Abstract
The WRN gene defective in the premature aging disorder Werner syndrome encodes a helicase/exonuclease. We examined the ability of WRN to rescue DNA damage sensitivity of a yeast mutant defective in the Rad50 subunit of Mre11-Rad50- Xrs2 nuclease complex implicated in homologous recombination repair. Genetic studies revealed WRN operates in a yEXO1-dependent pathway to rescue rad50 sensitivity to methylmethane sulfonate (MMS) and prevent mitotic catastrophe. WRN helicase, but not exonuclease, is required for MMS resistance. WRN missense mutations in helicase or RecQ C-terminal domains interfered with the ability of WRN to rescue rad50 MMS sensitivity. WRN does not rescue rad50 ionizing radiation (IR) sensitivity, suggesting that WRN, in collaboration with yEXO1, is tailored to relieve replicational stress imposed by alkylated base damage. WRN and yEXO1 are associated with each other in vivo. Purified WRN stimulates hEXO1 nuclease activity on DNA substrates associated with a stalled or regressed replication fork. We propose WRN helicase operates in an EXO1-dependent pathway to help cells survive replicational stress. In contrast to WRN, BLM helicase defective in Bloom’s syndrome failed to rescue rad50 MMS sensitivity, but partially restored IR resistance, suggesting a delineation of function by the human RecQ helicases.
Keywords: Werner syndrome, helicase, RecQ, rad50, replicational stress, genomic instability
1. INTRODUCTION
Werner syndrome (WS) is a genetic disorder characterized by premature aging, genomic instability, sensitivity to certain DNA damaging agents, elevated recombination, and replication defects [1–3]. The WRN gene encodes a RecQ helicase and exonuclease that is proposed to play a role in the regulation of recombination events, primarily homologous recombination (HR) [4]. It seems probable that WRN has a specialized function when the replication fork encounters a blocking lesion or alternate DNA structure. Through its catalytic activities and protein interactions in DNA replication, recombination, and repair, WRN protects the genome from deleterious mutagenic and recombinogenic events when the progression of the replication fork is perturbed. Although this model is attractive, it has been a formidable challenge for researchers to definitively characterize the molecular and cellular roles of WRN in a genetic pathway.
A number of laboratories have undertaken studies of WRN function using in vitro and in vivo approaches with model systems. Biochemical analyses of purified WRN protein demonstrate that WRN can unwind key DNA structures of HR (D-loop, Holliday Junction) and engage in physical/functional interactions with proteins involved in the progression and stabilization of the replication fork (e.g., Flap Endonuclease 1 (FEN-1), Replication Protein A (RPA), DNA polymerase δ, PCNA) as well as DNA recombination proteins (e.g., Rad52, Ku heterodimer, BRCA1) and telomere factors (TRF2, POT1) (for review, see [4]). Co-immunoprecipitation and affinity pull-down experiments have verified the association of WRN with these and other DNA repair/signaling proteins in human cells. Characterization of human WS cell lines stably transfected with WRN or mutant forms of the protein as well as cells depleted of WRN by RNA interference has provided biological evidence that WRN responds to replicational stress or DNA damage to maintain genomic stability [5, 6].
We developed a model genetic system in yeast to study human WRN functions in defined genetic pathways that are conserved in yeast and human. To investigate the role of WRN in replication, we examined its ability to rescue cellular phenotypes of a yeast dna2 mutant defective in a helicase–endonuclease that participates with FEN-1 in Okazaki fragment processing. Genetic experiments demonstrated that human WRN or a non-catalytic C-terminal domain fragment of WRN that interacts with FEN-1 rescued the dna2-1 mutant phenotypes of growth, cell cycle arrest and sensitivity to the replication inhibitor hydroxyurea (HU) or DNA damaging agent MMS [7]. Based on genetic and biochemical results, we proposed that the WRN–FEN-1 interaction has an important role in DNA replication intermediate processing. In a sgs1 top3 mutant background, WRN restored the slow growth phenotype and MMS sensitivity of the top3 mutant [8]. WRN expression did not affect growth rate of the wild-type parental strain or sensitivity to MMS. Moreover, WRN did not rescue the MMS or HU sensitivity of sgs1, indicating that WRN cannot directly replace Sgs1 in the sgs1 mutant background. The genetic interaction of WRN with Top3 suggests that WRN function in yeast is dependent on the genetic background [8].
To delineate the role of WRN in the DNA damage response, we have examined the ability of human WRN to rescue the DNA damage sensitivity of a yeast mutant defective in the Rad50 subunit of the Mre11-Rad50-Xrs2 (MRX) nuclease complex, that is implicated in double strand break (DSB) repair and a proper checkpoint response to DNA damage [9–12]. Mutations in Mre11 or the NBS1 human equivalent of Xrs2 lead to the autosomal recessive diseases ataxia telangiectasia-like disorder and Nijmegen breakage syndrome. Overexpression of the 5’ to 3’ exonuclease EXO1 can substitute for Mrx in processing of DSBs and partially rescues the MMS and IR sensitivity of mre11, rad50, or xrs2 mutants by specifically elevating repair through a homologous recombination pathway and not by nonhomologous end-joining [13–15]. Human Exonuclease 1 (hEXO1) is functionally similar to yeast EXO1 (yEXO1) based on the ability of hEXO1 to complement S. cerevisiae exo1 and the mutator phenotype of the yeast rad27 mutant [16]. hEXO1 interacts physically with WRN [17] and RECQ1 [18], thereby suggesting a role for EXO1 in the resolution of DNA structural intermediates formed during DNA repair or replication; however, the biological significance of the protein interaction is not yet known.
Exo1 and Sgs1, the sole RecQ homolog in S. cerevisiae, were shown to have a role in long range 5’ strand resection at double strand breaks at a step after initial trimming by the MRX/Sae2 complex ([19–21]. Using fission yeast as a model system, it was shown that in the absence of MRN, EXO1 becomes the major nuclease required for efficient meiotic recombination [22]. Recently, the Kowalczykowski lab demonstrated that the Bloom’s syndrome helicase (BLM) stimulates the nucleolytic activity of EXO1 on a linearized plasmid DNA molecule [23].
In the current study, we have demonstrated that WRN helicase, but not exonuclease activity, operates in an EXO1-dependent pathway to rescue the rad50 mutant sensitivity to MMS, a DNA damaging agent that introduces alkylated bases which have been shown to interfere with normal replication fork progression. Biochemical studies show that WRN stimulates EXO1 incision of DNA structures associated with stalled or regressed replication forks, suggesting a model to explain how WRN and EXO1 collaborate to confer cellular resistance to alkylation base damage. The ability of BLM but not WRN to partially restore the IR resistance of the rad50 mutant provides evidence for a delineation of function by the BLM and WRN RecQ helicases, one important for repair of direct strand breaks and the other important for the repair of replication forks blocked by alkylated base damage.
2. MATERIALS AND METHODS
2.1 Plasmid DNA constructs
Plasmids YEp195SpGAL-WRN or YEp195SpGAL-WRN940–1432, which express full-length WRN or WRN940–1432, respectively, under the control of a galactose-inducible promoter were made as described previously [7]. Site-directed mutations of WRN (E84A, K577M, R834C, and K1016A) were constructed using mutagenic primers (Table 1) and a standard protocol from Quickchange II XL site-directed mutagenesis kit (Stratagene) by Lofstrand labs (Gaithersburg, MD). The BLM expression plasmid pJK1 was kindly provided by Dr. Ian Hickson (University of Oxford). The BLM cDNA was PCR amplified from pJK1 using 5’GGACTAGTCTAACCATGTCAACTGAACCGGCTGCTGTTCCTCAAAATAAT3' and 5’AGCGACGCGTTCAGTGGTGGTGGTGGTGGTGTGAGAATGCATATGAAGGCTT3’ as forward and reverse primers, respectively. Amplification was carried out with PCR super mix HiFi (Invitrogen) for 25 cycles (denaturation at 94°C for 30 s per cycle, annealing at 60°C for 50 s per cycle, and elongation at 72°C for 5 min per cycle). The amplified product was cloned into the SpeI–MluI sites of vector YEp195SpGAL. Plasmid pESC-TRP was purchased from Strategene.
Table 1.
Oligonucleotides Used for WRN Site Site-directed Mutagenesis
| Residue Mutated | Primers used |
|---|---|
| E84A | 5’-GGG ATT TGA CAT GGC GTG GCC ACC ATT ATA C-3’ |
| 3’-GTA TAA TGG TGG CCA CGC CAT GTC AAA TCC C-5’ | |
| K577M | 5’-GGC AAC TGG ATA TGG AAT GAG TTT GTG CTT CC-3’ |
| 3’-GGA AGC ACA AAC TCA TTC CAT ATC CAG TTG CC-5’ | |
| R834C | 5’-AAT AAA GCT GAC ATT TGC CAA GTC ATT CAT TAC-3’ |
| 3’-GTA ATG AAT GAC TTG GCA AAT GTC AGC TTT ATT-5’ | |
| K1016A | 5’-CAG AGA GTT GGT GGG CGG CTT TTT CCC GTC AGC-3’ |
| 3’-GCT GAC GGG AAA AAG CCG CCC ACC AAC TCT CTG-5’ |
Nucleotide in bold indicates position of mutation
2.2 Yeast media and strains
Strains with wild-type RAD50 (WT; T334, genotype MATα ura3-52 leu2-3,112 Δtrp1::hisG reg1-501 gal1 pep4-3 prb1-1122) or a rad50 mutant (YLKL440; genotype T334, Δrad50::G418) have been characterized [13, 24]. Strains with wild-type RAD50 EXO1 (WT; VL6α, genotype MATα ura3-52 trp1(Δ63) lys2-801 his3-Δ200 met14 ade2-101) or a rad50 exo1 mutant (YLKL619; genotype VL6α, Δexo1::G418 Δrad50::hisG) and an exo1 mutant (YLKL546; genotype VL6Δ, Δexo1::G418) have been characterized [13, 25]. Yeast cultures were grown using standard protocol and transformations were performed using a Lithium Acetate-based protocol [26]. Transformed yeast strains were grown in Synthetic Complete (SC) media containing 2% glucose minus uracil (Ura).
2.3 Western blot analyses
Transformed rad50 strains were grown in SC glucose minus Ura at 30°C to an OD600 of 0.5. Cells (3 ml) were collected by centrifugation, washed with phosphate-buffered saline (PBS), lysed in alkaline lysis buffer [50 mM NaOH, pH 10.5, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2% SDS, 10% glycerol, 5% 2-mercaptoethanol and protease inhibitors (Roche Molecular Biochemicals)], boiled for 5 min, clarified by centrifugation, and neutralized with 1 M HCl. Proteins from equivalent amounts of cell lysate were resolved on 8–16% polyacrylamide SDS gels. Expression of WRN, WRN mutants or WRN940–1432 was determined by Western blot using a WRN mouse monoclonal antibody directed against an epitope in a purified C-terminal fragment of WRN [27] (1:1000, Spring Valley Labs). For an internal control, the blots were also probed with a monocloncal antibody against yeast 3-Phosphoglyceric Phosphokinase (yPGK) (1;5,000, Invitrogen). Expression of BLM was determined by Western blot using a rabbit anti-BLM antibody directed against a region between residue 1425 and the C-terminus (residue 1477) of human BLM (1:500, Bethyl Labs). For quantitative Western blot analysis, increasing concentrations of purified recombinant His-tagged full-length WRN, WRN940–1432, or purified recombinant His-tagged full-length BLM proteins were included on gels with yeast lysate samples.
2.4 Genetic rescue assays
rad50 cells transformed with YEp195SpGAL, YEp195SpGAL-WRN, YEp195SpGAL-WRN940–1432 or YEp195SpGAL-BLM were grown in SC glucose minus Ura at 30°C. Cultures were reinoculated in SC glucose minus Ura and grown at 30°C to early log phase (A600 of ~0.6 to 0.8). Ten-fold serial dilutions of these strains were plated onto SC glucose minus Ura plates with MMS at the indicated concentrations. Plates were incubated for 3 days at 30°C. As a control, the parental wild-type strain (T334) was transformed with YEp195SpGAL and treated as described above. The exo1 and rad50 exo1 strains transformed with YEp195SpGAL or YEp195SpGAL-WRN or parent (VL6α) strain transformed with YEp195SpGAL were grown in SC glucose minus Ura at 30°C to early log phase (A600 of ~0.6 to 0.8) and serial dilutions were plated on SC glucose minus Ura plates with MMS at the indicated concentrations.
For ionizing radiation (IR) experiments, rad50 cells transformed with YEp195SpGAL, YEp195SpGAL-WRN or YEp195SpGAL-BLM were grown to early log phase (A600 of ~0.6 to 0.8) and cultures diluted 1000-fold were γ-irradiated at the indicated doses (Fig. 1) using a Gammacell 40 (Nordon International, Inc.), a 137Cs source emitting at a fixed dose rate of 0.82 Gy/min. Cultures were subsequently plated onto SC glucose minus Ura plates, and incubated for 3 days at 30°C. As a control, wild-type parental strain (T334) was transformed with YEp195SpGAL and treated as described above.
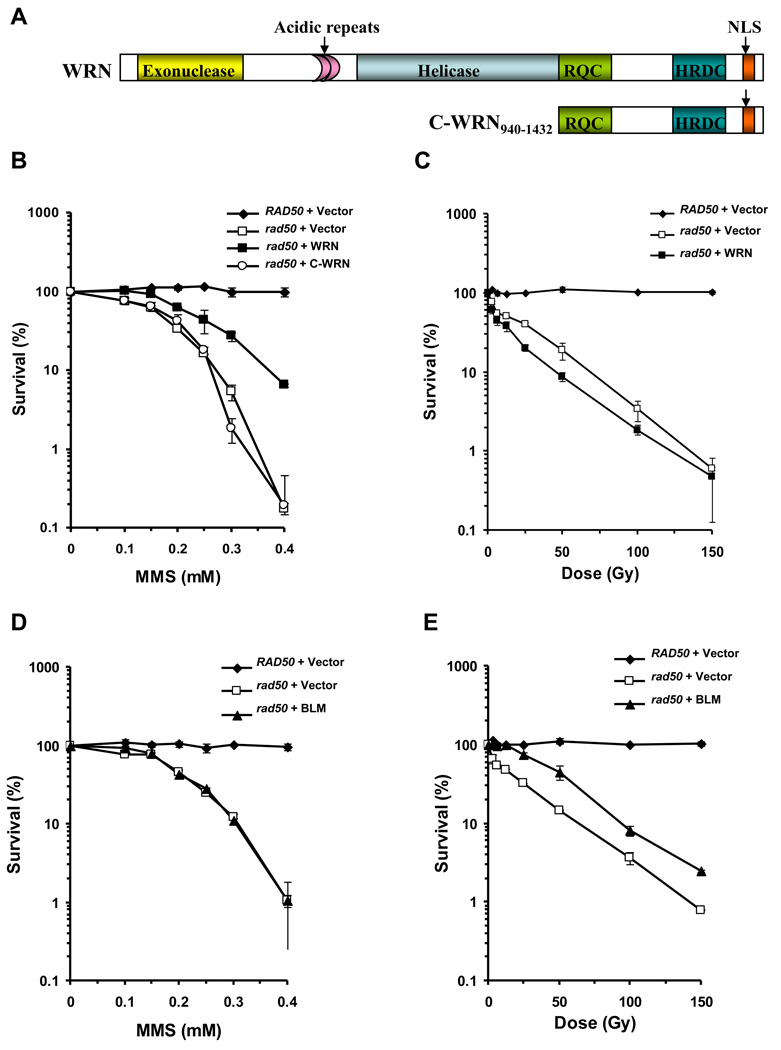
Figure 1. Effect of RecQ Helicases WRN and BLM on MMS and IR sensitivity phenotypes of rad50 mutant strain.
Panel A, Schematic representation of full-length WRN and C-terminal WRN940–1432 (C-WRN) proteins encoded by the respective yeast expression constructs. The WRN helicase and 3’ to 5’ exonuclease domains are shown, along with the conserved RecQ C-terminal (RQC) domain, Helicase and RNaseD C-terminal (HRDC) domain, perfect direct 27 amino acid acidic repeats, and the Nuclear Localization Sequence (NLS). Cultures of rad50 strain YLKL440 transformed with YEp195SpGAL, YEp195SpGAL–WRN, YEp195SpGAL–WRN940–1432 or YEp195SpGAL–BLM and the vector transformed wild-type parental strain were grown to early log phase (A600 of ~0.6 to 0.8), serially diluted 1000-fold, and either plated on SC glucose minus Ura plates at the indicated MMS concentration (Panel B and Panel D) or irradiated at the indicated doses, and plated on SC glucose minus Ura plates (Panel C and Panel E). Plates were incubated for 3 days at 30°C and colonies were counted. Survival at each MMS concentration or IR dose is expressed as a percent of colonies formed in absence of MMS or IR exposure for each transformant respectively.
2.5 Co-immunoprecipitation experiments
rad50 cells were co-transformed with YEp195SpGAL–WRN and pJAS-EXO1–FLAG (TRP, 2µ) expressing yeast Exo1p as a FLAG-tagged protein [29]. As a control rad50 cells were co-transformed either with YEp195SpGAL–WRN and pESC-TRP (empty FLAG tagged vector) or with YEp195SpGAL and pJAS-EXO1–FLAG. The transformed rad50 cells were grown in SC glucose minus Ura and Trp at 30°C to an OD600 of 0.5. Cells were pelleted and resuspended in SC raffinose minus Ura and Trp but containing 1% galactose or in SC glucose minus Ura and Trp containing 0.3 mM MMS and incubated at 30 °C for 6 h. For preparation of yeast cell lysates, cells were pelleted and washed once with cold sterile water, lysed in yeast lysis buffer (50 mM Tris-Cl pH 7.5, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS and Protease inhibitors (Roche Molecular Biochemicals)) with glass beads (Sigma) and clarified at 15,000 × g for 10 min at 4°C. The supernatants were then diluted in yeast lysis buffer, and 2 mg yeast lysate protein gently tumbled at 4°C for 4 h with anti-WRN (1:40, Santa Cruz Biotech) or anti-FLAG (1:80, Sigma) antibodies. Protein G–agarose beads (Amersham) were then added to the suspensions and incubation was performed overnight at 4 °C. The beads were washed four times with yeast lysis buffer supplemented with protease inhibitors and the immunoprecipitates were eluted by boiling in Laemmli buffer and resolved on 8–16% SDS–PAGE. Immunoprecipitated WRN and yEXO1-FLAG were detected by Western blotting using anti-WRN (BD Transduction) or anti-FLAG (Sigma) antibodies. The small difference in mobility of input and precipitated FLAG-yExo1 (Fig. 1A, 1D) is attributed to slightly different salt concentration in the precipitated sample compared to the input sample. The faint band in lane 5 of Fig. 4A (input) was not consistently observed and is likely due to a low level of cross-reactivity of the FLAG antibody with other protein in the input cell lysate. To determine if the interaction between WRN and yEXO1 was DNA-dependent, yeast lysates were first treated either with DNase I (0.5 µ/ml) or ethidium bromide (EtBr) (50 µ/ml) before co-immunoprecipitation.
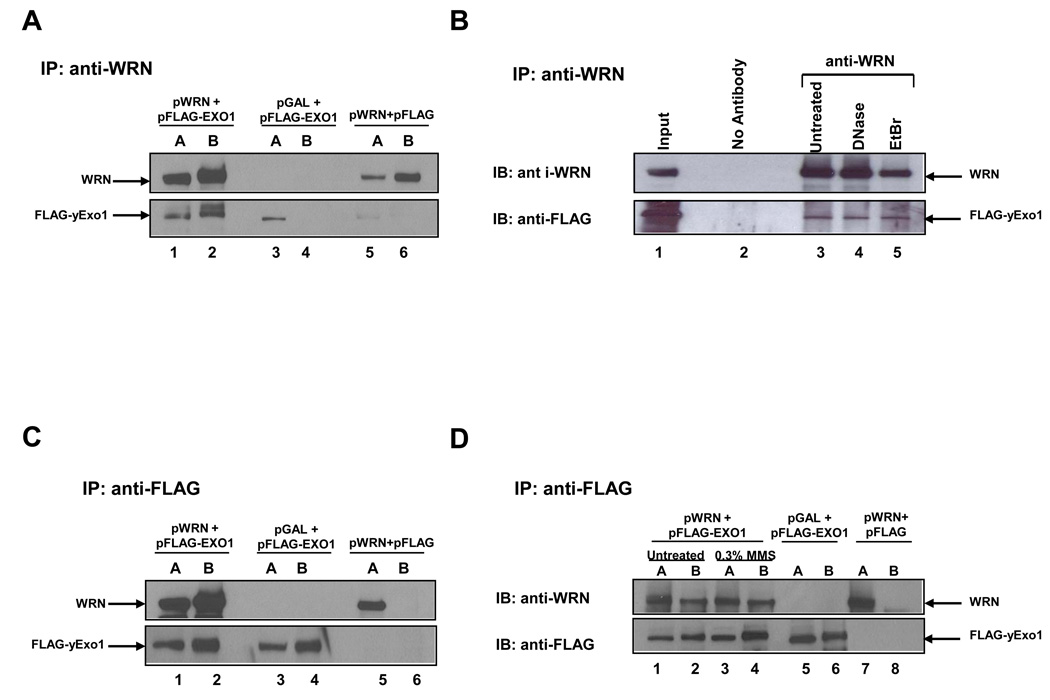
Figure 4. Physical interaction between WRN and yeast EXO1.
WRN and EXO1 were co-immunoprecipitated from whole cell lysates of rad50 cells co-transformed with YEp195SpGAL–WRN and pJAS-EXO1–FLAG, or YEp195SpGAL–WRN and pESC or YEp195SpGAL and pJAS-EXO1–FLAG using the anti-WRN (Panel A, B) or the anti- FLAG (Panel C and Panel D) antibody. The immunoprecipitated proteins were resolved by SDS-PAGE and probed with anti-WRN and anti-FLAG antibodies as described in Materials and Methods. Panel A, Lane 1, Yeast lysate input from cells expressing YEp195SpGAL–WRN and pJAS-EXO1–FLAG ; lane 2, immunoprecipitate from yeast lysate using anti-WRN antibody; lane 3, Yeast lysate input from cells expressing YEp195SpGAL and pJAS-EXO1–FLAG; lane 4, immunoprecipitate from yeast lysate using anti-WRN antibody; lane 5, Yeast lysate input from cells expressing YEp195SpGAL–WRN and pESC; lane 6, immunoprecipitate from yeast lysate using anti-WRN antibody. Panel B, lane 1, Yeast lysate input; lane 2, immunoprecipitate from yeast lysate using no antibody as control; lane 3, immunoprecipitate from yeast lysate using anti-WRN antibody except that yeast lysates were first treated either with DNase I (0.5 µg/ml) or ethidium bromide (EtBr) (50 µg/ml) before co-immunoprecipitation with anti-WRN antibody. Panel C, the same as Panel A except anti-FLAG antibody was used for co-immunoprecipitation. Panel D, WRN and EXO1 were co-immunoprecipitated from whole cell lysates of rad50 cells either untreated or treated with 0.3 mM MMS as described under Materials and Methods. lane 1, Yeast lysate input from untreated cells ; lane 2, immunoprecipitate from yeast lysate using anti-FLAG antibody; lane 3, Yeast lysate input from MMS treated cells; lane 4, immunoprecipitate from yeast lysate using anti-FLAG antibody; lane 5, Yeast lysate input from cells expressing YEp195SpGAL and pJAS-EXO1–FLAG; lane 6, immunoprecipitate from yeast lysate using anti-FLAG antibody; lane 7, Yeast lysate input from cells expressing YEp195SpGAL–WRN and pESC; lane 8, immunoprecipitate from yeast lysate using anti-FLAG antibody. In Panels A, C, and D, input and co-immunoprecipitate samples are indicated above the blot images as the letters A and B, respectively.
2.6 Proteins and DNA substrates
Hexahistidine-tagged recombinant human EXO1, overexpressed in insect cells and purified as described elsewhere [30], was kindly provided by Dr. T. Wilson (University of Maryland School of Medicine). Hexahistidine-tagged WRN was overexpressed in insect cells and purified as described elsewhere [31]. A recombinant hexahistidine-tagged carboxyl-terminal fragment of WRN (residues 940–1432, designated C-WRN940–1432) was overexpressed in Escherichia coli and purified as described previously [32].
Synthetic X12 Holliday Junction was prepared as previously described [31, 33]. The replication fork structure with a 5’ 32P label on the strand corresponding to the nascent lagging strand was prepared as previously described [33].
2.7 EXO1 incision assays
20-µl reactions contained 0.5 nM DNA substrate and the indicated concentrations of WRN and/or EXO1 in 30 mM HEPES pH 7.6, 5% glycerol, 40 mM KCl, 0.1 mg/ml bovine serum albumin, and 8 mM MgCl2. WRN was mixed with the substrate on ice prior to the addition of EXO1. Reactions were incubated at 37 °C for 15 min (unless indicated otherwise), terminated with the addition of 10 µl of formamide dye (80% formamide (v/v), 0.1% bromophenol blue, and 0.1% xylene cyanol), and heated to 95 °C for 5 min. Products were resolved on 20% polyacrylamide, 7 M urea denaturing gels. A PhosphorImager was used for detection, and the ImageQuant software (Molecular Dynamics) was used for quantification of the reaction products. The percent incision was calculated as described previously [34].
3. RESULTS
3.1 WRN rescues the MMS-sensitive survival phenotype of a yeast rad50 DNA repair mutant
A yeast rad50 mutant strain transformed with a multi-copy URA3 selectable plasmid encoding either full-length human WRN protein (residues 1–1432) or a WRN non-catalytic C-terminal fragment (residues 940–1432) (Fig. 1A) was tested for protein expression by Western blot analysis. Quantitative Western blot analyses using purified recombinant WRN or WRN940–1432 protein as standard indicated that 5,900 or 8,100 molecules/cell of WRN or WRN940–1432, respectively, were present (Supp. Data Fig. 1). In comparison, the level of endogenous WRN in HeLa cells was determined to be 89,000 molecules/cell [7], in agreement with published values for WRN copy number in other human cells [35, 36]. The rad50 strain transformed with empty vector (rad50/vector) did not express protein specifically recognized by WRN antibody (Supp. Data Fig. 1).
The rad50 mutant strain is characterized by sensitivity to the compound MMS which introduces base alkylation damage. To assess the effect of WRN expression on the MMS-sensitive phenotype of the rad50 strain, transformed rad50 cells were plated onto synthetic complete (SC) media lacking uracil (Ura) and containing the indicated concentrations of MMS (0–0.4 mM). An isogenic wild-type parent strain (T334) was included as a positive control in our experiments. As expected, rad50/vector cells were highly sensitive to MMS, displaying a reduction in cell growth at MMS concentrations as low as 0.1 mM, and approximately a 560-fold reduction at 0.4 mM (Fig. 1B). Mutant rad50 cells expressing WRN showed partial resistance to MMS in colony forming ability at all MMS concentrations tested compared to the rad50/vector cells (Fig. 1B). At an MMS concentration of 0.4 mM, the rad50/WRN cells were 40-fold more resistant to the drug compared to rad50/vector cells. Partial complementation of the rad50 MMS sensitivity was also observed when yeast EXO1 was overexpressed in the rad50 mutant [15]. We also tested the MMS sensitivity of a rad50 mutant strain expressing the C-terminal WRN940–1432 protein fragment that mediates the protein interaction with FEN-1 and EXO1 [37] and shown to be sufficient to rescue the replication and DNA repair phenotypes of a dna2-1 mutant [7]; however, the rad50/WRN940–1432 strain was as sensitive to MMS as rad50/vector (Fig. 1B). From these results, we conclude that full-length WRN could partially rescue the MMS sensitivity of the rad50 mutant, but the C-terminal WRN fragment could not, suggesting that additional domains of the WRN protein are required for cellular resistance of the rad50 mutant to MMS.
3.2 WRN does not rescue the IR-sensitive phenotype of the rad50 mutant
The sensitivity of rad50 mutants to IR reflects the importance of the Rad50- Mre11-Xrs2 nuclease complex in processing DNA strand breaks. We asked if WRN expression in the rad50 mutant might rescue its IR sensitivity, similar to what was observed for the MMS sensitivity of this strain. As expected, rad50/vector cells were sensitive to IR in a dose-dependent fashion (Fig. 1C). At 100 Gy IR, the percent viability was reduced 10-fold compared to the parent strain which was relatively resistant to IR. Expression of WRN in the rad50 mutant failed to rescue the IR sensitivity (Fig. 1C). These results demonstrate that the ability of WRN to rescue the MMS-sensitive phenotype of the rad50 mutant is specific and not applicable to IR exposure.
3.3 BLM fails to rescue rad50 MMS sensitivity but partially restores IR resistance
Since the genetic rescue studies are carried out in the background of an endogenous RecQ helicase (Sgs1), the ability of WRN to rescue the MMS sensitivity of the rad50 mutant may have simply been an effect of overexpression of a RecQ helicase that confers resistance. To address the specificity of the effect of human WRN expression on genetic rescue, we examined the ability of human BLM to affect the MMS- and IR-sensitive rad50 phenotypes. In these experiments, BLM protein expressed from YEp195SpGAL in the rad50 mutant background was confirmed by Western blot analysis (data not shown). Quantitative Western blot analyses using purified recombinant BLM protein as standard indicated that 8,100 BLM molecules/cell were present. To assess the effect of BLM expression on the MMS-sensitive phenotype of the rad50 strain, transformed rad50 cells were plated onto synthetic complete (SC) media lacking uracil (Ura) and containing the indicated concentrations of MMS (0–0.4 mM). An isogenic wild-type parent strain (T334) was included as a positive control in our experiments. In contrast to WRN, BLM completely failed to rescue MMS sensitivity of the rad50 mutant as evidenced by sensitivity comparable to rad50/vector (Fig. 1D). However, BLM expression in rad50 partially rescued IR sensitivity at all doses (Fig. 1E). Partial complementation of the rad50 IR sensitivity was also observed by overexpression of yEXO1 in the rad50 mutant [13]. These results and our previous observation that WRN cannot directly restore sgs1 MMS and HU resistance [8] suggest that the ability of WRN to rescue the rad50 MMS sensitivity is specific and not a general effect of overexpression of a RecQ helicase. Moreover, the ability of BLM but not WRN to restore IR resistance suggests that human BLM can function in the rad50 background, and that the two human RecQ helicases behave differently from one another in terms of their effects on rad50 phenotypes.
3.4 Genetic analyses of WRN variants to rescue the MMS sensitivity of rad50
To assess the functional requirements of WRN to rescue the MMS-sensitive phenotype of the rad50 mutant, we assessed the importance of WRN catalytic activities (helicase, exonuclease) for MMS resistance. To do this, we constructed WRN expression plasmids that encode the E84A exonuclease inactive mutant or K577M ATPase/helicase inactive mutant proteins (Fig. 2A). Plasmids encoding the WRN mutant alleles were transformed into the rad50 mutant as before and Western blot analysis confirmed expression of the mutant WRN proteins at levels comparable to wild-type WRN (Supp. Data Fig. 1). For an internal control, the blots were also probed with an antibody against yeast 3-Phosphoglyceric Phosphokinase (yPGK), which demonstrated equal loading of cell lysate protein. Cell survival analysis of rad50 transformants as a function of MMS concentration revealed that the WRN exonuclease dead mutant retained the ability to rescue the MMS sensitivity of the rad50 mutant comparable to wild-type WRN, whereas the WRN ATPase/helicase dead mutant failed to rescue the MMS sensitivity as shown by its similar level of survival to that of the rad50/vector cells at a given MMS concentration (Fig. 2B). Thus, the rescue of rad50 MMS sensitivity required WRN ATPase/helicase, but not exonuclease activity.
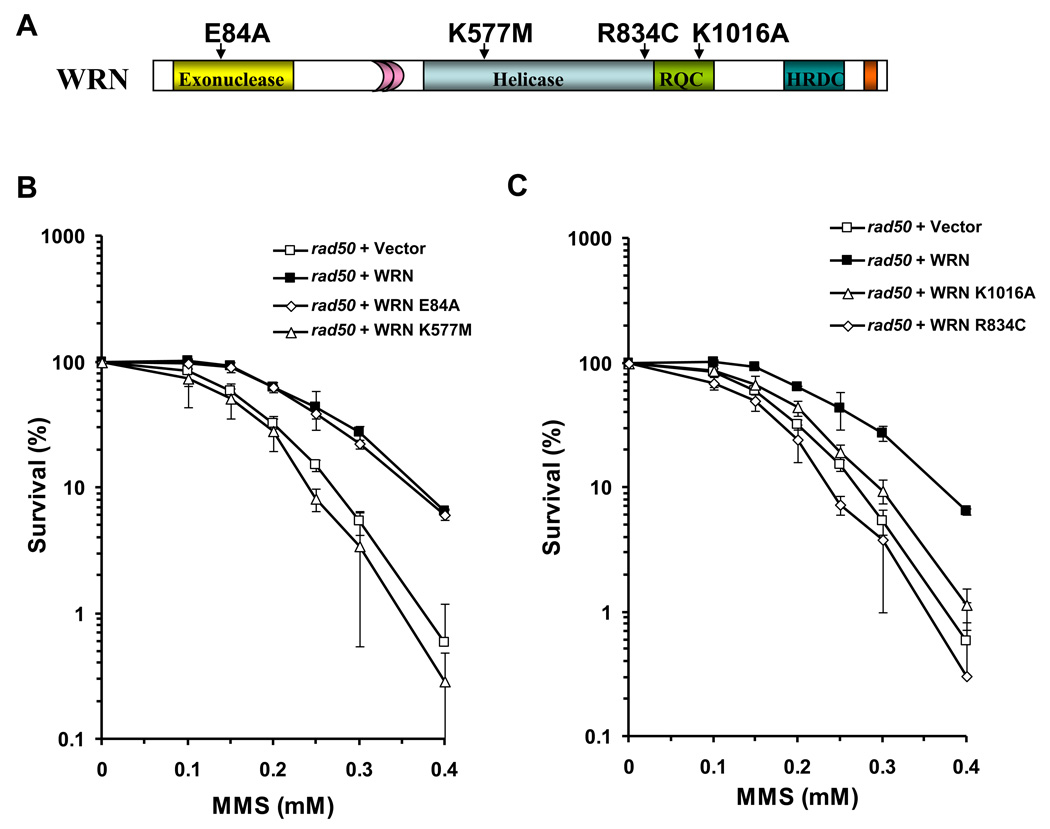
Figure 2. WRN ATPase/helicase, but not exonuclease activity, is required to rescue the MMS sensitivity of the rad50 mutant.
Panel A, Schematic representation of WRN with site-directed mutations encoded by the respective yeast expression constructs. Panels B and C, Logarithmically growing cultures of rad50 strains transformed with ATPase/helicase-dead (YEp195SpGAL–WRN K577M, Panel B), exonuclease-dead (YEp195SpGAL-WRN E84A, Panel B), RQC mutant (YEp195SpGAL-WRN K1016A, Panel C), or polymorphic mutant (YEp195SpGAL-WRN R834C, Panel C) were plated at a 1000-fold dilution on SC glucose minus Ura plates with the indicated MMS concentrations. Plates were incubated for 3 days at 30 °C and colonies were counted. Survival at each MMS concentration is expressed as a percent of colonies formed in absence of MMS for each transformant respectively. Percent survival with rad50 mutant expressing wild-type WRN or transformed with empty vector is included for comparison.
In addition to the helicase domain, a majority of RecQ helicases shares another conserved sequence designated the RecQ C-terminal (RQC) region [4]. The RQC region was first determined to contain a Zn2+-binding domain and a winged helix domain from the crystal structure of E. coli RecQ [40]. A structure of a recombinant human WRN protein fragment indicated that the WRN RQC region also forms a Zn2+-binding domain and winged helix domain [41]; furthermore the WRN RQC has been shown to bind DNA [27] and numerous other proteins (for a review, see [4]). To address the potential importance of the WRN RQC domain for its biological function, we examined the ability of a specific WRN RQC missense mutant, WRN-K1016A (Fig. 2A), to rescue the rad50 MMS sensitivity. The WRN-K1016A mutant was previously characterized and shown to have significantly reduced DNA binding and helicase activity on a variety of DNA structures (fork, D-loop, Holliday Junction) [42]; furthermore, the K1016A mutation of WRN significantly decreased its ability to stimulate the incision activity of FEN-1 despite its ability to physically interact with FEN-1 with a similar affinity as wild-type WRN [42]. As shown in Fig. 2C, WRN-K1016A poorly rescued the MMS sensitivity as demonstrated by its similar survival as rad50/vector throughout the MMS titration, despite its expression level comparable to WRN-WT (Supp. Data Fig 1). These results suggest that impairment of the DNA binding activity by the K1016A mutation, which also perturbs WRN helicase activity, interferes with its ability to rescue the rad50 mutant.
Disease-linked mutations in the WRN gene result in stop codons, splice site variants, and insertion/deletions giving rise to frameshift mutations [The International Registry of Werner syndrome (www.wernersyndrome.org)] [43]. However, WRN polymorphic variations resulting in amino acid substitutions have been identified in the human population. In certain cases, correlations have been made between certain WRN polymorphic variants and aging [44], myocardial infarction [45], or osteoporosis [46]. A WRN R834C polymorphism located in the central helicase core domain (Fig. 2A) expresses to nearly the same level as endogenous WRN; however, a cell line heterozygous for the R834C polymorphism was found to have ~50% of WRN helicase/exonuclease activity relative to the wild-type [47]. The purified recombinant WRN-R834C protein was shown to have dramatically reduced WRN ATPase, helicase, and helicase-coupled exonuclease activity [47]. To assess the biological effect of the WRN R834C polymorphic change, we used our established yeast genetic system. WRN-R834C, expressed in the rad50 mutant at a similar level as wild-type WRN (Supp. Data Fig 1), failed to rescue the MMS sensitivity as demonstrated by its poor survival, comparable to rad50/vector (Fig. 2C). These results suggest that the R834C polymorphism, similar to the K577M and K1016A mutants that interfere with WRN helicase activity, impair the ability of WRN to function in an appropriate response to alkylated base damage introduced by MMS in the rad50 mutant.
3.5 WRN rescue of rad50 MMS sensitivity requires EXO1
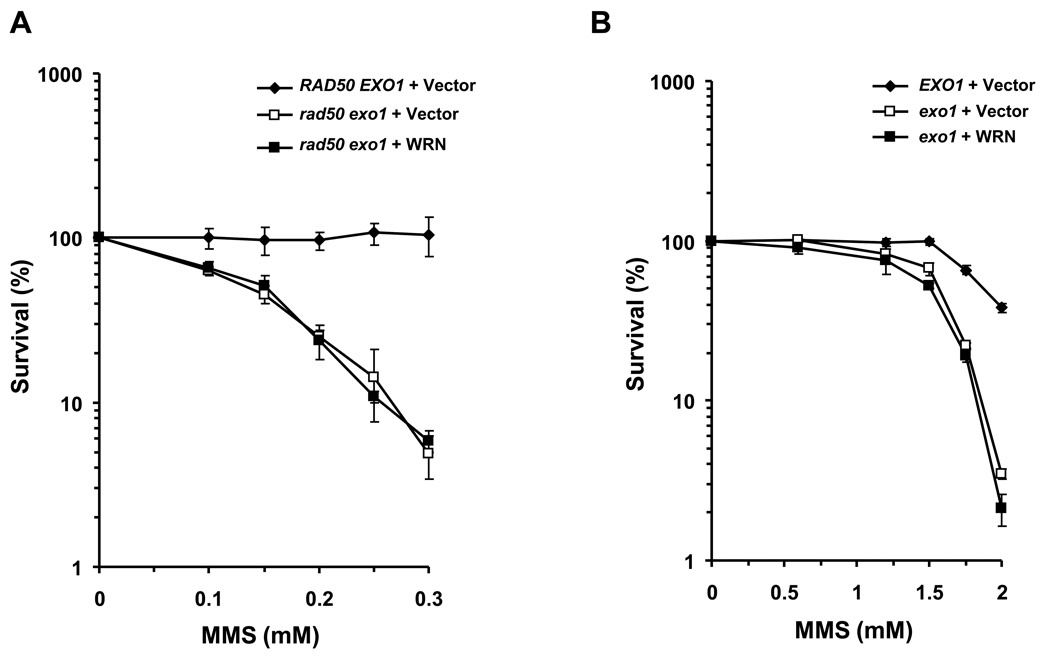
Expression of exogenously expressed EXO1 was previously shown to partially rescue the MMS sensitivity of the rad50 mutant [13–15]. It was proposed that EXO1 nucleolytic processing at the ends of double strand breaks that form at blocked replication forks facilitates an early step of end trimming that enables strand invasion of a single-stranded DNA molecule into homologous duplex in HR repair. However, more recent studies also suggest that EXO1 processes stalled replication forks and counteracts fork regression in checkpoint defective cells [48]. We hypothesized that WRN through its interaction with yEXO1 is responsible for the genetic rescue of the MMS phenotype of the rad50 mutant. In support of this, WRN expression in the rad50 exo1 double mutant did not restore any MMS resistance to the mutant strain (Fig. 3A). WRN also failed to rescue the MMS sensitivity of an exo1 mutant (Fig. 3B). Thus, WRN rescue of the MMS phenotype of a rad50 mutant is dependent on EXO1, indicating that WRN operates in an EXO1-dependent pathway.
Figure 3. WRN fails to rescue the MMS sensitivity of a rad50 exo1 double mutant.
Cultures of rad50 exo1 (Panel A) or exo1 (Panel B) strains transformed with YEp195SpGAL or YEp195SpGAL– WRN or wild-type parental cells transformed with YEp195SpGAL were grown in SC glucose minus Ura, serially diluted 1000-fold, and plated on SC glucose minus Ura plates with the indicated MMS concentrations. Plates were incubated for 3 days at 30°C and colonies formed were counted. Survival at each dose of MMS concentration is expressed as a percent of colonies formed in untreated cells for each transformant respectively.
3.6 Human WRN expressed in rad50 cells has the capability to interact with yeast EXO1
The requirement of EXO1 in order for WRN to rescue the rad50 mutant suggested to us that WRN might have the ability to interact with yeast EXO1 since the human and yeast versions of EXO1 are highly homologous [16, 49 50] and human EXO1 can substitute for yeast EXO1 in DNA repair, recombination, and mutation avoidance [16]. Therefore, we performed co-immunoprecipitation experiments from yeast whole cell lysates to explore the in vivo association of WRN with yEXO1. Yeast cells were co-transformed with the WRN expression vector and a FLAG-epitope-tagged yEXO1 expression construct. For controls, cells were co-transformed either with the WRN expression vector and an empty FLAG vector (pESC-TRP) or with YEp195SpGAL and a FLAG-epitope-tagged yEXO1 expression construct. An antibody against either the FLAG epitope or N-terminal epitope of WRN was used for the immunoprecipitation experiments from the yeast whole cell lysates. The anti-WRN antibody precipitated yEXO1–FLAG only from the lysates of the constructs expressing both WRN and yEXO1, whereas no yEXO1-FLAG was precipitated in the absence of WRN expression (Fig. 4A).
Co-immunoprecipitation of WRN and yEXO1-FLAG was resistant to either DNase I or ethidium bromide (EtBr) (Fig. 4B), suggesting that the association of the two proteins in the yeast cell lysate was not dependent on DNA. Furthermore, the anti-FLAG antibody precipitated yeast expressed WRN, whereas no WRN was precipitated in the absence of yEXO1-FLAG expression (Fig. 4C). The reciprocal co-immunoprecipitation of yEXO1 and WRN from the rad50 cell lysates provides further evidence that the proteins are associated with each other in a complex in vivo.
Since WRN partially rescued the rad50 MMS sensitivity phenotype, we wanted to determine if MMS treatment of the transformed rad50 cells affected the WRN: EXO1 interaction. To detect the effect of MMS treatment, yeast whole cell lysates were prepared from the cultures that were either untreated or treated with 0.3 mM MMS for 6 hrs. An antibody against the FLAG epitope was used for the immunoprecipitation experiments from the yeast whole cell lysates. The co-immunoprecipitation of WRN with EXO1 was increased 1.7-fold higher from lysates of rad50 cells treated with 0.3 mM MMS compared to lysates from untreated cells (Fig. 4D).
3.7 WRN stimulates EXO1 cleavage of DNA structures associated with a stalled or regressed replication fork
Recent evidence that EXO1 is recruited to stalled replication forks and counteracts fork reversal in checkpoint-defective cells [48] suggested to us that WRN may stimulate EXO1 processing of DNA structures associated with stalled replication forks that occur in MMS-treated cells. Firstly, WRN may collaborate with EXO1 to resect the newly synthesized lagging strand to prevent fork regression. Secondly, if fork regression occurs, WRN and EXO1 may function together on a reversed fork, the so-called chicken foot structure resembling a Holliday Junction. To determine whether WRN and EXO1 can process such DNA structures associated with stalled replication forks, we performed a series of biochemical experiments using purified recombinant wild-type WRN and EXO1 and synthetic replication fork or Holliday Junction structures.
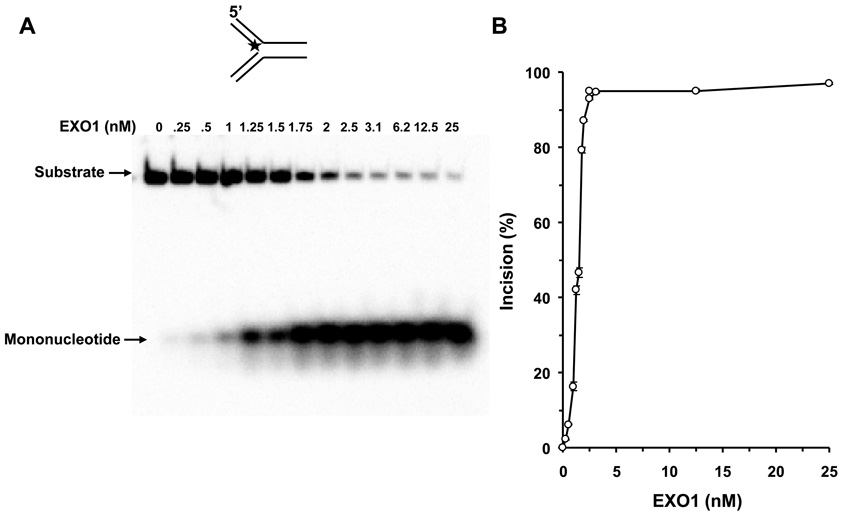
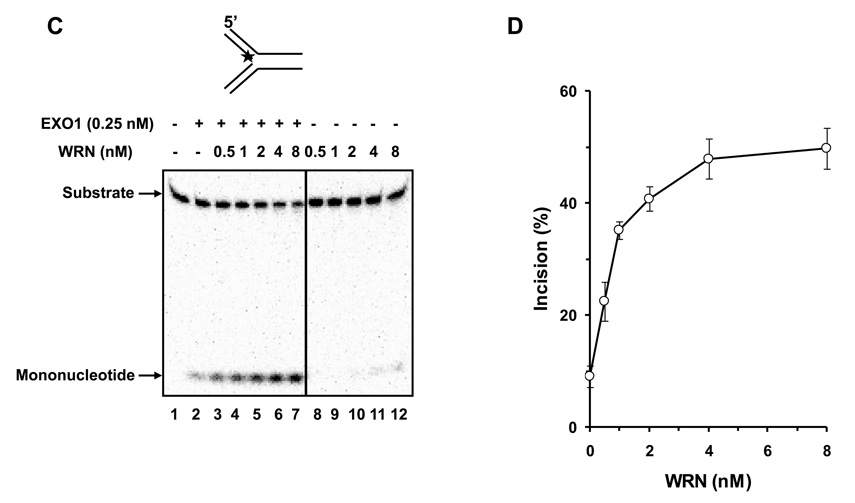
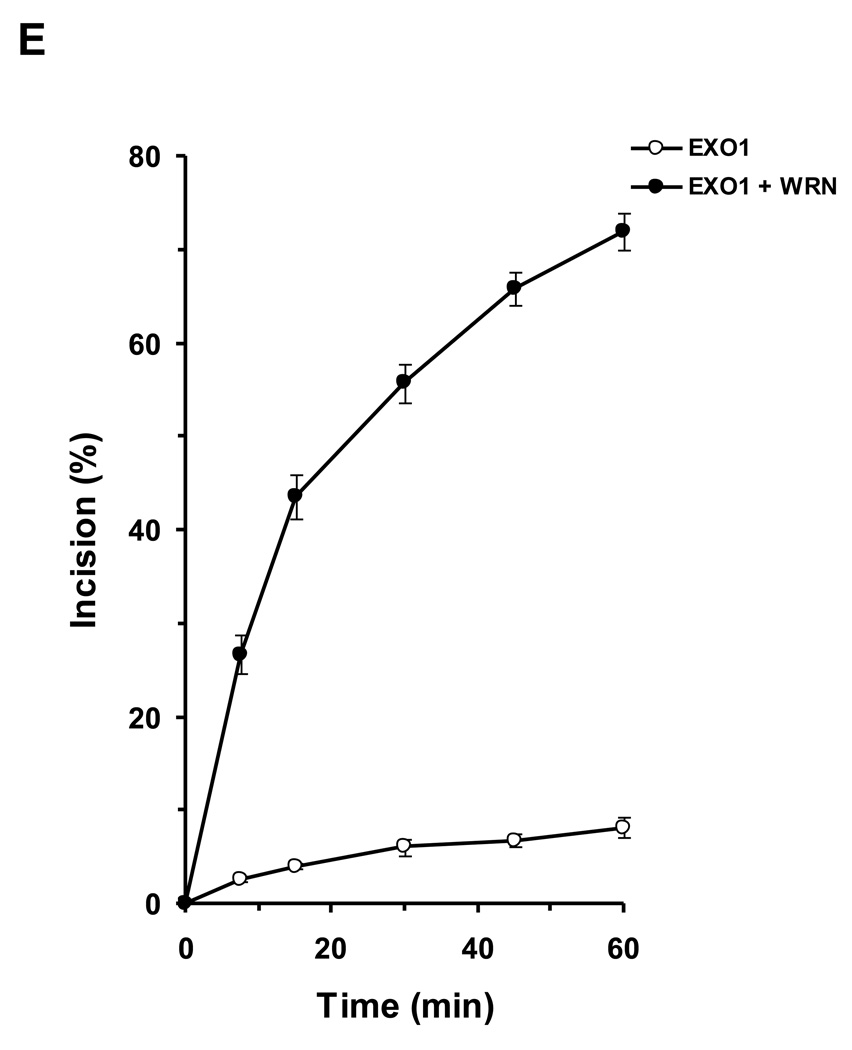
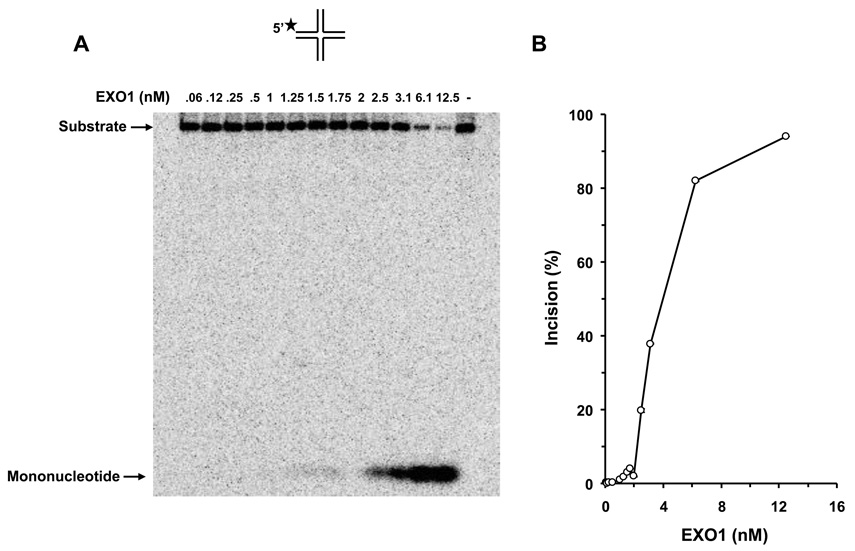
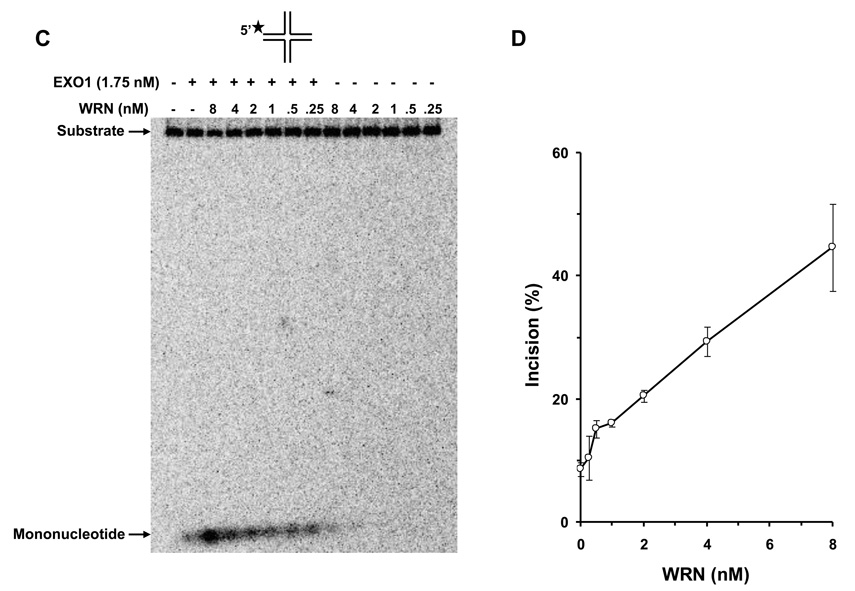
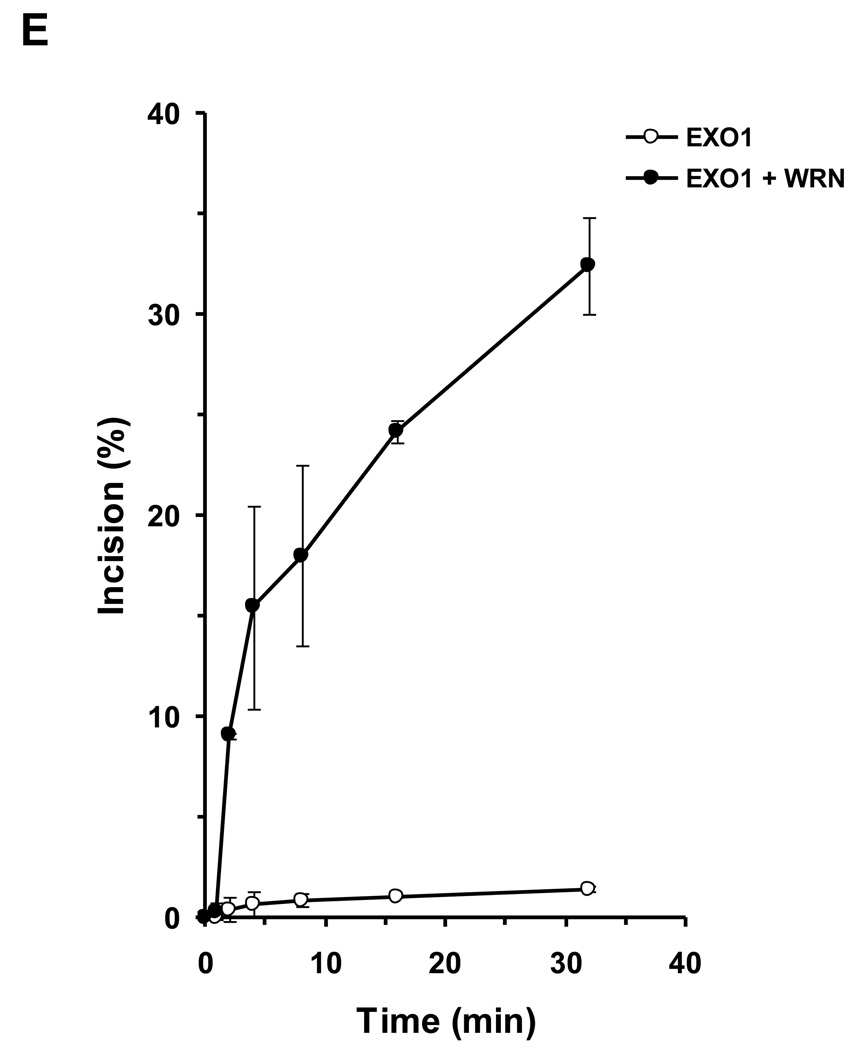
We tested human EXO1 on a replication fork structure with a 5’ 32P-label on the DNA strand representing the newly synthesized lagging strand. EXO1 exonuclease activity was dependent on EXO1 protein concentration from 0.25–2.5 nM where a plateau of ~92% incision was reached (Fig. 5A, 5B). In the presence of increasing WRN concentrations, EXO1 (0.25 nM) cleavage activity was stimulated proportionately (Fig. 5C, 5D). A 5.5-fold increase in EXO1 incision was observed at a concentration of 8 nM WRN monomer. In control reactions with only WRN, only a very small amount of 5’ mononucleotide was detected. A kinetic analysis of EXO1 incision in the presence or absence of WRN demonstrated that the rate of EXO1 incision was increased 11-fold by WRN (Fig. 5E).
Figure 5. WRN stimulates EXO1 incision of a replication fork structure.
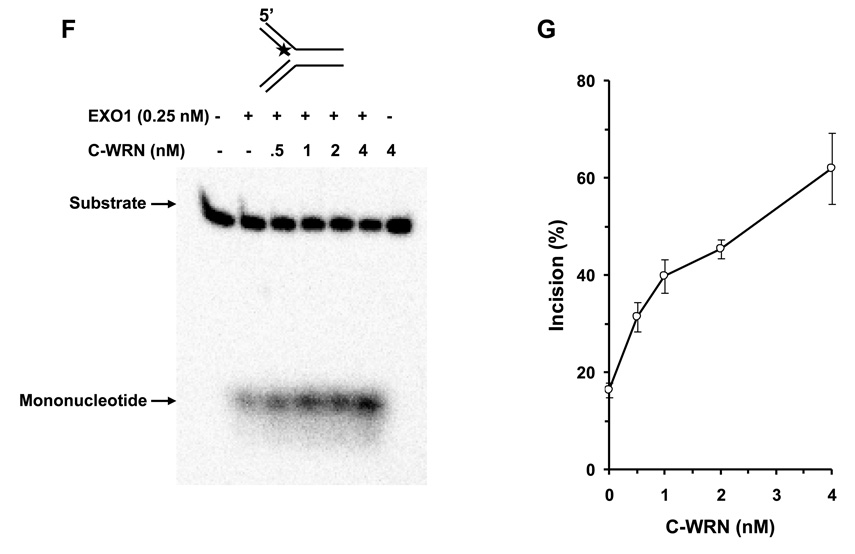
Panel A, 20-µl reactions containing 0.5 nM replication fork substrate and the indicated concentrations of EXO1 were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel B, percent incision from A (mean value of three experiments) with standard deviation (SD) indicated by error bars. Panel C, 20-µl reactions containing 0.5 nM replication fork substrate, 0.25 nM EXO1, and the indicated concentrations of WRN were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel D, percent incision from C (mean value of three experiments) with SD indicated by error bars. Panel E, Kinetics of EXO1 cleavage of the replication fork structure in the presence or absence of WRN. 140- µl reactions containing 0.5 nM replication fork substrate and 0.062 nM EXO1 were incubated at 37 °C under standard conditions, and aliquots were removed at 7.5, 15, 30, 45, and 60 min. The reactions in the presence of WRN contained 2 nM WRN monomer. Percent incision data for the reactions (mean value of three experiments) with SD indicated by error bars are shown. Open circles, EXO1; filled circles, EXO1 + WRN. Panel F, A non-catalytic domain of WRN that physically interacts with EXO1 is sufficient for stimulation of EXO-1 incision on the replication fork structure. 20-µl reactions containing 0.5 nM replication fork substrate, 0.25 nM EXO1, and the indicated concentrations of C-WRN were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel G, percent incision from F (mean value of three experiments) with SD indicated by error bars.
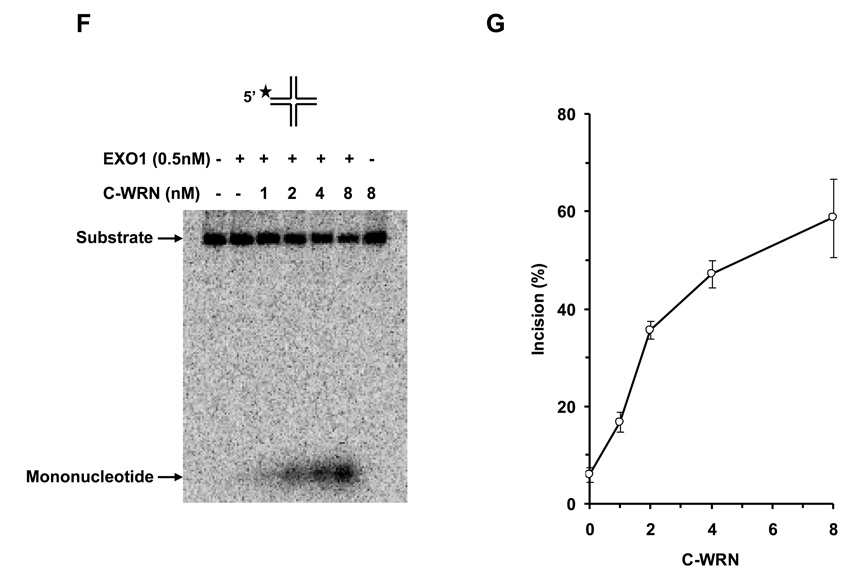
To address if the WRN protein interaction with EXO1 is sufficient for stimulation of EXO1 exonuclease activity on the replication fork, we performed EXO1 assays with WRN940–1432 (C-WRN), a non-catalytic C-terminal fragment of WRN that mediates the physical interaction with EXO1 [17]. As shown in Fig. 5F and 5G, C-WRN stimulated EXO1 incision in a protein concentration dependent manner. At 4 nM C-WRN, EXO1 incision was stimulated approximately 4-fold in the 15 min reaction. Thus, the stimulation of EXO1 exonuclease activity by C-WRN was comparable to that of the full-length WRN protein.
Next we examined the activity of EXO1 on a four-stranded Holliday Junction structure resembling a fully regressed replication fork. EXO1 incision on this structure displayed a linear dependence on EXO1 protein concentration from 2–6 nM where ~80% of the DNA substrate was cleaved (Fig. 6A, 6B). Using a limiting amount of EXO1 (1.75 nM), EXO1 incison activity was stimulated in a dose-dependent manner by WRN throughout the WRN titration range (0.25–8 nM) (Fig. 6C, 6D). At 8 nM WRN monomer, EXO1 incision was stimulated 5.2-fold. In control reactions with only WRN, little to no 5’ mononucleotide was detected. A kinetic analysis of EXO1 incision of the Holliday Junction structure in the presence or absence of WRN demonstrated that the rate of EXO1 incision was increased 23-fold by WRN (Fig. 6E).
Figure 6. WRN stimulates EXO1 incision of a regressed replication fork chicken foot structure.
Panel A, 20-µl reactions containing 0.5 nM regressed fork Holliday Junction substrate and the indicated concentrations of EXO1 were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel B, percent incision from A (mean value of three experiments) with SD indicated by error bars. Panel C, 20-µl reactions containing 0.5 nM regressed fork substrate, 1.75 nM EXO1, and the indicated concentrations of WRN were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel D, percent incision from C (mean value of three experiments) with SD indicated by error bars. Panel E, Kinetics of EXO1 cleavage of the regressed fork structure in the presence or absence of WRN. 140-µl reactions containing 0.5 nM regressed fork substrate and 0.5 nM EXO1 were incubated at 37 °C under standard conditions, and aliquots were removed at 2, 4, 8, 16, and 32 min. The reactions in the presence of WRN contained 8 nM WRN monomer. Percent incision data for the reactions (mean value of three experiments) with SD indicated by error bars are shown. Open circles, EXO1; filled circles, EXO1 + WRN. Panel F, A non-catalytic domain of WRN that physically interacts with EXO1 is sufficient for stimulation of EXO-1 incision on the Holliday Junction structure. 20-µl reactions containing 0.5 nM Holliday Junction substrate, 0.5 nM EXO1, and the indicated concentrations of C-WRN were incubated at 37 °C for 15 min under standard conditions as described under Materials and Methods. A phosphorimage of a typical gel is shown. Panel G, percent incision from F (mean value of three experiments) with SD indicated by error bars.
We next asked if the non-catalytic C-WRN fragment was sufficient for stimulation of EXO1 on the Holliday Junction structure. C-WRN stimulated EXO-1 incision on the Holliday Junction in a protein concentration dependent manner (Fig. 6F, 6G). At 8 nM C-WRN, EXO1 incision was stimulated approximately 12-fold in the 15 min reaction. From the biochemical studies, we conclude that EXO1 can incise DNA structures associated with a stalled or regressed replication fork, and WRN stimulates the EXO1 catalyzed reactions through its protein interaction.
4. DISCUSSION
WRN functions in an EXO1-dependent manner to rescue rad50 MMS sensitivity. Physiologically relevant expression levels of WRN helped to rescue the survival phenotypes of the rad50 mutant. Inability of WRN to rescue IR sensitivity indicates that WRN does not operate in the repair of direct strand breaks in the rad50 mutant background. WRN rescue of MMS sensitivity provides new insight to an earlier observation that yEXO1overexpression partially restored MMS resistance of rad50 or mre11 mutants [15]. WRN stimulates EXO1 incision of DNA structures associated with stalled or regressed replication forks, leading us to propose that WRN in collaboration with EXO1 is relied upon to prevent deleterious events at DNA replication forks impeded by base alkylation DNA damage.
WRN functional requirements for rescue of rad50 MMS sensitivity are distinct from that for rescue of the dna2-1 mutant in which WRN non-catalytic C-terminal domain was sufficient [7]. WRN ATPase/helicase activity operating in an EXO1- dependent pathway is responsible for rescue of rad50 to MMS-induced DNA damage, but not IR-induced damage. Thus, catalytic requirements of WRN necessary for a proper response to DNA damage can be genetically separated in the rad50 mutant background. Although biochemical studies with purified proteins and model DNA substrates demonstrated that the interaction of the C-terminal WRN domain with EXO1 is the minimal requirement for stimulation of EXO1 nuclease activity, the genetic requirement for an intact WRN ATPase/helicase domain to rescue MMS sensitivity of the rad50 mutant suggests that the situation in vivo is likely to be more complex. It is conceivable that the C-terminal domain fragment of WRN is sufficient to stimulate human EXO1 in vitro, but that the full-length WRN is required for stimulation of yeast EXO1 in vivo. Possibly, ATPase-dependent translocation to the site of MMS-induced damage or unwinding of blocked replication fork structures is important for WRN to collaborate with EXO1 and help cells cope with replicational stress. In support of a direct role of WRN to deal with replication fork problems, the Monnat lab showed WRN is required for normal replication fork progression after MMS-induced DNA damage or hydroxyurea-induced fork arrest [6].
Exo1 and yeast Sgs1 function in alternative pathways for long range 5’ strand resection at a restriction endonuclease-induced DSB [20]. Cells deficient in the MRX complex are impaired in the initiation of 5’ resection [21]. It has been shown that 5’ resection in rad50 sgs1 and rad50 exo1 double mutant is even more delayed than in rad50 single mutant, suggesting that in the absence of MRX complex both Sgs1 and Exo1 can still initiate limited DSB end processing. Our results implicate WRN helicase activity operates in the rad50 background in an Exo1-dependent pathway to confer MMS resistance. This is in contrast to Sgs1 in yeast and BLM in human cells that function in parallel with Exo1 to promote DSB resection [19]. Our studies thus suggest the delineation of WRN and BLM functions in DNA damage repair pathway. Consistent with the inability of helicase/ATPase dead WRN to rescue MMS sensitivity, the helicase domain of Sgs1 is required for resection of DNA ends generated by the trimming function of the MRX/Sae2 at a DSB [20, 21].
BLM expression in the rad50 mutant partially restored IR resistance but not MMS resistance, consistent with the possibility that BLM helps cells to cope with strand breaks, but not alkylated base damage induced by MMS. BLM stimulates EXO1 nucleolytic activity on a linearized plasmid DNA molecule [23]. A delineation of WRN and BLM function is suggested by their differential effects on rad50 MMS and IR sensitivity.
Establishment of a model genetic system for WRN enabled us to investigate the biological consequences of an engineered mutant (K1016A) in the conserved RQC domain as well as a naturally occurring polymorphism (R834C) in the helicase core domain. The R834C polymorphism resulted in a loss of WRN function in DNA damage resistance drawing further attention to this allele as potentially contributing to age-related disease. Thus yeast is a useful model system for studying WRN function in conserved genetic pathways that confer resistance to DNA damaging agents. Moreover, yeast genetics can be used to screen other naturally occurring WRN polymorphisms and engineered WRN domain mutants to decipher WRN functions in genome stability.
Dependence of WRN on EXO1 to rescue rad50 MMS-sensitivity suggested that enhancement of endogenous EXO1 cleavage of DNA structural intermediates associated with stalled replication forks is responsible for genetic complementation. Previously, we showed that WRN interacts with hEXO1 in vivo and WRN stimulates EXO1 incision of nicked duplex and 5’ flap substrates [17]. While these substrates represent key intermediates of mismatch repair and Okazaki fragment processing, respectively, we wanted to investigate the effect of WRN on EXO1 processing of DNA structures associated with stalled replication forks. Demonstration that EXO1 is recruited to stalled replication forks in checkpoint defective rad53 mutants [48] and acts to counteract reversed fork accumulation by generating ssDNA intermediates [48] suggested that WRN might collaborate with EXO1 to process a stalled replication fork intermediate. On a replication fork structure, WRN stimulated EXO1 5’ incision of what would be the nascent lagging strand arm. In a model for EXO1-mediated processing of stalled forks, EXO1 resects the newly synthesized lagging strand, thereby resolving the sister chromatid junction and preventing reversal at collapsed replication forks [48].
An alternative strategy for EXO1 to act with WRN at stalled replication forks is presented when fork regression occurs. Accumulation of chicken foot structures through fork reversal produces abnormal replication intermediates that can be processed by recombination pathways leading to genomic instability in certain mutant backgrounds. WRN stimulated EXO1 5’ incision of the synthetic Holliday Junction, suggesting that the two proteins may collaborate to process chicken foot structures that form as a consequence of fork regression. Altogether, our results unmask a previously unappreciated role of EXO1 in processing stalled and regressed replication forks that form in DNA repair deficient cells. WRN expression in the rad50 background may help to insure appropriate processing of stalled or regressed replication fork structures.
How are these results consistent with characteristics of mutant WRN cells? WS cells display a reduced rate of repair, elevated apoptotic cell death and increased DNA strand breaks after replication arrest by hydroxyurea treatment or DNA damage induced in S phase [53]. Expression of a HJ resolvase rescues both the recombination defect and cell survival following DNA damage in WS cells [54], suggesting that inappropriate processing of stalled replication fork intermediates directly contributes to aberrant homologous recombination characteristic of WS cells. Moreover, suppression of RAD51-dependent recombination significantly improved survival of WS cells following DNA damage [54]. Replication defects in WS cell lines point to an underlying deficiency when fork progression is abnormal. Co-localization of WRN with arrested replication forks in response to hydroxyurea treatment [55] or DNA damage [31] is consistent with its role in response to replicational stress. Asymmetry of DNA replication fork progression in WS cells suggests that WRN acts to prevent collapse of forks or to resolve junctions at stalled forks and that loss of this capacity may be a contributory factor in premature ageing [56]. Collectively, our findings implicate a role of WRN helicase acting with EXO1 in a conserved pathway to process DNA structures associated with stalled or blocked replication forks.
The genetic results provide new insight to the molecular and cellular roles of WRN helicase in DNA metabolism. In conclusion, WRN helicase function is required in an EXO1-dependent pathway to maintain genomic stability after replicational stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Teresa M. Wilson (University of Maryland School of Medicine, Baltimore, MD) for purified recombinant human EXO1 and Dr. Robert M. Liskay (University of Oregon Health and Science University, Portland, OR) for the FLAG-epitope-tagged yEXO1 expression construct. We are grateful to Dr. Ian Hickson (University of Oxford, Oxford, UK) for the BLM cDNA plasmid pJK1. Yeast strains were kindly provided by Dr. L. Kevin Lewis (Texas State University, San Marcos, TX).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat. Rev. Mol. Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: Cancer and aging through the prism of human RecQ helicases. Mech. Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng WH, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, Shiloh Y, Bohr VA. WRN Is Required for ATM Activation and the S-Phase Checkpoint in Response to Interstrand Crosslink-induced DNA Double Strand Breaks. Mol. Biol. Cell. 2008;19:3923–3933. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma S, Sommers JA, Brosh RM., Jr In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication. Hum. Mol. Genet. 2004;13:2247–2261. doi: 10.1093/hmg/ddh234. [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal M, Brosh RM., Jr WRN helicase defective in the premature aging disorder Werner Syndrome genetically interacts with Topoisomerase 3 and restores the top3 slow growth phenotype of sgs1 top3. Aging. 2009;1:219–233. doi: 10.18632/aging.100020. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 11.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 12.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 13.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human Exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J. Biol. Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 17.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human Exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J. Biol. Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 18.Doherty KM, Sharma S, Uzdilla L, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr RECQ1 helicase interacts with human mismatch repair factors that regulate gentic recombination. J.Biol.Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- 19.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farah JA, Cromie GA, Smith GR. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9356–9361. doi: 10.1073/pnas.0902793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis LK, Kirchner JM, Resnick MA. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol. Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larionov V, Kouprina N, Nikolaishvili N, Resnick MA. Recombination during transformation as a source of chimeric mammalian artificial chromosomes in yeast (YACs) Nucleic Acids Res. 1994;22:4154–4162. doi: 10.1093/nar/22.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 27.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome pretein contains three structure specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 28.Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- 29.Tran PT, Erdeniz N, Dudley S, Liskay RM. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair. 2002;1:895–912. doi: 10.1016/s1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 30.McDowell HD, Carney JP, Wilson TM. Inhibition of the 5' to 3' exonuclease activity of hEXO1 by 8-oxoguanine. Environ. Mol. Mutagen. 2008;49:388–398. doi: 10.1002/em.20398. [DOI] [PubMed] [Google Scholar]
- 31.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branch-migrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper MP, Machwe A, Orren DK, Brosh RM, Jr, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 33.Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 34.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human Flap Endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, Seki M, Enomoto T, Goto M, Ellis NA, Ide T, Furuichi Y, Sugimoto M. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 36.Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S, Sommers JA, Brosh RM., Jr Processing of DNA replication and repair intermediates by the concerted action of RecQ helicases and Rad2 structure-specific nucleases. Protein Pept. Lett. 2008;15:89–102. doi: 10.2174/092986608783330369. [DOI] [PubMed] [Google Scholar]
- 38.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 39.Myung K, Kolodner RD. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4500–4507. doi: 10.1073/pnas.062702199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu JS, Feng H, Zeng W, Lin GX, Xi XG. Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18379–18384. doi: 10.1073/pnas.0509380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JW, Kusumoto R, Doherty KM, Lin GX, Zeng W, Cheng WH, von Kobbe C, Brosh RM, Jr, Hu JS, Bohr VA. Modulation of Werner syndrome protein function by a single mutation in the conserved RecQ domain. J. Biol. Chem. 2005;280:39627–39636. doi: 10.1074/jbc.M506112200. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von HA, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum. Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro E, Edland SD, Lee L, Ogburn CE, Deeb SS, Brown G, Panduro A, Riestra R, Tilvis R, Louhija J, Penttinen R, Erkkola R, Wang L, Martin GM, Oshima J. Polymorphisms at the Werner locus: II. 1074Leu/Phe, 1367Cys/Arg, longevity, and atherosclerosis. Am. J. Med. Genet. 2000;95:374–380. [PubMed] [Google Scholar]
- 45.Ye L, Miki T, Nakura J, Oshima J, Kamino K, Rakugi H, Ikegami H, Higaki J, Edland SD, Martin GM, Ogihara T. Association of a polymorphic variant of the Werner helicase gene with myocardial infarction in a Japanese population. Am. J. Med. Genet. 1997;68:494–498. doi: 10.1002/(sici)1096-8628(19970211)68:4<494::aid-ajmg30>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 46.Ogata N, Shiraki M, Hosoi T, Koshizuka Y, Nakamura K, Kawaguchi H. A polymorphic variant at the Werner helicase (WRN) gene is associated with bone density, but not spondylosis, in postmenopausal women. J. Bone Miner. Metab. 2001;19:296–301. doi: 10.1007/s007740170013. [DOI] [PubMed] [Google Scholar]
- 47.Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J. Biol. Chem. 2004;279:55499–55505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 48.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 49.Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 50.Wilson DM, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic. Acids. Res. 1998;26:3762–3768. doi: 10.1093/nar/26.16.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasko BM, Holland CL, Resnick MA, Lewis LK. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst) 2009;8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson K, Asawachaicharn N, Galloway DA, Grandori C. c-Myc accelerates S-Phase and requires WRN to avoid replication stress. PLoS. ONE. 2009;4:e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat JRJ. Homologous recombination resolution defect in Werner syndrome. Mol. Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Reports. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.