were measured using a gel shift assay on native PAGE. A. WT EF-Tsmt (upper panel) or EF-Tsmt R325W (lower panel) were incubated with the indicated amount of EF-Tumt. The proteins were separated on native-PAGE and visualized by SYPRO Ruby. The position of EF-Tsmt, EF-Tumt:EF-Tsmt complex and EF-Tumt are indicated with arrows. B. The efficiency of the complex formation in A was assessed by the amount of free EF-Tsmt remaining. The relative amount of free EF-Tsmt in each panel was normalized to a value of 1 for EF-Tsmt. EF-Tsmt is shown in closed circles, and EF-Tsmt R325W is shown in closed squares.
The effect observed with the R325 mutation is in agreement with previous mutational data as described in the introduction. The data provided here and the mutational studies carried out previously indicate that the interactions of residues in the β-sheet of subdomain C and other nearby residues with EF-Tumt are essential for the stable binding of EF-Tumt with EF-Tsmt.
3.2 Biochemical consequences of the R336Q mutation in EF-Tumt
Arg336 in EF-Tumt is located in domain II in a pocket where the 5′ end of the aa-tRNA is bound. The adjacent residue (Arg335) in EF-Tumt was previously found to be important for the activity of EF-Tumt in both ternary complex formation and A-site binding with mitochondrial Phe-tRNAPhe [22]. Previous work on the lethal R336Q mutation in EF-Tumt indicated that this mutation did not affect the structural integrity of EF-Tumt, as demonstrated by CD spectroscopy [10]. However, the mutation drastically reduced the ability of EF-Tumt to promote polymerization using mitochondrial [14C]Phe-tRNA on E. coli ribosomes [10]. The R336Q mutation was not predicted to influence the interaction of EF-Tumt with EF-Tsmt, because the primary interaction with EF-Tsmt occurs via interactions involving domains I and III [4]. The mutation was also not expected to interfere with GTP binding, which is localized to domain I.
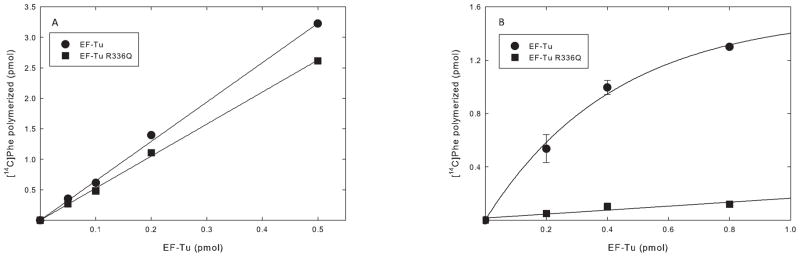
The effects of the R336Q mutation on the activity of EF-Tumt were examined further using E. coli and mitochondrial ribosomes and aa-tRNAs. Surprisingly, when EF-Tumt R336Q was tested in poly(U)-directed polymerization using E. coli [14C]Phe-tRNA and E. coli 70S ribosomes, it was as active as the wild-type factor in this assay (Figure 5A). However, in the homologous system using mitochondrial [14C]Phe-tRNA and ribosomes, EF-Tumt R336Q was almost inactive in polypeptide chain elongation (Figure 5B). Previous work [10] indicated that EF-Tumt R336Q lacked activity with mitochondrial [14C]Phe-tRNA when tested on E. coli ribosomes, suggesting that the lack of activity in the mitochondrial system was not a function of the ribosomes used but, rather, due to a failure of the mutated factor to interact with mitochondrial Phe-tRNA. Mitochondrial tRNAs are less structurally stable than E. coli tRNAs [12], and additional stabilizing interactions between EF-Tumt and the mitochondrial aa-tRNA may be important in forming the ternary complex.
Figure 5.
Poly(Phe) polymerization with EF-Tumt and EF-Tumt R336Q. The abilities of EF-Tumt and its R336Q derivative to function in a poly(Phe) polymerization assay was tested as described in Materials and Methods. A. Poly(Phe) polymerization assay using E. coli 70S ribosomes and E. coli [14C]Phe-tRNA. EF-Tumt is shown in closed circles and EF-Tumt R336Q in closed squares. B. Poly(Phe) polymerization assay using mitochondrial 55S ribosomes and native bovine mitochondrial [14C]Phe-tRNA. EF-Tumt is shown in closed circles and EF-Tumt R336Q is shown in closed squares.
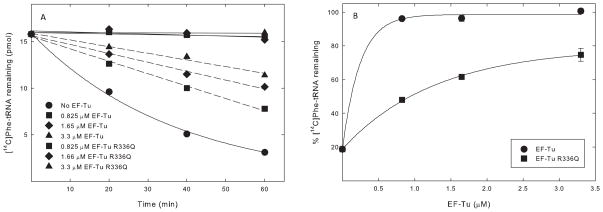
The poly(Phe) polymerization assay does not directly measure the interaction between EF-Tumt and aa-tRNA. This interaction can be measured directly using one of two ternary complex formation assays: the hydrolysis protection assay and the RNase protection assay. The hydrolysis protection assay measures the ability of EF-Tumt to protect the aa-tRNA bond from spontaneous hydrolysis. Since the mutated EF-Tumt was active using E. coli Phe-tRNA in polymerization, its interaction with bacterial Phe-tRNA was tested directly. The spontaneous hydrolysis of E. coli Phe-tRNA was apparent (Figure 6A, closed circles). When bound to EF-Tumt, the Phe-tRNA bond was clearly protected from hydrolysis (Figure 6A, solid lines). However, when the R336Q derivative of EF-Tumt was used in place of the normal factor, substantially less protection of the Phe-tRNA bond was observed (Figure 6A, dashed lines). At sufficiently high concentrations of EF-Tumt R336Q, a considerable number of ternary complexes were formed (Figure 6B, closed squares), indicating that R336Q has some ability to bind E. coli Phe-tRNA. This data suggests that, although the mutated protein is deficient in binding to E. coli Phe-tRNA, the reduced affinity is not sufficient to prevent its efficient use in polymerization (Figure 5).
Figure 6.
Activities of EF-Tumt and EF-Tumt R336Q in ternary complex formation measured using a non-enzymatic hydrolysis protection assay. A. Amount of [14C]Phe-tRNA protected from hydrolysis by increasing concentrations of EF-Tumt (solid lines) and EF-Tumt R336Q (dashed lines) as a function of time. B. Relative amount of [14C]Phe-tRNA at time 60 min in Figure 5A is plotted against the EF-Tumt concentration.
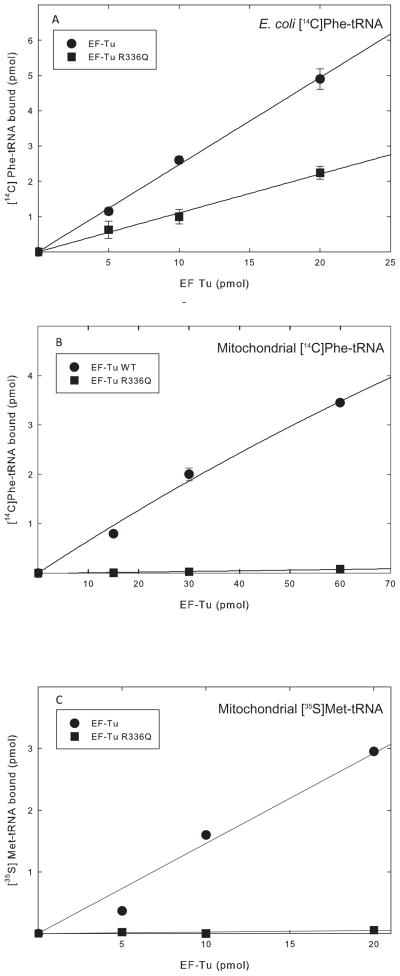
To verify the results obtained from the hydrolysis protection experiments and to examine the interaction with mitochondrial aa-tRNAs, the RNase protection assay was used. This assay measures the ability of EF-Tumt to protect the acceptor stem of the aa-tRNA from degradation by RNase A. Because the RNase A digestion of the tRNA is done in 30 s, it provides a snapshot of the interaction between the two molecules. When the binding of EF-Tumt to E. coli [14 C]Phe-tRNA was tested, the R336Q variant showed a decrease in binding of just over 50% (Figure 7A), indicating that the mutation reduces but does not abolish the ability of EF-Tumt to form a ternary complex with bacterial aa-tRNAs. These results agree with the observation above, that the reduction in ternary complex formation observed is not sufficient to compromise the activity of this factor in polymerization (Figure 5). It is important to note that the binding of the aa-tRNA to EF-Tumt is probably not the rate limiting step in polypeptide chain elongation. Rather, interactions with the ribosome, including the release of inorganic phosphate from EF-Tumt following GTP hydrolysis, appear to be rate limiting [24].
Figure 7.
Activities of EF-Tumt and EF-Tumt R336Q in ternary complex formation measured using an RNase protection assay. EF-Tumt is shown in closed circles and EF-Tumt R336Q is shown in closed squares. A. Ternary complex formation using E. coli [14C]Phe-tRNA. B. Ternary complex formation with native bovine mitochondrial [14C]Phe-tRNA. C. Ternary complex formation using human mitochondrial [35S]Met-tRNA.
Previous work indicated that EF-Tumt R336Q did not bind mitochondrial Ser-tRNA effectively in the ternary complex [10]. This observation is in sharp contrast to the significant level of ternary complex formation observed with the E. coli Phe-tRNA (Figure 6) but in agreement with the lack of polymerization observed with mitochondrial Phe-tRNA (Figure 5). To explore whether the failure of EF-Tumt to use mitochondrial Phe-tRNA in polymerization arises from a failure of ternary complex formation, the RNase protection assay was carried out with mitochondrial [14C]Phe-tRNA (Figure 7B). EF-Tumt was quite active in forming a ternary complex with native bovine mitochondrial Phe-tRNA. However, the R336Q variant did not bind mitochondrial Phe-tRNA detectably (Figure 7B). This observation agrees with previous work and explains the lack of activity of the mutated protein in poly(Phe)-directed polymerization with mitochondrial Phe-tRNA on mitochondrial 55S ribosomes (Figure 5).
To address the question of whether other mitochondrial aa-tRNAs also fail to bind the R336Q variant, ternary complex formation assays were carried out using human mitochondrial [35S]Met-tRNA (Figure 6C). This tRNA was transcribed in vitro and has been shown to fold properly and to bind EF-Tumt [12]. In this assay, EF-Tumt readily formed a ternary complex with mitochondrial [35S]Met-tRNA. However, once again EF-Tumt R336Q was unable to bind mitochondrial [35S]Met-tRNA, indicating that the mutated protein is probably unable to form ternary complexes with most or all mitochondrial aa-tRNAs.
It is perplexing that the mutated variant of EF-Tumt can bind E. coli Phe-tRNA but not mitochondrial aa-tRNAs. This discrepancy must stem from the innate differences between the mitochondrial and bacterial tRNAs. Canonical tRNAs fold into a distinct L-shape stabilized by strong G-C base pairs in the stems of the tRNA and tertiary interactions involving many invariant or semi-invariant residues [25]. In contrast, mitochondrial tRNAs are characterized by weaker stems with numerous A-U base pairs or mismatches. They often lack the conserved residues that create important tertiary interactions in other tRNAs [26–28]. Although mitochondrial tRNAs can fold into an L-shape, the interactions that stabilize the elbow region are weaker [29]. Mitochondrial tRNAs observed on 55S ribosomes have an L-shape but show a “caved in” feature at the corner of the L [29]. These unusual structural features of mitochondrial tRNAs may lead to a requirement for interactions with EF-Tumt that are not essential for the binding of canonical aa-tRNAs.
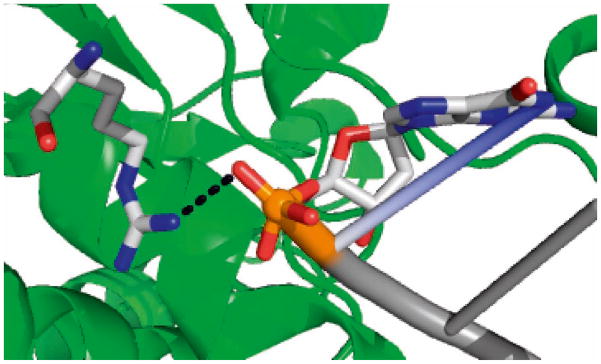
Previous data indicate that mitochondrial aa-tRNAs must be phosphorylated at their 5′ ends in order to bind effectively to EF-Tumt [12]. The crystal structure of the Thermus thermophilus ternary complex indicates that Arg300 (corresponding to Arg336 in EF-Tumt) forms a salt bridge with the phosphate group at the 5′ end of the aa-tRNA (Figure 8) [6]. Replacement of the Arg with Gln in the mutated EF-Tumt could disrupt this interaction, since the side chain of Gln is shorter and may not be within bridging distance of the phosphate backbone. This interaction is not critical for the formation of the ternary complex with E. coli aa-tRNAs, in agreement with data showing that a 5′-phosphate is not important for ternary complex formation in prokaryotes [30]. However, the data presented here indicate that the interaction between Arg336 and the 5′ phosphate of mitochondrial aa-tRNAs is critical for ternary complex formation in the mitochondrial translational system. The interaction between Arg336 and the 5′ phosphate of aa-tRNAs could also be important following ternary complex formation, since the aa-tRNA in the ternary complex is substantially distorted upon the ribosome binding, allowing it to interact simultaneously with the mRNA codon and the EF-Tumt [31].
Figure 8.
Interaction of T. thermophilus EF-Tu with aa-tRNA. This image was obtained from the crystal structure of the T. thermophilus Phe-tRNA:EF-Tu:GDPNP ternary complex (PDB 1TTT). The residue homologous to R336 is shown with its nitrogen atoms in blue. The phosphate of G1 of the Phe-tRNA is shown in orange, with its oxygen atoms shown in red. The tRNA is shown in dark gray.
4. Conclusions
Numerous mutations in mitochondrial tRNAs and rRNAs have been reported that lead to diseases such as Myoclonic Epilepsy and Ragged Red Muscle Fibers (MERRF), and Leber Hereditary Optic Neuropathy (LHON), and Leigh Syndrome (Mitomap). However, mutations in nuclear-encoded proteins required for the translation of mitochondrial mRNAs are rarer. In addition to the mutations in EF-Tumt and EF-Tsmt investigated in this report, several other mutations in proteins involved in mitochondrial translation have been reported. Lethal mutations of mitochondrial elongation factor G1 were discovered in a patient with had lactic acidosis, fatal encephalopathy, and early-onset Leigh syndrome [9]. An additional mutation was previously reported in the coding sequence for EF-G1 that led to deficiencies in oxidative phosphorylation and to liver dysfunction [32]. A fatal mutation in mitochondrial small ribosomal protein MRPS16 was found in a patient with neonatal lactic acidosis [33], and a mutation in the small subunit ribosomal protein MRPS22 was also lethal [34]. Both ribosomal protein mutations lead to defects in the assembly of the small subunit [35].
In this paper, we have analyzed the mechanisms by which mutations in two elongation factors (EF-Tumt and EF-Tsmt) cause fatal defects in mitochondrial translation. Oxidative phosphorylation requires the assembly of the electron transport chain and ATP synthase, which contain protein components synthesized by the mitochondrial translational machinery. A defect in mitochondrial translation caused by the inability of EF-Tsmt R325Q to bind to EF-Tumt likely leads to the defects in oxidative phosphorylation seen in the clinical manifestation of the EF-Tsmt mutation.
The R336Q variant of EF-Tumt is inactive in mitochondrial polypeptide chain elongation as a result of its inability to bind mitochondrial aa-tRNAs. Failure of ternary complex formation leads to defective mitochondrial protein synthesis, causing a decrease in oxidative phosphorylation. One direct clinical phenotype of the EF-Tumt R336Q mutation is lactic acidosis [9], which most likely resulted from the buildup of lactic acid from pyruvate due to a failure in the synthesis of the respiratory chain.
Acknowledgments
Funding for this project was provided by National Institutes of Health grant GM32734 to LLS and Grant-in-Aid for Young Scientists (B) from JSPS, and the Naito Memorial Grant for Mother Researchers to NT. The funding sources had no role in this project.
Abbreviations
- EF-Tumt
mitochondrial elongation factor Tu
- EF-Tsmt
mitochondrial elongation factor Ts
- aa-tRNA
aminoacyl-tRNA
- EF-G1mt
mitochondrial elongation factor G1
Footnotes
In the original paper [3], the position identified as mutated is listed as Arg333. This position corresponds to the residue in isoform 2 of human EF-Tsmt (P-43897-2). However, there is no direct experimental evidence for the expression of this isoform. Rather, isoform 1 (P-43897-1) has been chosen as the canonical sequence. In humans, the mutation in isoform 1 is in position 312 (R312W). There is direct experimental evidence for isoform 1 in the bovine system. The mutation created here is in isoform 1 of Bos taurus EF-Tsmt and corresponds to residue 325 (R325W). The mature forms of human and Bos taurus EF-Tsmt are over 92% identical.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sprinzl M. Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem Sci. 1994;19:245–250. doi: 10.1016/0968-0004(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 2.Miller D, Weissbach H. Factors involved in the transfer of Aminoacyl-tRNA to the ribosome. In: Weissbach H, Pestka S, editors. Molecular Mechanisms of Protein Biosynthesis. Academic Press; New York: 1977. pp. 323–373. [Google Scholar]
- 3.Smeitink JAM, Elpeleg O, Antonicka H, Diepstra H, Saada A, Smits P, Sasarman F, Vriend G, Jacob-Hirsch J, Shaag A, Rechavi G, Welling B, Horst J, Rodenburg RJ, van den Heuvel B, Shoubridge EA. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EF-Ts. Am J Hum Genet. 2006;79:869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeppesen MG, Navratil T, Spremulli LL, Nyborg J. Crystal structure of the bovine mitochondrial elongation factor Tu:Ts complex. J Biol Chem. 2005;280:5071–5081. doi: 10.1074/jbc.M411782200. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Spremulli LL. Roles of residues in mammalian mitochondrial elongation factor Ts in the interaction with bacterial and mitochondrial elongation factor Tu. J Biol Chem. 1998;273:28142–28148. doi: 10.1074/jbc.273.43.28142. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 7.Cai YC, Bullard JM, Thompson NL, Spremulli LL. Interaction of mammalian mitochondrial elongation factor EF-Tu with guanine nucleotides. Protein Sci. 2000;9:1791–1800. [PMC free article] [PubMed] [Google Scholar]
- 8.Cai YC, Bullard JM, Thompson NL, Spremulli LL. Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNA and elongation factor Ts. J Biol Chem. 2000;275:20308–20314. doi: 10.1074/jbc.M001899200. [DOI] [PubMed] [Google Scholar]
- 9.Valente L, Tiranti V, Marsano RM, Malfatti E, Fernandez-Vizarra E, Mereghetti P, De Gioia L, Burlina A, Castellan C, Comi G, Salvasta S, Ferrero I, Zeviani M. Infantile encephalopathy and defective mtDNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EF-Tu. Am J Hum Genet. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valente L, Shigi N, Suzuki T, Zeviani M. The R336Q mutation in human mitochondrial EFTu prevents the formation of an active mt-EFTu.GTP.aa-tRNA ternary complex. Biochim Biophys Acta. 2009;1792:791–795. doi: 10.1016/j.bbadis.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Schwartzbach C, Spremulli LL. Interaction of animal mitochondrial EF-Tu:EF-Ts with aminoacyl-tRNA, guanine nucleotides and ribosomes. J Biol Chem. 1991;266:16324–16330. [PubMed] [Google Scholar]
- 12.Jones CN, Jones CI, Graham WD, Agris PF, Spremulli LL. A disease-causing point mutation in human mitochondrial tRNAMet results in tRNA misfolding leading to defects in translational initiation and elongation. J Biol Chem. 2008;283:34445–34456. doi: 10.1074/jbc.M806992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullard JM, Cai YC, Spremulli LL. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol. 1999;288:567–577. doi: 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- 14.Remold-O’Donnell E, Thach RE. A new method for the purification of initiation factor F2 in high yield, and an estimation of stoichiometry in the binding reaction. J Biol Chem. 1970;245:5737–5742. [PubMed] [Google Scholar]
- 15.Spremulli LL. Large-scale isolation of mitochondrial ribosomes from mammalian tissues. In: Leister D, Herrmann J, editors. Mitochondria: Practical Protocols, vol. 372, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2007. pp. 265–275. [DOI] [PubMed] [Google Scholar]
- 16.Woriax V, Burkhart W, Spremulli LL. Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim Biophys Acta. 1995;1264:347–356. doi: 10.1016/0167-4781(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 17.Xin H, Woriax VL, Burkhart W, Spremulli LL. Cloning and expression of mitochondrial translational elongation factor Ts from bovine and human liver. J Biol Chem. 1995;270:17243–17249. doi: 10.1074/jbc.270.29.17243. [DOI] [PubMed] [Google Scholar]
- 18.Xin H, Leanza K, Spremulli LL. Expression of bovine mitochondrial elongation factor Ts in Escherichia coli and characterization of the heterologous complex formed with prokaryotic elongation factor Tu. Biochim Biophys Acta. 1997;1352:101–112. doi: 10.1016/s0167-4781(97)00003-1. [DOI] [PubMed] [Google Scholar]
- 19.Schwartzbach C, Farwell M, Liao HX, Spremulli LL. Bovine mitochondrial initiation and elongation factors. Methods Enzymol. 1996;264:248–261. doi: 10.1016/s0076-6879(96)64025-7. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsuki T, Sakurai M, Sato A, Watanabe K. Characterization of the interaction between the nucleotide exchange factor EF-Ts from nematode mitochondria and elongation factor Tu. Nucleic Acids Res. 2002;30:5444–5451. doi: 10.1093/nar/gkf679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suematsu T, Sato A, Sakurai M, Watanabe K, Ohtsuki T. A unique tRNA recognition mechanism of Caenorhabditis elegans mitochondrial EF-Tu2. Nucleic Acids Res. 2005;33:4683–4691. doi: 10.1093/nar/gki784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter SE, Spremulli LL. Mutagenesis of Arg335 in bovine mitochondrial elongation factor Tu and the corresponding residue in the Escherichia coli factor affects interactions with mitochondrial aminoacyl-tRNAs. RNA Biol. 2004;2:95–102. doi: 10.4161/rna.1.2.1034. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Sun V, Spremulli LL. Role of domains in Escherichia coli and mammalian mitochondrial elongation factor Ts in the interaction with elongation factor Tu. J Biol Chem. 1997;272:21956–21963. doi: 10.1074/jbc.272.35.21956. [DOI] [PubMed] [Google Scholar]
- 24.Kothe U, Rodnina MV. Delayed release of inorganic phosphate from elongation factor tu following gtp hydrolysis on the ribosome. Biochemistry. 2006;45:12767–12774. doi: 10.1021/bi061192z. [DOI] [PubMed] [Google Scholar]
- 25.Dirheimer G, Keith G, Dumas P, Westhof E. Primary, secondary and tertiary structures of tRNAs. In: RajBhandary U, Soll D, editors. tRNA:Structure, biosynthesis and function. ASM Press; Washington, D.C: 1995. pp. 93–126. [Google Scholar]
- 26.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1999;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakita K, Watanabe W, Yokogawa T, Kumazawa Y, Nakamura S, Ueda T, Watanabe K, Nishikawa K. Higher-order structure of bovine mitochondrial tRNAPhe lacking the ‘conserved’ GG and TYCG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 1994;22:347–353. doi: 10.1093/nar/22.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 30.Sprinzl M, Graeser E. Role of the 5′-terminal phosphate of tRNA for its function during protein biosynthesis elongation cycle. Nucleic Acids Res. 1980;8:4737–4744. doi: 10.1093/nar/8.20.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coenen MJH, Antonicka H, Ugalde C, Sasarman F, Rossi R, Heister JGAM, Newbold RF, Trijbels FJMF, van den Heuvel LP, Shoubridge EA, Smeitink JAM. Mutant mitochondrial Elongation Factor G1 and combined oxidative phosphorylation deficiency. New Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- 33.Miller C, Saada A, Shaul N, Shabtai N, Ben-Shalom E, Shaag A, Hershkowitz E, Elpeleg O. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 34.Saada A, Shaag A, Arnon S, Dolfin T, Miller C, Fuchs-Telem D, Lombes A, Elpeleg O. Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J Med Genet. 2007;44:784–786. doi: 10.1136/jmg.2007.053116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haque MdE, Grasso D, Miller C, Spremulli LL, Saada A. The effect of mutated mitochondrial ribosomal proteins S16 and S22 on the assembly of the small and large ribosomal subunits in human mitochondria. Mitochondrion. 2008;8:254–261. doi: 10.1016/j.mito.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]