Abstract
Identification of de novo minisatellite mutations in the offspring of parents exposed to mutagenic agents offers a potentially sensitive measure of germ line genetic events induced by ionizing radiation and genotoxic chemicals. Germ line minisatellite mutations (GMM) are usually detected by hybridizing Southern blots of unamplified size-fractionated genomic DNA with minisatellite probes. However, this consumes a relatively large amount of DNA, requires several steps and may lack sensitivity. We have developed a polymerase chain reaction (PCR)-based GMM assay, which we applied to the hypermutable minisatellite, CEB1. Here, we compare the sensitivity and specificity of this assay with the conventional Southern hybridization method using DNA from 10 spouse pairs, one parent of each pair being a survivor of cancer in childhood, and their 20 offspring. We report that both methods have similar specificity but that the PCR method uses 250 times less DNA, has fewer steps and is better at detecting GMM with single repeats provided that specific guidelines for allele sizing are followed. The PCR GMM method is easier to apply to families where the amount of offspring DNA sample is limited.
Introduction
Although mammalian germ cells are highly sensitive to the mutagenic effects of environmental and therapeutic genotoxic agents such as chemicals and ionizing radiation (1,2), no human germ cell mutagen has yet been confirmed (1,3,4). Furthermore, radiotherapy for cancer in childhood does not seem to increase the risk of malformations in the offspring of irradiated children, though other genetic effects have not been ruled out (5,6). However, the potential health impact in humans of radiation- and genotoxin-induced germ line genetic effects is an important and contentious issue, notably because fixation of mutations in the germ line could have long-term health consequences such as birth defects, de novo genetic diseases and chromosomal abnormality syndromes in offspring (1,3,5,6). Failure to detect mutations in the offspring of parents exposed environmentally or therapeutically during the critical gametogenic time window may be due to low mutation frequency or to limitations of the molecular technology (1,7).
One molecular technology that may help to overcome the limitations of detecting the low frequency of germ line mutations in protein coding genes is the analysis of mutations in tandem DNA repeat loci known as minisatellites (8). In a number of studies, Dubrova et al. (9–12) reported a statistically significantly increased germ line mutation rate in certain hypervariable minisatellite loci, following parental exposure to ionizing radiation, though other studies have failed to replicate these findings (13–15). The advantage of the germ line minisatellite mutation (GMM) assay is that the de novo and induced mutation frequencies are ∼1000-fold higher than in structural genes, requiring much smaller populations to detect significant genotoxic effects (16,17). However, because GMM are usually detected as allele size expansions and contractions resulting from the addition or loss of repeat units, analysis has largely been confined to Southern blots of size-fractionated unamplified genomic DNA hybridized with minisatellite probes (9–12). This approach has two limitations. First, it requires a relatively large amount of DNA (5 μg) making it difficult to apply to families where blood volumes, as a source of DNA, especially from children, may be small. Second, the number of steps needed to resolve alleles makes the method time consuming. To overcome this, we recently developed a direct GMM analysis approach for the CEB1 locus, based on polymerase chain reaction (PCR) amplification, size fractionation by electrophoresis and direct staining of alleles (18). We applied the PCR-based GMM approach to a case–control comparison of GMM frequency in childhood leukaemia (18), but it was not clear whether the GMM detected by PCR were identical to those detected by Southern hybridization. Here, we compare the sensitivity and specificity of the Southern and PCR-based GMM detection methods using the same DNA samples from 10 spouse pairs and their 20 offspring.
Materials and methods
Genomic DNA samples
Blood samples were obtained from 10 spouse pairs and 20 offspring as part of a study investigating the impact of radiotherapy for childhood and young adult cancer on reproductive outcomes in cancer survivors [(19); www.gcct.org]. One of each of the spouse pairs is a survivor of cancer in childhood. Eight of the spouse pairs had two children, one had one child and one had three children. The 10 families are included in an ongoing study of the GMM rate in childhood and adolescent cancer survivors and their families (15). All cancer survivors had been treated with radiotherapy for cancer prior to the conception of their offspring. Genomic DNA samples were extracted using standard techniques (FlexiGene; QIAGEN, Crawley, West Sussex, UK). The 10 families included in this study were selected (by G.S.R.) to have a range of CEB1 allele and GMM sizes and included four families in which GMM had not been detected by Southern blotting. GMM analysis by PCR was undertaken (by M.C.) without prior knowledge of Southern GMM results. Replicate PCR analysis (by A.O.) was carried out on samples with known GMM.
GMM analysis by Southern blotting
GMM analysis was carried out by the Southern hybridization method described previously (15), with modifications. Five micrograms of genomic DNA was digested with the Alu I restriction enzyme (New England Biolabs, Hitchin, Hertfordshire, UK) and electrophoresed on 30 cm 0.8% agarose gels in 1× Tris-Borate-EDTA (TBE) buffer, containing 0.5 μg/ml ethidium bromide, for 18 h at 125 V. The size-fractionated DNA was denatured, neutralized and transferred to a nylon membrane (Magnacharge; Genetic Research Instrumentation, Braintree, Essex, UK) where it was fixed by ultraviolet (UV) cross linking. The CEB1 minisatellite probe was biotin labelled using BioPrime DNA labeling system (Invitrogen, Paisley, Scotland) and hybridized to immobilized sample DNA on Southern blots. Hybridization was detected using a KPL Detector AP Chemiluminescent Blotting Kit (Insight Biotechnology Limited, Wembley, Middlesex, UK). Each blot was scored visually by two independent assessors and digitally using Phoretix 1D software (Non-Linear Dynamics, Newcastle upon Tyne, UK) using a 1-kb ladder (Promega, Southampton, Hampshire, UK) for size reference across the well resolved 1- to 23-kb region. Criteria for identification of mutations were taken from previously published studies (9,13). A mutation was considered to be a band (allele) present in the offspring, but absent from both parents, and larger or smaller than the parental progenitor allele by at least one bandwidth. Any small or suspected mutations were run on a second gel for a longer time period to resolve the size difference between parental and offspring bands.
GMM analysis by PCR
GMM analysis by PCR was carried out as previously described (18) but using different primers and amplification conditions. Genomic DNA samples (1 μl/20 ng) were amplified using a single pair of modified CEB1 primers, CEB1-F-CB (22 bp) (CCT-TCT-CCC-TGT-AAC-CAG-TTA-C) and CEB1-R-CB (21 bp) (TGA-GAG-TCG-GCC-GTG-AAG-AAT) using a PCR Express Thermal cycler (Hybaid Limited, Ashford, Middlesex, UK). PCR mixtures contained CEB1-F-CB and CEBI-R-CB primers (2 μl and 0.5 μM each), 10× high-fidelity PCR buffer at a 1× final concentration (2 μl, Invitrogen), deoxynucleotide triphosphates (1 μl; Promega) at a final concentration of 0.5 mM, H2O (11 μl; Sigma–Aldrich), MgSO4 (0.8 μl, 50 mM) at a final concentration of 2 mM and 1 U of Platinum Taq DNA polymerase High Fidelity (0.2 μl; Invitrogen Ltd). A negative control consisting of PCR mix without DNA was included to detect contamination. A positive control consisting of 20 ng DNA from the human lymphoblastoid cell line, LV012, which has two CEB1 alleles (2.76 kb and 4.01 kb), was used to check for PCR amplification. PCRs were carried out in 0.2-ml thin-walled PCR tubes (STARLAB GMBH, Ahrensburg, Germany) under cycling conditions that included an initial denaturation step (2 min at 94°C), 10 cycles of amplification (94°C/10 s denaturation, 61°C/30 s annealing and 68°C/12 min elongation) and finally 20 cycles under the same conditions, except that the elongation step was extended by 20 s per cycle. PCRs were then incubated for a further 12 min at 68°C as a final elongation step. To check for optimum amplification, PCR products were electrophoresed on 15-cm agarose gels (0.8%; Combrex BioScience, Rockland, ME, USA) in 1× TBE buffer at 80 V for 4 h. Samples from the families and 1 kb plus DNA ladders (Invitrogen) were size fractionated on 40-cm agarose gels (0.8%) at 80 V for 24 h to obtain optimum allele separation and high resolution. Gels were stained using SYBR Gold nucleic acid stain (Invitrogen) in 1× TBE (1:16 000 dilution) buffer for 40 min and bands visualized by UV transillumination on a Bio-Rad Gel Doc 2000. Images were digitized in real-time and alleles sized by allowing for the gel retardation factor (Rf) across the gel and measuring the distance in millimeters between the base of the lower and upper 1-kb size markers closest to the allele being measured and then measuring the distance of the allele between the two markers to obtain a size estimate in base pairs. GMM were defined as expansions or contractions of at least 39-bp difference, or one repeat unit, in size from the parental progenitor allele (20).
Parentage analysis
To exclude non-parentage as a confounder of GMM, the AmpFlSTR COfiler PCR Amplification Kit (Applied Biosystems, Warrington, UK) was used to confirm kinship by amplifying six tetranucleotide short tandem repeat loci (D3S1358, D16S539, TH01, TPOX, CSF1PO and D7S820) plus a segment of the sex-specific amelogenin locus. Semi-automated analysis of PCR products was carried out on an ABI 310 genetic analyser, data files being analysed using Genotyper 2.X software (Applied Biosystems) to complete the DNA profiling. A mismatch between parent and offspring at two or more loci is considered to be non-paternity or non-maternity. None of the families included in the present study showed evidence of non-parentage.
Results
Since the detection of CEB1 GMM using Southern hybridization (15) requires an amount of DNA that can exclude offspring (such as those with cytopaenia) from whom a limited amount of DNA can be obtained, we developed a sensitive and direct PCR-based genotyping assay that requires much less DNA to visualize alleles and germ line mutations (18). However, the PCR primers that we described previously (18) gave variable results, probably due to the 5°C melting temperature (Tm) difference between the forward and reverse primers and the low Mg2+ concentration (∼0.9 mM) in the PCR amplification mixture. Accordingly, we redesigned and optimized a new pair of primers, CEB1-F-CB at positions 12913–12934 and CEB1-R-CB at positions 13386–13406 (see Materials and methods) in the CEB1 reference sequence (AF048727; National Center for Biotechnology Information nucleotide database: http://www.ncbi.nlm.nih.gov/nuccore/2935483). These were designed having regard to similar size (21–22 bp), guanine-cytosine content (forward: 50% and reverse: 52.4%) and Tm (forward: 54.8°C and reverse: 54.4°C), using sequences likely to have a reduced risk of hairpin formation. The new primers were optimized over a Tm range of 53–65°C, a final Mg2+ concentration range of 1–2.25 mM, a Taq polymerase concentration of 0.75 or 1 U/20 μl and 8 or 12 min extension. Addition of bovine serum albumin (0.01–0.1 μg/μl) and dimethyl sulphoxide (2.5–7.5% v/v) was found not to improve amplification, and they were excluded from the modified PCR. Using the optimized PCR protocol (see Materials and methods), CEB1-F-CB and CEB1-R-CB were tested for reliability, reproducibility, allele sizing and segregation and GMM detection using DNA from lymphoblastoid cell lines prepared from a three generation family (details to be published elsewhere).
Using the optimized PCR-based GMM detection assay, we compared the specificity and sensitivity of the conventional Southern hybridization method (15) with the PCR method. Since these two methods have not, as far as we are aware, previously been compared using the same samples, we tested 10 spouse pairs and a total of 20 offspring for CEB1 GMM. The spouse pairs and their offspring were selected based on prior results of the Southern hybridization assay obtained at the Westlakes Research Institute (15), from a larger series included in the Genetic Consequences of Cancer Treatment study (19). PCR-based genotyping was carried out in Manchester on samples coded by family identifier, initially without knowledge of GMM status by Southern hybridization but subsequently selected for replication analysis based on GMM detected by PCR.
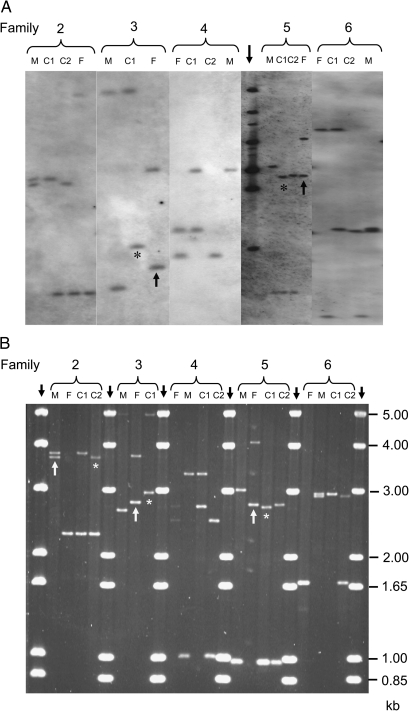
Data from the two assays were assembled into the allele sizes and GMM scores shown in Table I. Both methods were repeated on samples where alleles were expected but missing, if bands were faint and thus difficult to score or if germ line mutations required confirmation. Thus, Southern hybridization was perfomed once in three families (Families 2, 9 and 10), twice in two families (1, 3 and 4), three times in three families (5, 7 and 8) and four times in one family (6). For PCR-based genotyping, samples from Families 1–6 were amplified on two separate occasions and both run on short (15 cm) gels and once on a long (40 cm) gel. PCR products for families 7–10 were run only on long gels. PCRs on Families 1 (with a 54-bp paternal GMM), 7 and 8 (both with missing parental alleles which precluded offspring genotyping) were repeated. Figure 1 shows representative Southern and PCR images for families 2–6 and were compiled independently by the two laboratories. Family samples were run on separate gels using the Southern method, but on the same gel for the PCR method. GMM were detected by Southern hybridization (Figure 1A) in the offspring of two families: Child 1 of Family 3 and Child 1 of Family 5. By PCR, GMM were detected in three families: Child 2 of Family 2, Child 1 of Family 3 and Child 1 of Family 5 (Figure 1B).
Table I.
Comparison of Southern hybridization and PCR genotyping for the detection of CEB1 germ line minisatellite mutations in 10 spouse pairs and 20 offspring
| Family no. | Parent/child | Southern-based CEB1 genotyping |
PCR-based CEB1 genotyping |
||||||
| Allele 1a (kb) | Allele 2 (kb) | GMM status | GMMb size (bp) | Allele 1a (kb) | Allele 2 (kb) | GMM status | GMMb size (bp) | ||
| 1 | Father | 4.955 | 3.831 | 3.919 | 2.876 | ||||
| Mother | 5.044 | 4.006 | 4.125 | 2.941 | |||||
| Child 1 | 3.982 | 2.303 | Paternal contraction | −1528 | 2.941 | 2.165 | Paternal contraction | −711 | |
| Child 2 | 5.044 | 4.972 | No GMM detected | — | 4.125 | 3.973 | Paternal expansion | [+54]c | |
| 2 | Father | 8.200 | 2.629 | 6.526 | 2.357 | ||||
| Mother | 4.923 | 4.866 | 3.833 | 3.736 | |||||
| Child 1 | 4.920 | 2.636 | No GMM detected | — | 3.842 | 2.357 | No GMM detected | — | |
| Child 2 | 4.855 | 2.634 | No GMM detected | — | 3.779 | 2.357 | Maternal expansion | [+43] | |
| 3 | Father | 4.871 | 3.719 | 3.846 | 2.829 | ||||
| Mother | 5.970 | 3.431 | 5.100 | 2.714 | |||||
| Child 1 | 5.970 | 4.052 | Paternal expansion | +333 | 5.100 | 3.000 | Paternal expansion | +171 | |
| 4 | Father | 3.551 | 3.075 | 2.807 | 2.592 | ||||
| Mother | 4.526 | Not visualized | 3.421 | 1.042 | |||||
| Child 1 | 4.529 | 3.545 | No GMM detected | — | 3.421 | 2.786 | No GMM detected | — | |
| Child 2 | 3.069 | Not visualized | No GMM detected | — | 2.571 | 1.042 | No GMM detected | — | |
| 5 | Father | 5.023 | 3.676 | 4.171 | 2.824 | ||||
| Mother | 4.184 | 1.233 | 3.092 | 0.983 | |||||
| Child 1 | 3.592 | 1.231 | Paternal contraction | −84 | 2.785 | 0.983 | Paternal contraction | −39 | |
| Child 2 | 3.683 | 1.228 | No GMM detected | — | 2.829 | 0.983 | No GMM detected | — | |
| 6 | Father | 4.883 | 2.067 | 8.400 | 1.705 | ||||
| Mother | 4.030 | 3.970 | 3.000 | 2.957 | |||||
| Child 1 | 4.887 | 4.037 | No GMM detected | — | 8.400 | 3.000 | No GMM detected | — | |
| Child 2 | 3.978 | 2.066 | No GMM detected | — | 2.964 | 1.710 | No GMM detected | — | |
| 7 | Father | 8.632 | 2.378 | Not visualized | 2.118 | ||||
| Mother | 4.911 | 2.074 | 3.817 | 1.739 | |||||
| Child 1 | 8.756 | 2.074 | Paternal expansion | +124 | Not visualized | 1.747 | Not informative | ||
| Child 2 | 8.614 | 2.069 | No GMM detected | — | Not visualized | 1.739 | Not informative | ||
| 8 | Father | 8.440 | 5.024 | 6.454 | 4.071 | ||||
| Mother | 10.093 | Not visualized | Not visualized | 5.155 | |||||
| Child 1 | 10.093 | 5.040 | No GMM detected | — | Not visualized | 4.122 | Not informative | ||
| Child 2 | 10.096 | 8.456 | No GMM detected | — | Not visualized | 6.500 | Not informative | ||
| 9 | Father | 2.443 | 1.576 | 2.185 | 1.189 | ||||
| Mother | 8.206 | 4.269 | 6.286 | 3.167 | |||||
| Child 1 | 8.226 | 2.438 | No GMM detected | — | 6.286 | 2.185 | No GMM detected | — | |
| Child 2 | 8.414 | 2.762 | Paternal expansion | +319 | 6.426 | 2.396 | Paternal expansion | +211 | |
| Maternal expansion | +208 | Maternal expansion | +140 | ||||||
| Child 3 | 4.274 | 2.433 | No GMM detected | — | 3.161 | 2.185 | No GMM detected | — | |
| 10 | Father | 4.208 | 2.020 | 3.107 | 1.587 | ||||
| Mother | 4.666 | 3.676 | 3.555 | 2.435 | |||||
| Child 1 | 4.672 | 2.447 | Paternal expansion | +427 | 3.555 | 2.159 | Paternal expansion | +572 | |
| Child 2 | 4.080 | 3.689 | Paternal contraction | −128 | 2.971 | 2.435 | Paternal contraction | −136 | |
Parental GMM progenitor alleles marked in bold type.
GMM ± base pairs expansion or contraction.
Bracketed results indicate one repeat GMM not confirmed in replicated PCR.
Fig. 1.
(A and B) Representative gel images obtained for Families 2–6 using the (A) Southern and (B) PCR-based CEB1 genotyping assays. The two sets of images were assembled independently by the two laboratories, the Southern images from five separate assays and the PCR images from a single gel. Family members are denoted as: F, father; M, mother; C1, Child 1; C2, Child 2. The vertical black downward arrows show the position of 1-kb allele-sizing ladders; this only approximates to allele sizes in the Southern images of Families 2, 3, 4 and 6 because assays were carried out on separate occasions. The asterisk shows germ line mutant bands and the vertical black upward arrows , the appropriate parental progenitor alleles.
By Southern hybridization, eight GMM with an average size of 393 bp (∼10 repeats) were detected in 7 of the 20 offspring, seven being paternal and one being maternal (Table I). Five of the eight GMM were expansion mutations of which one was maternal. All three contraction GMM were paternal. In Family 9, Child 2 had both a paternal and a maternal GMM. In family 10, both children had a paternal GMM, one being derived from each of the father's alleles. By PCR, nine GMM in eight offspring were detected with an average size of 230 bp (∼5.75 repeats) of which eight (four paternal expansions, three paternal contractions and one maternal expansion) had the same polarity (expansion/contraction) as those identified by Southern hybridization. One paternal expansion GMM detected by Southern hybridization in Child 1 of Family 7 was not detected by PCR because the paternal progenitor allele could not be resolved. Two GMM, each approximately one repeat unit in size and identified by PCR, were not detected by Southern hybridization. One was a 54-bp paternal expansion GMM in Child 2 of Family 1 and the other was a 43-bp maternal expansion GMM in Child 2 of Family 2. A third PCR-detected GMM, a 39-bp contraction mutation in Child 1 of Family 5, was also detected by Southern hybridization.
By Southern hybridization, the mean paternal allele size was 4.671 kb and the mean maternal allele size was 4.053 kb, with one non-visualized paternal and two non-visualized maternal alleles. By PCR, the average paternal allele size was 3.451 kb and the average maternal allele size was 3.321 kb, with one non-visualized paternal and one maternal allele. This represents size differences between paternal and maternal alleles detected by Southern hybridization and PCR of 602 bp and 1350 bp, respectively. In general, parental allele sizes were smaller by PCR than Southern hybridization, the exception being paternal Allele 1 in Family 6.
Three of the GMM detected by PCR involved one repeat unit (Table I). Since this is at the limit of measurement sensitivity, we carried out a replication study to determine whether GMM detection at this limit was reproducible. The first experiment involved DNA from Family 1 with a one repeat paternal expansion in Child 2. DNA samples from each family member were amplified in 10 separate reactions and amplified samples run in family groups on three separate gels. Table II shows the results of allele size measurements on the 10 replicated Family 1 samples, with the mean allele sizes for each gel, and the cumulative mean for the 10 replicates. Two points emerge from these results: (i) sample replication both within and between gels gave very consistent allele size results and (ii) while the larger of the two GMM in Family 1 was confirmed (Table I), the smaller one-repeat GMM was not. This prompted us to test the two other families with one-repeat GMM (Families 2 and 5, Table I). This time, PCRs were replicated three times and tested on the same gel. As Table II shows, the one repeat maternal expansion GMM in Child 2 of Family 2 was not confirmed, whereas the one repeat paternal contraction GMM in Child 1 of Family 5 was confirmed and is in agreement with the Southern hybridization results. In light of these results, we re-examined the method of PCR allele sizing and concluded that the measurement of band sizes on 40-cm gels using 12-cm photographic images from a thermal printer that were used to generate the data in Table I can lead to false positives. To overcome this, PCR images from 40-cm gels were saved as TIFF files, imported into Microsoft Word and printed as 24-cm images using a laser printer. Alleles were sized on the printed image as described in the Materials and methods by allowing for the Rf across the gel.
Table II.
Results of replicate PCR-based CEB1 genotyping of three families
| Allele no. | Family 1a |
||||||||||||||||
| Gel no. 1 |
Gel no. 2 |
Gel no. 3 |
|||||||||||||||
| Replicate no.b |
Replicate no. |
Replicate no. |
|||||||||||||||
| 1 | 2 | 3 | Mean | SD | 4 | 5 | 6 | Mean | SD | 7 | 8 | 9 | 10 | Mean | SD | ||
| Father | 1 | 3.974 | 3.974 | 3.974 | 3.974 | 0.000 | 3.950 | 3.950 | 3.976 | 3.959 | 0.015 | 3.952 | 3.952 | 3.952 | 3.952 | 3.952 | 0.000 |
| 2 | 2.904 | 2.904 | 2.911 | 2.906 | 0.004 | 2.897 | 2.897 | 2.915 | 2.903 | 0.010 | 2.903 | 2.903 | 2.903 | 2.903 | 2.903 | 0.000 | |
| Mother | 1 | 4.143 | 4.143 | 4.143 | 4.143 | 0.000 | 4.172 | 4.172 | 4.172 | 4.172 | 0.000 | 4.133 | 4.133 | 4.133 | 4.133 | 4.133 | 0.000 |
| 2 | 2.964 | 2.964 | 2.964 | 2.964 | 0.000 | 2.966 | 2.966 | 2.983 | 2.972 | 0.010 | 2.984 | 2.984 | 2.968 | 2.984 | 2.980 | 0.008 | |
| Child 1 | 1 | 2.964 | 2.964 | 2.964 | 2.964 | 0.000 | 2.966 | 2.966 | 2.983 | 2.972 | 0.010 | 2.984 | 2.984 | 2.968 | 2.984 | 2.980 | 0.008 |
| 2 | 2.179 | 2.179 | 2.179 | 2.179 | 0.000 | 2.172 | 2.172 | 2.172 | 2.172 | 0.000 | 2.177 | 2.177 | 2.177 | 2.177 | 2.177 | 0.000 | |
| GMM (bp)b | −725 | −725 | −732 | −727.3 | 4.041 | −725 | −725 | −743 | −731 | 10.392 | −726 | −726 | −726 | −726 | −726 | 0.000 | |
| Child 2 | 1 | 4.143 | 4.143 | 4.143 | 4.143 | 0.000 | 4.172 | 4.172 | 4.172 | 4.172 | 0.000 | 4.133 | 4.133 | 4.133 | 4.133 | 4.133 | 0.000 |
| 2 | 3.974 | 3.989 | 3.974 | 3.979 | 0.009 | 3.950 | 3.950 | 3.976 | 3.959 | 0.015 | 3.952 | 3.952 | 3.952 | 3.952 | 3.952 | 0.000 | |
| GMM (bp) | — | — | — | — | — | — | — | — | — | — | |||||||
| Allele no. | Family 1 |
Family 2a |
Family 5a |
||||||||||||||
| Mean of gel no. |
Overall (n = 10) |
Replicate no. |
Replicate no. |
||||||||||||||
| 1 | 2 | 3 | Mean | SD | 1 | 2 | 3 | Mean | SD | 1 | 2 | 3 | Mean | SD | |||
| Father | 1 | 3.974 | 3.959 | 3.952 | 3.961 | 0.012 | NA | 6.500 | 6.500 | NA | 0.000 | 4.214 | 4.214 | 4.214 | 4.214 | 0.000 | |
| 2 | 2.906 | 2.903 | 2.903 | 2.904 | 0.005 | 2.360 | 2.360 | 2.360 | 2.360 | 0.000 | 2.877 | 2.877 | 2.877 | 2.877 | 0.000 | ||
| Mother | 1 | 4.143 | 4.172 | 4.133 | 4.148 | 0.017 | 3.882 | 3.882 | 3.882 | 3.882 | 0.000 | 3.103 | 3.103 | 3.103 | 3.103 | 0.000 | |
| 2 | 2.964 | 2.972 | 2.980 | 2.973 | 0.010 | 3.794 | 3.794 | 3.794 | 3.794 | 0.000 | 0.992 | 0.992 | 0.992 | 0.992 | 0.000 | ||
| Child 1 | 1 | 2.964 | 2.972 | 2.980 | 2.973 | 0.010 | 3.882 | 3.882 | 3.882 | 3.882 | 0.000 | 2.825 | 2.825 | 2.825 | 2.825 | 0.000 | |
| 2 | 2.179 | 2.172 | 2.177 | 2.176 | 0.003 | 2.360 | 2.360 | 2.360 | 2.360 | 0.000 | 0.992 | 0.992 | 0.992 | 0.992 | 0.000 | ||
| GMM (bp) | −727.3 | −731 | −726 | −727.9 | 5.705 | −52 | −52 | −52 | −52 | 0.000 | |||||||
| Child 2 | 1 | 4.143 | 4.172 | 4.133 | 4.148 | 0.017 | 3.794 | 3.794 | 3.794 | 3.794 | 0.000 | 2.877 | 2.877 | 2.877 | 2.877 | 0.000 | |
| 2 | 3.979 | 3.959 | 3.952 | 3.962 | 0.015 | 2.360 | 2.360 | 2.360 | 2.360 | 0.000 | 0.992 | 0.992 | 0.992 | 0.992 | 0.000 | ||
| GMM (bp) | — | — | — | — | — | — | — | — | |||||||||
NA, no amplification.
Families numbered as in Table I. Numbers in bold type are parental progenitor and mutated child alleles. Numbers in bold italics are parent and child alleles originally thought to have been mutated (see Table I).
Replicates are PCRs on different DNA samples. Allele sizes are in kilobase pairs; germ line mutation sizes are in base pairs. Dashes (—) indicate mutation shown in Table I, but not detected in replicate.
Discussion
GMM analysis by Southern hybridization of unamplified DNA using minisatellite probes is currently the method of choice for analysing sub-chromosomal mutations following germ cell exposure to radiation and genotoxins (9–12). The high frequency of spontaneous GMM and the increased rate of induced GMM makes it possible to carry out epidemiological studies of the effects of radiation and genotoxins using family series of modest size to achieve sufficient statistical power. However, this method requires a relatively large amount of DNA per sample and may preclude the use of DNA from buccal cells or small sample volumes from cytopaenic or young patients.
To overcome the sample amount limitation, we devised a direct PCR-based GMM detection method, which we used to compare parental CEB1 GMM frequencies in a case–control study of childhood leukaemia (18). Since the PCR method requires only 20-ng DNA per sample to visualize CEB1 alleles and GMM, it is some 250 times more sensitive than Southern hybridization. In our previous case–control study of childhood leukaemia (18), DNA samples from buccal cells produced satisfactory genotypes (18). However, exacting Mg2+ requirements and differences in primer Tm values made it difficult to obtain consistent amplification, necessitating frequent replication of PCRs (18). In the present study, we addressed this problem using redesigned CEB1 primers, which were carefully optimized and exhaustively tested for reproducibility using DNA from a three generation leukaemia family until they gave consistent genotyping results. Nonetheless, even though the PCR assay would appear to be much more sensitive than the Southern method, the question whether PCR genotyping has the same GMM detection specificity as Southern hybridization remained. In the present study, we demonstrate, albeit in a small series of families, that the two methods have virtually the same specificity, while the PCR method with qualifications that we discuss below is much more sensitive.
The Southern hybridization and PCR GMM assays differ in respect of the size of the alleles detected. In the small series of families presented here, we found that CEB1 alleles >8.5 kb were difficult to amplify by PCR, with the result that one GMM detected by Southern hybridization (in Child 1 of Family 7, Table I) could not be detected by PCR. However, of the 40 CEB1 alleles in 10 spouse pairs, only two (5%) were not resolved, with the result that two families were not informative for CEB1 GMM. In their study of the germ line minisatellite mutation rate in the parents of children exposed to contamination from the Chernobyl nuclear power station, Dubrova et al. (9) resolved CEB1 alleles over a 3.5–22 kb range using Southern hybridization, but their CEB1 allele size distribution profile (9) shows that only a very small minority of alleles are >8.0 kb. Kiuru (20), also using Southern hybridization, quotes a CEB1 allele size range of 0.5–12 kb. Dubrova et al. (10) report a mean parental control CEB1 allele size of 3.354 kb using the Southern method, which compares with a mean parental allele size of 4.362 kb by the Southern and 3.386 kb by the PCR method in the present study. Our PCR amplification results using the redesigned primers presented here suggest that we can detect an allele size range from 0.98 to 8.5 kb, which is well within maximum frequency of CEB1 allele sizes detected by Dubrova et al. (9). The problem of resolving small minisatellite alleles by Southern hybridization and their detection by PCR has been reported before (21,22). It is possible that CEB1 alleles >8.5 kb could be amplified if fewer PCR cycles were used. However, in preliminary experiments, PCR product concentrations after 26 cycles are too low to detect by visible SYBR—Gold staining though they can be detected by Southern hybridization, suggesting that this approach could be used in families with unresolved alleles.
Our results suggest that PCR can detect GMM as small as one repeat (39 bp) but that small differences in the migration rate of the same parent and child alleles can lead to false positives. Based on the results of the replication studies, we suggest that one repeat GMM should be scored by repeating PCRs and using the modified allele-sizing method that we describe in the Results.
One advantage of Southern hybridization over PCR is that GMM at several loci can be screened by stripping and re-hybridizing blots with different probes. Southern analysis of up to eight loci also allows sample identity to be confirmed, thus dispensing with separate tests of non-parentage (15). However, automated sample replication makes it possible to amplify several minisatellites in parallel. With further technical refinements, using dye-labelled primers, and a more sensitive allele detection system, it may be possible to multiplex the PCR assay so that GMM in several loci can be detected simultaneously.
In summary, our results suggest that Southern hybridization and PCR-based GMM analysis have similar specificities, but that PCR is more sensitive than Southern hybridization in requiring much less DNA. We suggest that GMM detected by PCR involving single repeats are checked by replicate PCRs, using the modified allele-sizing approach described in the Results. In view of the problem of resolving alleles >8.5 kb by PCR, it may be necessary in future to devise a detection method based on Southern hybridization of sub-visible PCR products so that the complete GMM spectrum can be evaluated as a dosimeter of environmental and therapeutic germ line genotoxic effects.
Funding
UK Department of Health Radiation Protection Research Programme (contract no. RRX110) to G.M.T.; National Institutes of Health, USA (RO1 CA 104666) through Vanderbilt Medical Centre, USA.
Acknowledgments
We thank Laura Gordon for secretarial support.
Conflict of interest statement: None declared.
References
- 1.Wyrobek AJ, Mulvihill JJ, Wassom JS, et al. Assessing human germ-cell mutagenesis in the post-genome era: a celebration of the legacy of William (Bill) Lawson Russell. Environ. Mol. Mutagen. 2007;48:71–95. doi: 10.1002/em.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sankaranaryanan K, Wassom JS. Reflections on the impact of advances in the assessment of genetic risks of exposure to ionizing radiation on international radiation protection recommendations between the mid-1950s and the present. Mutat. Res. 2008;658:1–27. doi: 10.1016/j.mrrev.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Shelby MD. Human germ cell mutagens. Environ. Mol. Mutagen. 1994;23(Suppl. 24):30–34. doi: 10.1002/em.2850230609. [DOI] [PubMed] [Google Scholar]
- 4.Olshan AF. Lessons learned from epidemiologic studies of environmental exposure and genetic disease. Environ. Mol. Mutagen. 1995;25(Suppl. 26):74–80. doi: 10.1002/em.2850250611. [DOI] [PubMed] [Google Scholar]
- 5.Winther JF, Boice JD, Jr., Frederiksen K, Bautz A, Mulvihil JJ, Stovall M, Olsen JH. Radiotherapy for childhood cancer and risk of congenital malformations in offspring: a population-based cohort study. Clin. Genet. 2009;75:50–56. doi: 10.1111/j.1399-0004.2008.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winther JF, Boice JD, Jr., Mulvihill JJ, Stovall M, Frederiksen K, Tawn EJ, Olsen JH. Chromosomal abnormalities among offspring of childhood-cancer survivors in Denmark: a population-based study. Am. J. Hum. Genet. 2004;74:1282–1285. doi: 10.1086/421473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elespuru RK, Sankaranarayanan K. New approaches to assessing the effects of mutagenic agents on the integrity of the human genome. Mutat. Res. 2007;616:83–89. doi: 10.1016/j.mrfmmm.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Bouffler SD, Bridges BA, Cooper DN, Dubrova Y, McMillan TJ, Thacker J, Wright EG, Waters R. Assessing radiation-associated mutational risk to the germline: repetitive DNA sequences as mutational targets and biomarkers. Radiat. Res. 2006;165:249–268. doi: 10.1667/rr3506.1. [DOI] [PubMed] [Google Scholar]
- 9.Dubrova YE, Nesterov VN, Krouchinsky NG, Ostapenko VA, Neumann R, Neil DL, Jeffreys AJ. Human minisatellite mutation rate after the Chernobyl accident. Nature. 1996;380:683–686. doi: 10.1038/380683a0. [DOI] [PubMed] [Google Scholar]
- 10.Dubrova YE, Nesterov VN, Krouchinsky NG, Ostapenko VA, Vergnaud G, Giraudeau F, Buard J, Jeffreys AJ. Further evidence for elevated human minisatellite mutation rate in Belarus eight years after the Chernobyl accident. Mutat. Res. 1997;381:267–278. doi: 10.1016/s0027-5107(97)00212-1. [DOI] [PubMed] [Google Scholar]
- 11.Dubrova YE, Grant G, Chumak AA, Stezhka VA, Karakasian AN. Elevated minisatellite mutation rate in the post-Chernobyl families from Ukraine. Am. J. Hum. Genet. 2002;71:801–809. doi: 10.1086/342729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrova YE, Bersimbaev RI, Djansugurova LB, Tankimanova MK, Mamyrbaeva ZZ, Mustonen R, Lindholm C, Hulten M, Salomaa S. Nuclear weapons tests and human germline mutation rate. Science. 2002;295:1037. doi: 10.1126/science.1068102. [DOI] [PubMed] [Google Scholar]
- 13.Kiuru A, Auvinen A, Luokkamaki M, et al. Hereditary minisatellite mutations among the offspring of Estonian Chernobyl cleanup workers. Radiat. Res. 2003;159:651–655. doi: 10.1667/0033-7587(2003)159[0651:hmmato]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Kodaira M, Izumi S, Takahashi N, Nakamura N. No evidence of radiation effect on mutation rates at hypervariable minisatellite loci in the germ cells of atomic bomb survivors. Radiat. Res. 2004;162:350–356. doi: 10.1667/rr3243. [DOI] [PubMed] [Google Scholar]
- 15.Rees GS, Trikic MZ, Winther JF, et al. A pilot study examining germline minisatellite mutations in the offspring of Danish childhood and adolescent cancer survivors treated with radiotherapy. Int. J. Radiat. Biol. 2006;82:153–160. doi: 10.1080/09553000600640538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bois P, Jeffreys AJ. Minisatellite instability and germline mutation. Cell. Mol. Life Sci. 1999;55:1636–1648. doi: 10.1007/s000180050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubrova YE. Long-term genetic effects of radiation exposure. Mutat. Res. 2003;544:433–439. doi: 10.1016/j.mrrev.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Davies BG, Hussain A, Ring SM, Birch JM, Eden TO, Reeves M, Dubrova YE, Taylor GM. New germline mutations in the hypervariable minisatellite CEB1 in the parents of children with leukaemia. Br. J. Cancer. 2007;96:1265–1271. doi: 10.1038/sj.bjc.6603706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boice JD, Jr., Tawn EJ, Winther JF, et al. Genetic effects of radiotherapy for childhood cancer. Health Physics. 2003;85:65–80. doi: 10.1097/00004032-200307000-00013. [DOI] [PubMed] [Google Scholar]
- 20.Kiuru A. Molecular biology methods in assessing radiation-induced hereditary risk in humans. 2004 Thesis. Department of Biosciences, Division of Genetics, University of Helsinki, Finland. [Google Scholar]
- 21.Armour JAL, Crosier M, Jeffreys AJ. Human minisatellite alleles detectable only after PCR amplification. Genomics. 1992;12:116–124. doi: 10.1016/0888-7543(92)90413-m. [DOI] [PubMed] [Google Scholar]
- 22.Alonso S, Castro A, Fernandez-Fernandez I, de Pancorbo MM. Short alleles revealed by PCR demonstrate no heterozygote deficiency at minisatellite lcoi D1S7, D7S21, and D12S11. Am. J. Hum. Genet. 1997;60:417–425. [PMC free article] [PubMed] [Google Scholar]