Abstract
DNA–protein cross-links (DPCs) are caused by a large number of human carcinogens and anti-cancer drugs. However, cellular processes involved in decreasing a burden of these genotoxic lesions remain poorly understood. Here, we examined the impact of nucleotide excision repair (NER), which is a principal repair pathway for bulky DNA adducts, and the main cellular reducers on removal of chromium(VI)-induced DPC. We found that standard and ascorbate-restored cultures of isogenic XPA-null (NER deficient) and XPA-complemented human fibroblasts had very similar repair of Cr–DPC (60–65% average DPC removal after 24 h). However, XPA absence caused depletion of G1 and accumulation of G2 cells at low Cr(VI) doses, suggesting that Cr–DPC were not a significant cause of cell cycle perturbations. Interestingly, although pro-oxidant metabolism of Cr(VI) in glutathione-depleted cells generated significantly fewer DPC, they were repair resistant irrespective of the NER status of cells. Inhibition of proteasome activity by MG132 abolished DPC repair in both XPA-null and XPA-complemented cells. XPA loss caused two to three times higher initial DPC formation, demonstrating the importance of NER in removal of the precursor lesions. Our results indicate that human NER is not involved in removal of Cr–DPC containing non-histone proteins but it acts as a defence mechanism against these large lesions by preventing their formation. Therefore, individual differences in NER activity are expected to alter sensitivity but not persistence of DPC as a biomarker of hexavalent Cr.
Introduction
Reactive by-products of cellular metabolism and exogenous carcinogens cause damage to both proteins and DNA. These damages are usually viewed as separate forms of cellular injury, such as proteotoxicity and genotoxicity. However, there are a large number of toxic agents that induce lesions by covalently linking proteins and DNA to form DNA–protein cross-links (DPC). Carcinogenic metals, aldehydes and platinum-based anti-cancer drugs are examples of DPC inducers (1–4). In general, DPC are expected to be formed by all bifunctional chemicals, which include several major anti-cancer drugs commonly described as DNA cross-linkers. Despite their early discovery, DPC remain arguably the most poorly understood class of DNA damage with respect to their biological properties and repair mechanisms. The determination of the mechanisms by which DPC are repaired can also help elucidate their toxicological significance through the use of genetic approaches to dissect the relative roles of various forms of DNA damage, which unavoidably arise in cells treated with bifunctional carcinogens and drugs. Studies with prokaryotic UvrABC nuclease in vitro and various Escherichia coli mutants showed that bacterial nucleotide excision repair (NER) was capable of DPC excision, albeit with rapidly declining activity for cross-links with larger proteins (5,6). In vitro mammalian NER showed the ability to excise small oxanine-DPC but not other types of cross-links (7–9). Involvement of specific repair processes for chromosomal DPC has so far been examined only for formaldehyde-induced DNA–histone cross-links. Time-course studies with formaldehyde-treated human (10,11) and yeast (12) cells showed no effect of NER on DPC removal although NER-deficient mutants displayed lower survival. While it is tempting to view various DPC as members of a single class of DNA lesions, cleared by identical repair mechanisms and causing similar genotoxic consequences, even a currently limited set of studies on DPC mutagenicity (13–15) have already revealed a complex situation where the site of cross-linking and the type of cross-linked protein can have a major effect.
Chromium(VI), a widespread human carcinogen (16), is the most potent inducer of DPC among toxic metals. The ability of Cr(VI) to form DPC has been demonstrated in various cultured cells and in vivo (1,17–19). Cr-induced DPC have been implicated in repression of inducible gene expression (20) and are believed to cause gross chromosomal abnormalities (21). Formation of DPC and other forms of DNA damage by Cr(VI) occurs as a result of its reduction to Cr(III) by cellular ascorbate (Asc) and small thiols (22). DPC isolated from Cr(VI)-treated mammalian cells contained non-histone proteins, most of which had larger molecular weights than histones (1). Mammalian cells are proficient in eliminating DPC after low but not very high doses of Cr(VI) (23). DPC measurements have been used for the assessment of human exposure to toxic forms of Cr (19,24,25), but the value of these biomonitoring findings is limited in part due to poorly understood responsiveness and persistence of DPC as a biomarker.
In this work, we investigated factors influencing repair of chromosomal DPC induced by Cr(VI). We were particularly interested in examining removal of cellular Cr–DPC by NER, as this repair process is capable of excising a wide range of bulky and helix-distorting DNA lesions (26), including Cr–DNA adducts with and without cross-linked amino acids (27,28). Chromosomal DPC containing non-histone proteins have not yet been investigated in an isogenic human system for NER-dependent repair. Because of their formation mechanism (29), Cr–DPC is also a well-suited model for testing the role of NER in preventing DPC formation.
Materials and methods
Materials
L-ascorbic acid (Asc, 99.9% pure), bovine serum albumin (99% pure), L-buthionine-(S,R)-sulfoximine (BSO, >97% pure), glutathione (GSH, >98% pure), sodium dodecyl sulphate (SDS, 95% pure), dehydro-L-(+)-ascorbic acid dimer (DHA), proteinase K, ascorbate oxidase and all salts, solvents and electrophoresis reagents were from Sigma (St Louis, MO, USA). K2CrO4 (ACS reagent, >99% pure) was from Aldrich (Milwaukee, WI). MG132 (>98% pure) was supplied by Calbiochem (Gibbstown, NJ, USA). Tissue culture media were from Gibco BRL (Gaithersburg, MD, USA). Foetal bovine serum was purchased from Gemini Bio-Products. PicoGreen and 1,2-diamino-4,5-dimethoxybenzene dihydrochloride dyes were obtained from Molecular Probes (Eugene, OR, USA).
Cells and treatment conditions
SV40-immortalized GM04312C (XP-A) and GM15876A (XPA+) human fibroblasts were purchased from Coriell Repository (Camden, NJ, USA). XP-A cells were derived from a female donor with severe ultraviolet (UV) sensitivity due to homozygous G-to-C transversion in intron 3 of the XPA gene, resulting in the loss of the canonical 3′-splice site and XPA expression. XPA+ line was created by stable transfection of XP-A cells with the full-length cDNA of the XPA gene expressed under control of β-actin promoter (30). Both cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% foetal bovine serum and 1% penicillin–streptomycin. Cells were grown at 37°C with 95% air and 5% CO2 concentrations. To avoid a potential impact of different batches and age of serum on treatments with Cr(VI), all exposures were done in serum-free media for 3 h. Cr(VI) doses were selected based on their ability to produce sufficiently high initial levels of DPC for repair studies during 24 h. To deplete cellular GSH, cells were pre-incubated with 0.1 mM BSO for 24 h prior to Cr(VI) exposures. Cellular Asc levels were restored by incubations with 1 mM DHA for 90 min in a Krebs–N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer supplemented with 0.5 mM glucose (31). In the experiments examining the role of proteasome activity, 0.5 μM MG132 was added immediately after Cr(VI) treatments.
Cell survival
For assessment of clonogenic survival, cells were seeded onto 100-mm dishes 1 day prior to treatments with UVC (UltraLum irradiator, model UVC-18, λmax = 254 nm). Three dishes were used for each dose. Ten to fourteen days after exposure, colonies were fixed with methanol and stained with the Giemsa solution. Cytotoxicity of Cr(VI) treatments was determined using the CyQuant assay (Invitrogen) according to the manufacturer's instructions.
Western blotting
Cells were collected by scraping, washed twice with cold phosphate-buffered saline (PBS) and resuspended in a lysis buffer containing 50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 1% NP40, 0.1% SDS, 5 mM ethylenediaminetetraacetic acid, 2 mM Na3VO4, 10 mM Na2P2O7, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail. Lysates were incubated on ice for 10 min and then cell debris was spun down at 10 000g for 5 min at 4°C. Proteins were separated by SDS–polyacrylamide gel electrophoresis and electrotransferred to an immunoblot polyvinylidene difluoride membrane (Bio-Rad). Protein bands were visualized using horseradish peroxidase-conjugated secondary antibodies (Upstate) and an enhanced chemiluminescence kit (Amersham).
DPC measurements
The number of DPC was measured by a K/SDS precipitation assay (32), as modified previously (10). This frequently used method is based on the selective precipitation of protein-linked DNA fragments from cellular SDS lysates by the addition of K+ ions. The percentage of the total chromosomal DNA that is precipitated in the presence of K+/SDS serves as a quantitative measure of DNA–protein cross-linking. A combination of the size of DNA fragments and the percentage of cross-linked DNA permits calculations of the number of DPC per basepair DNA (10). Cross-links per 108 bp = CF × 108/L, where CF = −ln (1 − cross-linked fraction) and represents the number of DPC per DNA fragment. The cross-linked fraction is the ratio of SDS-precipitable DNA to total DNA. L is the weighted average length of DNA fragments produced during shearing in the K/SDS precipitation assay. L is measured in DNA base pairs. The size of the DNA fragments produced during the precipitation of DPC was measured by agarose gel electrophoresis.
Determination of cellular Asc
Asc was measured by high-performance liquid chromatography (HPLC) using a sensitive fluorescent detection of its specific conjugate with 1,2-diamino-4,5-dimethoxybenzene dihydrochroride (31). Cells were collected by trypsinization, washed three times with cold PBS (1100g for 5 min at 4°C) and resuspended in a solution containing 50 mM methanesulfonic acid and 5 mM diethylenetriaminepentaacetic acid. Samples were subjected to two cycles of freezing (−80°C) and thawing (37°C), and Asc was recovered in the supernatant after centrifugation at 12 000g for 10 min at 4°C. Pellets were resuspended in a 1% SDS and 50 mM NaOH solution for determination of protein concentrations. Derivatization reactions contained 10 μl of cell extracts and 90 μl of a dye solution containing 0.2 U/ml ascorbate oxidase, 50 mM Na acetate (pH 6.2) and 0.5 mM 1,2-diamino-4,5-dimethoxybenzene and were incubated for 4 h at room temperature in the dark. The Asc-derived conjugate was separated on an Ultrasphere ODS column (5 μm, 250 × 4.6 mm) using isocratic elution with 75% 50 mM phosphoric acid (pH 2.0) and 25% acetonitrile and detected by its characteristic fluorescence at 458 nm after excitation with 371 nm light (Shimadzu RF-10AxL detector).
Measurements of cellular GSH
GSH concentrations were determined by HPLC with fluorescent detection as described previously (33). In brief, cells were collected by trypsinization, washed three times with cold PBS and resuspended in a solution containing 40 mM methanesulfonic acid and 1 mM diethylenetriaminepentaacetic acid. Cells were lysed by two cycles of freezing/thawing followed by centrifugation at 12 000g for 10 min at 4°C. The GSH-containing supernatants were reacted with 2 mM monobromobimane and the fluorescent GSH–monobromobimane conjugate was detected using Shimadzu LC-10ADvp HPLC equipped with Ultrasphere ODS column (5 μm, 250 × 4.6 mm) and RF-10AxL fluorescence detector.
Flow cytometry
Cells were collected by trypsinization, washed twice with cold PBS and resuspended in 50 μl of cold deionized water. Fixation/permeabilization was performed by addition of 200 μl of cold 70% EtOH followed by incubation on ice for 20 min. Cells were spun down and washed twice with 200 μl of cold PBS with 3% foetal bovine serum. Finally, the samples were resuspended in a solution containing 0.45 mg/ml propidium iodide and 0.5 mg/ml ribonuclease A. After 40 min incubation at 37°C, cells were analyzed on a Becton Dickinson FACSCaliber using Cell Quest software.
Cellular uptake of Cr(VI)
Cellular Cr concentrations were measured by a nitric acid extraction procedure described in detail previously (34). Cell monolayers were washed twice with warm PBS and collected by trypsinization. After additional two washes with cold PBS, cells were resuspended in 5% nitric acid. Samples were frozen at −80°C, heated at 50°C for 1 h and then placed on ice for 30 min. Supernatants obtained after centrifugation at 10 000g for 10 min at 4°C were diluted 2.5 times with deionized water and used for Cr measurements. The pellet was dissolved in 0.5 mM NaOH at 37°C for 30 min and used for protein measurements. The amount of Cr in the supernatants was determined by graphite furnace atomic absorption spectroscopy employing Zeeman background correction (Perkin-Elmer GF-AAS, model 41022L).
Measurements of Cr–DNA adducts
Cells were washed with PBS, collected by trypsinization and then resuspended in 50 μl of 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0) followed by addition of 0.5 ml of a solution containing 25 mM MOPS (pH 7.0), 0.5% Triton X-100 and 0.1 mg/ml RNase A (35). After 30-min incubation at 37°C, SDS was added to a 0.5% final concentration; samples were sheared five times with a 25G needle and digested with 1 mg/ml proteinase K (60 min, 37°C). DNA was extracted twice with phenol, once with chloroform and precipitated overnight at 4°C with two volumes of cold ethanol. DNA pellets were washed twice with cold 70% ethanol, air-dried and dissolved in deionized water overnight. DNA-bound Cr was measured by graphite furnace atomic absorption spectroscopy.
Statistical analysis
Statistical significance was evaluated using the Student's t-test. Differences with P-values <0.05 were considered to be statistically significant.
Results
Cellular responses in XPA-null cells
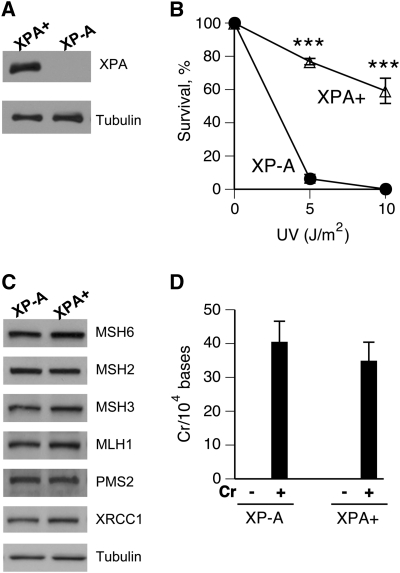
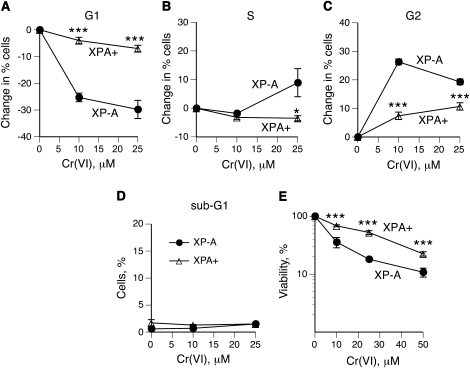
We chose to examine the role of human NER in repair of Cr(VI)-induced DPC using a pair of isogenic human fibroblasts with (XPA+) and without (XP-A) XPA expression (30). XPA is an absolutely essential component for both global genome and transcription-coupled NER branches (26). Unlike many other NER factors, XPA is not known to participate in transcription or other DNA repair processes, such as base excision or recombination repair. Expression of XPA protein in the XPA+ line was confirmed by western blotting and by dramatically increased resistance to killing by UVC radiation (Figure 1A and B). Cytotoxicity and genotoxicity of Cr(VI) are strongly influenced by cellular levels of mismatch repair proteins (36,37) and to a lesser extent, by the oxidative DNA damage repair protein XRCC1 (34,38). We found that XP-A and XPA+ cells contained identical levels of XRCC1 and five major mismatch repair proteins (Figure 1C) previously shown to affect various aspects of Cr(VI) toxicity (36,37). Thus, XPA status had no apparent effect on the NER-unrelated repair systems impacting cellular responses to Cr(VI). XP-A and XPA+ cells also displayed no significant differences (P = 0.39) in the amount of DNA-bound Cr (Figure 1D). However, FACS analysis of cells collected 24 h after Cr(VI) treatment revealed a much greater sensitivity of XP-A cells to cell cycle perturbations relative to NER-proficient XPA+ cells (Figure 2A–C). XP-A cells showed severe depletion of G1 and accumulation of G2/M populations with low Cr(VI) damage and increased S phase arrest at higher damage. In contrast, XPA+ cells exhibited only minimal cell cycle changes. Within the same dose range and post-exposure time, both cell lines displayed no evidence of apoptotic DNA fragmentation (Figure 2D). Assessment of cytotoxicity by the CyQuant assay, which similarly to clonogenic assay integrates both cell death and growth arrest, showed that XPA absence was clearly detrimental for cell viability after Cr(VI) exposures (Figure 2E). We have previously found that clonogenic survival of Cr(VI)-treated XP-A cells was also lower than that in XPA+ cells (27). The degree of cytotoxicity determined by the CyQuant assay was less severe in comparison to the clonogenic toxicity, which in part reflects increased cytotoxicity of Cr(VI) in low-density cultures used in clonogenic experiments (31). Collectively, FACS and cytotoxicity measurements showed that NER was clearly an important defence mechanism against Cr(VI)-induced DNA damage impacting normal cell cycle progression and cell viability. Given their very large size, unrepaired DPC could be a potential cause of enhanced toxic responses in XP-A cells.
Fig. 1.
Characterization of XP-A and XPA+ cells. (A) XPA western blot in extracts from XPA-null (XP-A cells) and XPA-complemented XP-A fibroblasts (XPA+ cells). (B) Clonogenic survival of XP-A and XPA+ fibroblasts treated with UVC (λmax = 254 nm). Data are means ± SDs from two independent experiments with three dishes per dose each. ***P < 0.001 relative to XP-A cells. (C) Western blots demonstrating expression of various DNA repair proteins in XP-A and XPA+ cells. (D) Cr–DNA binding in XP-A and XPA+ cells treated with 20 μM Cr(VI) for 3 h and collected immediately. The amount of DNA-bound Cr was determined by graphite furnace atomic absorption spectroscopy. Data are means ± SDs from four independent DNA samples.
Fig. 2.
Cell cycle changes and survival of Cr(VI)-treated XP-A and XPA+ cells. Cell cycle distribution was determined by FACS of propidium iodide-stained nuclei. Data are means ± SDs from three independent measurements. Statistics: *P < 0.05 and ***P < 0.001 relative to XP-A cells. (A) Changes in percentage of cells in G1 phase. (B) Changes in percentage of cells in S phase. (C) Changes in percentage of cells in G2/M phase. (D) Percentage of cells with subdiploid amount of DNA. (E) Cytotoxicity of Cr(VI) treatments at 48 h post-exposure (means ± SDs, n = 4).
DPC repair in standard cultures of cells
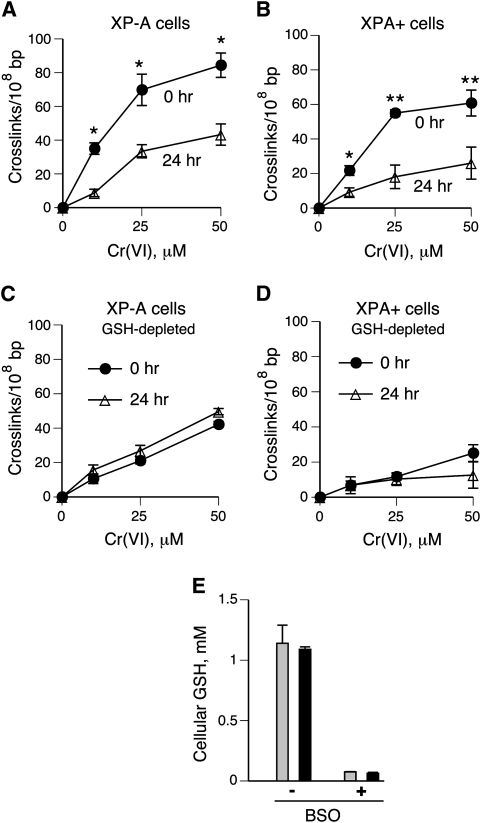
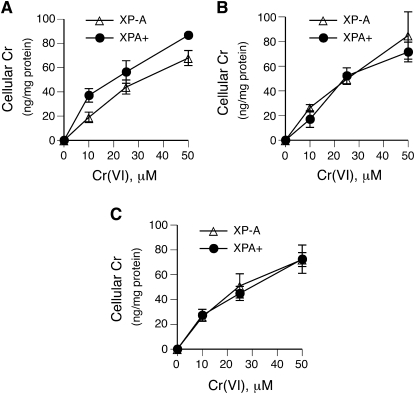
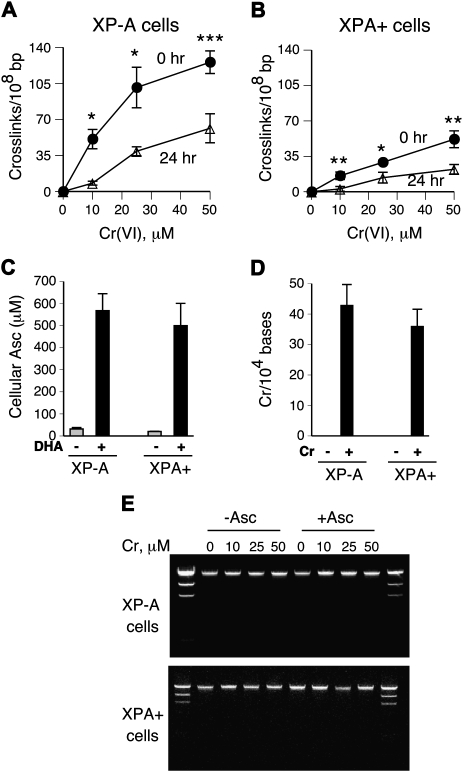
Cr(VI) is a pro-carcinogen which exhibits its genotoxicity during reduction to Cr(III). In human cells maintained under standard tissue culture conditions, GSH is a predominant Cr(VI) reducer (22). The formation of DPC by Cr(VI) in normally grown XP-A and XPA+ fibroblasts yielded similarly shaped dose–response curves (Figure 3A and B). Both cell lines displayed nearly identical decreases in the number of DPC during a 24-h long recovery after Cr(VI) exposure (59 ± 12% decrease in XP-A versus 61 ± 5% in XPA+ cells), indicating that NER had no significant role in DPC repair (Figure 3A and B). In contrast to repair, the absence of XPA had a detectable effect on the initial formation of DPC at all three Cr(VI) doses. After adjustments for modest differences in Cr(VI) uptake (Figure 5A), the initial levels of DPC in XP-A cells were on average 2.2 times higher in comparison to XPA+ cells (P < 0.001).
Fig. 3.
Formation and repair of DPC in normal and GSH-depleted cells. DPC were measured by the K-SDS precipitation assay in cells collected immediately (0 h) and 24 h after 3-h long exposures to Cr(VI). Cr-induced DPC values are background subtracted and represent means ± SDs from three independent experiments in triplicates. Statistics: *P < 0.05 and **P < 0.01 relative to 24 h samples. (A) DPC levels in XP-A cells grown under standard tissue culture conditions. (B) DPC in XPA+ cells grown under standard conditions. (C) DPC in XP-A cells pre-treated with 0.1 mM BSO for 24 h prior to Cr(VI) treatments. (D) DPC in XPA+ cells pre-treated with 0.1 mM BSO for 24 h prior to Cr(VI) treatments. (E) Cellular levels of GSH in control and 0.1 mM BSO pre-treated cells (grey bars, XP-A cells; black bars, XPA+ cells). GSH concentrations were measured by HPLC detection of the specific GSH–monobromobimane conjugate.
Fig. 5.
Cr(VI) uptake measurements. Data are means ± SDs from three independent samples. (A) Control cells. (B) Cells pre-treated with 0.1 mM BSO for 24 h. (C) Cells pre-incubated with 1 mM DHA for 90 min.
Formation and repair of DPC in GSH-depleted cells
To investigate repair of DPC induced by GSH-independent Cr(VI) metabolism, cells were pre-incubated for 24 h with 0.1 mM BSO to inhibit GSH synthesis. XP-A and XPA+ cells had identical initial GSH concentrations (1.1 mM) and showed similar decreases in GSH content following BSO treatments (16-fold decrease on average) (Figure 3E). GSH depletion strongly diminished the formation of DPC by Cr(VI) in both cell lines (2.9 ± 0.6-fold in XP-A and 3.4 ± 1-fold in XPA+) (Figure 3C and D). As in standard cultures, the absence of NER activity in XP-A cells resulted in a higher initial DPC burden relative to XPA+ cells (25 μM: 1.8 ± 0.4-fold and P = 0.03; 50 μM: 1.7 ± 0.3-fold and P = 0.03). These differences in cross-linking did not result from alterations in Cr(VI) uptake, as BSO-treated XP-A and XPA+ cells had equal cellular concentrations of Cr, which were comparable to their GSH-containing controls (Figure 5A and B). However, in contrast to standard cultures, DPC generated in GSH-deficient XP-A and XPA+ cells showed no detectable repair during 24-h recovery (Figure 3C and D).
Effects of Asc supplementation and proteasome inhibition
Asc plays a key role in metabolism of Cr(VI) in tissues (39) due to its >10 times higher rates of reduction over GSH (40). Primary and immortalized human cells grown in standard tissue culture are severely deficient in Asc (31,35,41), reflecting the absence of vitamin C in the majority of synthetic media, its low concentration in serum and relatively rapid loss in oxygenated solutions containing even trace amounts of the redox-active metals iron and copper. Under our growth conditions, intracellular Asc concentrations in 1-day old cultures of XP-A and XPA+ cells were in the range of 20–30 μM (Figure 4C). Cells in tissues experiencing Cr(VI)-induced toxic effects contain 0.5–2 mM Asc (42). Pre-incubation of XP-A and XPA+ cells for 90 min with the oxidized form of vitamin C, DHA, raised their Asc concentrations to ∼0.5 mM (Figure 4C). Examination of DPC after Asc normalization showed that the initial formation of cross-links was on average 3.1 times higher in XP-A versus XPA+ cells (Figure 4A and B). Significantly increased DPC formation in XP-A cells was observed at all three doses: 3.6 ± 1.5-fold and P = 0.04 for 10 μM; 3.1 ± 0.6-fold and P = 0.02 for 25 μM and 2.5 ± 0.6-fold and P = 0.02 for 50 μM. Cr(VI) uptake in both cell lines was practically identical and comparable to standard cultures (Figure 5C). XP-A cells contained ∼19% more DNA-bound Cr, but this small difference was not statistically significant (P = 0.34) due to experimental variability (Figure 4D). Analysis of DNA fragmentation during DPC measurements in control and Asc-supplemented cells showed the expected presence of a single DNA band of ∼22 kb in each sample (Figure 4E), which demonstrates similar technical characteristics of different samples and the absence of apoptotic DNA breakage.
Fig. 4.
Effects of Asc supplementation on DPC formation and repair. Cells were pre-loaded with 0.5 mM Asc during a 90-min long incubation with 1 mM DHA prior to Cr(VI) exposures. DPC data are means ± SDs from three independent experiments in triplicates. Statistics: *P < 0.05, **P < 0.01 and ***P < 0.001 relative to 24 h samples. (A and B) DPC in Asc-supplemented XP-A and XPA+ cells, respectively. (C) Cellular levels of Asc in control and 1 mM DHA-treated XP-A and XPA+ fibroblasts. Asc concentrations were measured by HPLC. (D) Cr–DNA binding in Asc-preloaded XP-A and XPA+ cells treated with 20 μM Cr(VI) for 3 h and collected immediately. (E) Ethidium bromide-stained agarose gels of DNA fragments generated during DPC measurements.
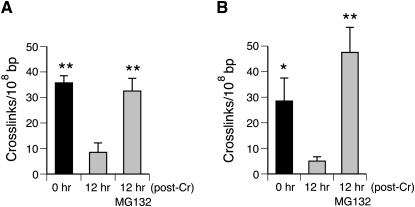
In contrast to the formation, average rates of DPC repair in XP-A and XPA+ cells were very similar (66 ± 14 and 64 ± 13% repair after 24 h, respectively) (Figure 4A and B). Although removal of DPC in cells treated with the lowest dose of 10 μM Cr(VI) was moderately faster than for higher doses, the rates of repair remained the same for XP-A and XPA+ cells (84.1 and 82.3%, respectively). Thus, irrespective of the predominant Cr(VI) activation pathway, NER does not influence the overall rate of removal of DNA-cross-linked proteins in chromate-treated human cells. An alternative DPC clearing mechanism to oligonucleotide excision could involve degradation of cross-linked proteins by proteasomal or other proteolytic activities. To test a potential role of proteasomes in removal of Cr–DPC, we examined the levels of remaining cross-links after 12-h recovery in the absence and presence of the proteasome inhibitor MG132 (Figure 6A and B). We found that a non-toxic dose of 0.5 μM MG132 completely blocked repair of DPC in both XP-A and XPA+ cells.
Fig. 6.
Inhibition of proteasomes suppresses DPC repair. Cells were exposed to Cr(VI) for 3 h and collected immediately (0 h) or following a 12-h recovery period in the presence or absence 0.5 μM MG132. Data are means ± SDs (n = 6). (A) DPC repair in XP-A cells. (B) DPC repair in XPA+ cells. Statistical significance relative to the 12-h samples without MG132: *P < 0.05 and **P < 0.01.
Discussion
Repair of Cr-induced DPC
NER is a versatile DNA repair system that is capable of removing a wide assortment of DNA lesions (26). Preferred NER substrates are typically bulky DNA adducts and helix-distorting modifications. The bulkiness of the attached group can stimulate mammalian NER independently of structural changes in the DNA duplex (8,27). Thus, it was conceivable that human NER could also be capable of removing Cr(VI)-induced DPC. Smaller amino acid–Cr–DNA cross-links have already been established as substrates for removal by human NER (27). However, we found that repair of Cr(VI)-induced DPC in NER-proficient and -deficient human fibroblasts was essentially identical. The overall rate of DPC removal in XP-A and XPA+ cells was comparable to that measured in rat kidney and lung but lower than in liver (17). Inhibition of 26S proteasomes by MG132 completely abolished DPC repair in cells irrespective of their NER status. We have previously proposed that DPC repair in human cells is a two-step process involving the initial digestion of cross-linked proteins by nuclear proteasomes followed by removal of the remaining small peptide–DNA cross-links by NER (10). In agreement with this model, subsequent in vitro NER assays with mammalian extracts showed lack of excision for DPC with 16 and 37 kDa proteins whereas shorter peptide–DNA cross-links with the same chemical linkages or protease-digested DPC were very good NER substrates (8,9). It should be noted that residual peptide–DNA adducts are not detectable by our DPC assay. Increased sensitivity of cell cycle to Cr(VI)-induced perturbations in standard cultures of XPA-null cells despite their normal DPC repair clearly demonstrated the importance of NER in removal of genotoxic Cr–DNA damage. Thus, it appears that abundant Cr–DNA adducts (27) but not DPC are likely to be the main cause of impaired cell cycle progression in XP-A cells. This conclusion is not contradicted by the fact that XP-A cells had 2.2 times higher yield of DPC because cell cycle perturbations caused in these cells by 10 μM Cr(VI) were much more severe than cell cycle changes in XPA+ cells treated with 25 μM Cr(VI) (Figure 2).
In contrast to standard and Asc-supplemented cultures, we did not detect DPC repair in GSH-depleted XP-A and XPA+ cells. These findings indicate that Cr(VI) metabolism by the secondary reducing systems (22) likely led to the inactivation of DPC repair. Loss of DPC repair in normal cells has been previously found only after very large doses of Cr(VI) (23). The absence of GSH strongly sensitizes cells to Cr(VI)-induced oxidative stress associated with increased production of hydrogen peroxide and reactive Cr intermediates (34,43). In normal cultured cells, induction of the same degree of oxidative stress is achieved only with much higher Cr(VI) concentrations (34). Thus, it is possible that inhibition of DPC repair in GSH-depleted cells treated with low-moderate doses and in normal cells with high Cr(VI) doses is caused by the same mechanisms. The repression of DPC repair could result from oxidation of critical SH- and other protein groups by reactive products of Cr(VI) metabolism (44,45). The type of cells can also influence the sensitivity of DPC repair to inhibitory influences since Cr(VI)-resistant A549 cells (43,46) were able to partially remove Cr(VI)-induced DPC under GSH-depleted conditions (29). It should be noted, however, that BSO-treated A549 cells retained 2.2 times higher GSH concentrations in comparison to XP-A/XPA+ cells. A549 cells also contain 1.9 times higher amounts of cysteine (G. Quievryn and A. Zhitkovich, unpublished results), which is another Cr(VI)-reducing thiol in human cells (22).
NER and formation of DPC
While the absence of NER had no effect on repair of DPC, their formation in XP-A cells was significantly elevated, particularly in Asc-restored cells. This finding implicates NER in repair of DNA lesions that act as DPC precursors. Formation of DPC by Cr(VI) proceeds through a three-step process involving reduction to Cr(III), rapid Cr(III)–DNA adduction and finally, a rate-limiting reaction of protein conjugation (29). DPC precursors, binary Cr(III)–DNA adducts, are substrates for human NER (27), which explains increased DPC yields in NER-negative cells. Thus, even though NER plays no detectable role in repair of DPC, it protects cells against these large DNA lesions by inhibiting their formation. Overall, of the three mechanisms that cells can employ to decrease DPC burden, namely diminishing formation, removing attached proteins and repairing residual cross-links, the first and the last approaches are NER dependent. This conclusion should apply to many other cross-linking agents that form DPC via capture of proteins by DNA adducts. However, when DPC are caused through attacking of DNA by adducted/oxidized proteins (47,48), the role of NER should be limited to only removal of products of proteolytic degradation of DPC.
Cr–DNA adducts are a heterogeneous group of DNA modifications largely comprised of various Cr–DNA cross-links with GSH, Asc and selected amino acids (22). Binary Cr–DNA conjugates are estimated to represent only a small fraction of total Cr–DNA adducts in cells. A strongly inhibited DPC production in Asc-normalized XPA+ cells despite a marginal 19% decrease in the amount of total Cr–DNA binding suggests that DPC-forming binary Cr–DNA adducts are probably better NER substrates than more abundant small Cr–DNA cross-links.
DPC as biomarkers of Cr(VI) exposure
Although DPC measurements have long been used for the assessment of Cr(VI) exposure (19,21,24,25,49), the factors impacting responsiveness and persistence of this biomarker remained largely unknown. Our findings indicate that the slope of DPC formation as a function of internal Cr(VI) dose will be increased (higher sensitivity) under conditions that suppress NER. Human exposure to Cr(VI) is frequently accompanied by co-exposure to Ni(II) and Co(II) ions (50), which are both known NER inhibitors (51–53). Co-exposure to agents producing DNA lesions that act as very good NER substrates (i.e. UV-DNA cross-links, polycyclic aromatic hydrocarbon–DNA adducts) should competitively suppress the removal of Cr–DNA adducts and promote a higher yield of Cr–DPC. However, once DPC are formed, their stability and consequently the duration of Cr(VI) exposure they can integrate should be NER independent. It would be interesting to investigate whether co-exposure factors with strong pro-oxidant properties can also cause inhibition of Cr–DPC repair, as was observed in GSH-depleted cells.
Conclusions
In contrast to a common view of NER as being capable of excising a broad spectrum of bulky lesions, our results showed that removal of chromosomal DPC in Cr(VI)-treated human cells was NER independent. Clearing of Cr–DPC was blocked by inhibition of a proteasomal activity, implicating proteolysis of cross-linked proteins in DPC repair. NER was important for preventing DNA–protein cross-linking via removal of the precursor DNA lesions. Thus, even though NER does not act on DPC directly, it protects cells against these large genotoxic lesions by diminishing their formation.
Funding
National Institute of Environmental Health Sciences (R01 ES008786, P42 ES013660).
Acknowledgments
Conflict of interest statement: None declared.
References
- 1.Costa M. DNA-protein complexes induced by chromate and other carcinogens. Environ. Health Perspect. 1991;92:45–52. doi: 10.1289/ehp.919245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa M, Zhitkovich A, Harris M, Paustenbauch D, Gargas M. DNA-protein crosslinks produced by various chemicals in cultured human lymphoma cells. J. Toxicol. Environ. Health. 1997;50:433–449. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- 3.Taioli E, Zhitkovich A, Toniolo P, Bernstein J, Blum R, Costa M. DNA-protein crosslinks as a biomarker of cis-platinum activity in cancer patients. Oncol. Rep. 1996;3:439–441. [PubMed] [Google Scholar]
- 4.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl Acad. Sci. USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano T, Morishita S, Katafuchi A, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 7.Nakano T, Katafuchi A, Matsubara M, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc. Natl Acad. Sci. USA. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O'Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 10.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 11.Speit G, Schütz P, Merk O. Induction and repair of formaldehyde-induced DNA-protein crosslinks in repair-deficient human cell lines. Mutagenesis. 2000;15:85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- 12.de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair. 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguaninetransferase. J. Biol. Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- 14.Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loecken EM, Dasari S, Hill S, Tabb DL, Guengerich FP. The bis-electrophile diepoxybutane cross-links DNA to human histones but does not result in enhanced mutagenesis in recombinant systems. Chem. Res. Toxicol. 2009;22:1069–1076. doi: 10.1021/tx900037u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa M, Klein CB. Toxicity and carcinogenicity of chromium compounds in humans. Crit. Rev. Toxicol. 2006;36:155–163. doi: 10.1080/10408440500534032. [DOI] [PubMed] [Google Scholar]
- 17.Tsapakos MJ, Hampton TH, Wetterhahn KE. Chromium(VI)-induced DNA lesions and chromium distribution in rat kidney, liver, and lung. Cancer Res. 1983;43:5662–5667. [PubMed] [Google Scholar]
- 18.Izzotti A, Bagnasco M, Camoirano A, Orlando M, De Flora S. DNA fragmentation, DNA-protein crosslinks, postlabeled nucleotidic modifications, and 8-hydroxy-2'-deoxyguanosine in the lung but not in the liver of rats receiving intratracheal instillations of chromium(VI). Chemoprevention by oral N-acetylcysteine. Mutat. Res. 1998;400:233–244. doi: 10.1016/s0027-5107(98)00028-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhitkovich A, Voitkun V, Kluz T, Costa M. Utilization of DNA-protein crosslinks as a biomarker of chromium exposure. Environ. Health Perspect. 1998;106:969–974. doi: 10.1289/ehp.98106s4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnekenburger M, Talaska G, Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol. Cell. Biol. 2007;7:7089–7101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa M, Zhitkovich A, Toniolo P. DNA-protein crosslinks in welders: molecular implications. Cancer Res. 1993;53:460–463. [PubMed] [Google Scholar]
- 22.Zhitkovich A. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI) Chem. Res. Toxicol. 2005;18:3–11. doi: 10.1021/tx049774+. [DOI] [PubMed] [Google Scholar]
- 23.Sugiyama M, Patierno SR, Cantoni O, Costa M. Characterization of DNA lesions induced by CaCrO4 in synchronous and asynchronous cultured mammalian cells. Mol. Pharmacol. 1986;29:606–613. [PubMed] [Google Scholar]
- 24.Werfel U, Langen V, Eickhoff I, Schoonbrood J, Vahrenholtz C, Brauksiepe A, Popp W, Norpoth K. Elevated DNA single-strand breakage frequencies in lymphocytes of welders exposed to chromium and nickel. Carcinogenesis. 1998;19:413–418. doi: 10.1093/carcin/19.3.413. [DOI] [PubMed] [Google Scholar]
- 25.Medeiros MG, Rodrigues AS, Batoreu MC, Laires A, Rueff J, Zhitkovich A. Elevated levels of DNA-protein crosslinks and micronuclei in peripheral lymphocytes of tannery workers exposed to trivalent chromium. Mutagenesis. 2003;18:19–24. doi: 10.1093/mutage/18.1.19. [DOI] [PubMed] [Google Scholar]
- 26.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien TJ, Brooks BR, Patierno SR. Nucleotide excision repair functions in the removal of chromium-induced DNA damage in mammalian cells. Mol. Cell. Biochem. 2005;279:85–95. doi: 10.1007/s11010-005-8225-0. [DOI] [PubMed] [Google Scholar]
- 29.Macfie A, Hagan E, Zhitkovich A. Mechanism of DNA-protein cross-linking by chromium. Chem. Res. Toxicol. 2010;23:341–347. doi: 10.1021/tx9003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy DD, Saijo M, Tanaka K, Kraemer KH. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis. 1995;16:1557–1563. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds M, Zhitkovich A. Cellular vitamin C increases chromate toxicity via a death program requiring mismatch repair but not p53. Carcinogenesis. 2007;28:1613–1620. doi: 10.1093/carcin/bgm031. [DOI] [PubMed] [Google Scholar]
- 32.Zhitkovich A, Costa M. A simple, sensitive assay to detect DNA-protein-crosslinks in intact cells and in vivo. Carcinogenesis. 1992;13:1485–1489. doi: 10.1093/carcin/13.8.1485. [DOI] [PubMed] [Google Scholar]
- 33.Quievryn G, Goulart M, Messer J, Zhitkovich A. Reduction of Cr(VI) by cysteine: significance in human lymphocytes and formation of DNA damage in reactions with variable reduction rates. Mol. Cell. Biochem. 2001;222:107–118. [PubMed] [Google Scholar]
- 34.Messer J, Reynolds M, Stoddard L, Zhitkovich A. Causes of DNA single-strand breaks during reduction of chromate by glutathione in vitro and in cells. Free Radic. Biol. Med. 2006;40:1981–1992. doi: 10.1016/j.freeradbiomed.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 35.Quievryn G, Messer J, Zhitkovich A. Carcinogenic chromium(VI) induces cross-linking of vitamin C to DNA in vitro and in human lung A549 cells. Biochemistry. 2002;41:3156–3167. doi: 10.1021/bi011942z. [DOI] [PubMed] [Google Scholar]
- 36.Peterson-Roth E, Reynolds M, Quievryn G, Zhitkovich A. Mismatch repair proteins are activators of toxic responses to chromium-DNA damage. Mol. Cell. Biol. 2005;25:3596–3607. doi: 10.1128/MCB.25.9.3596-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds MF, Peterson-Roth EC, Jonhston T, Gurel VM, Menard HL, Zhitkovich A. Rapid DNA double-strand breaks resulting from processing of Cr-DNA crosslinks by both MutS dimers. Cancer Res. 2009;69:1071–1079. doi: 10.1158/0008-5472.CAN-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant HE, Ying S, Helleday T. Homologous recombination is involved in repair of chromium-induced DNA damage in mammalian cells. Mutat. Res. 2006;599:116–123. doi: 10.1016/j.mrfmmm.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Standeven AM, Wetterhahn KE. Ascorbate is the principal reductant of chromium(VI) in rat lung ultrafiltrates and cytosols, and mediates chromium-DNA binding in vitro. Carcinogenesis. 1992;13:1319–1324. doi: 10.1093/carcin/13.8.1319. [DOI] [PubMed] [Google Scholar]
- 40.Quievryn G, Peterson E, Messer J, Zhitkovich A. Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry. 2003;42:1062–1070. doi: 10.1021/bi0271547. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds M, Stoddard L, Bespalov I, Zhitkovich A. Ascorbate acts as a highly potent inducer of chromate mutagenesis and clastogenesis: linkage to DNA breaks in G2 phase by mismatch repair. Nucleic Acids Res. 2007;35:465–476. doi: 10.1093/nar/gkl1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kojo S. Vitamin C: basic metabolism and its function as an index of oxidative stress. Curr. Med. Chem. 2004;11:1041–1064. doi: 10.2174/0929867043455567. [DOI] [PubMed] [Google Scholar]
- 43.Martin BD, Schoenhard JA, Hwang JM, Sugden KD. Ascorbate is a pro-oxidant in chromium-treated human lung cells. Mutat. Res. 2006;610:74–84. doi: 10.1016/j.mrgentox.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 44.Myers JM, Myers CR. The effects of hexavalent chromium on thioredoxin reductase and peroxiredoxins in human bronchial epithelial cells. Free Radic. Biol. Med. 2009;47:1477–1485. doi: 10.1016/j.freeradbiomed.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumner ER, Shanmuganathan A, Sideri TC, Willetts SA, Houghton JE, Avery SV. Oxidative protein damage causes chromium toxicity in yeast. Microbiology. 2005;151:1939–1948. doi: 10.1099/mic.0.27945-0. [DOI] [PubMed] [Google Scholar]
- 46.Dubrovskaya VA, Wetterhahn KE. Effects of Cr(VI) on the expression of the oxidative stress genes in human ling cells. Carcinogenesis. 1998;19:1401–1407. doi: 10.1093/carcin/19.8.1401. [DOI] [PubMed] [Google Scholar]
- 47.Voitkun V, Zhitkovich A. Analysis of DNA-protein crosslinking activity of malondialdehyde in vitro. Mutat. Res. 1999;424:97–106. doi: 10.1016/s0027-5107(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 48.Luxford C, Dean RT, Davies MJ. Induction of DNA damage by oxidised amino acids and proteins. Biogerontology. 2002;3:95–102. doi: 10.1023/a:1015228001561. [DOI] [PubMed] [Google Scholar]
- 49.Kuykendall JR, Miller KL, Mellinger KN, Cain AV. Waterborne and dietary hexavalent chromium exposure causes DNA-protein crosslink (DPX) formation in erythrocytes of largemouth bass (Micropterus salmoides) Aquat. Toxicol. 2006;78:27–31. doi: 10.1016/j.aquatox.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic and chromium. Chem. Res. Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartwig A, Schwerdtle T. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol. Lett. 2002;127:47–54. doi: 10.1016/s0378-4274(01)00482-9. [DOI] [PubMed] [Google Scholar]
- 52.Kopera E, Schwerdtle T, Hartwig A, Bal W. Co(II) and Cd(II) substitute for Zn(II) in the zinc finger derived from the DNA repair protein XPA, demonstrating a variety of potential mechanisms of toxicity. Chem. Res. Toxicol. 2004;17:1452–1458. doi: 10.1021/tx049842s. [DOI] [PubMed] [Google Scholar]
- 53.Hu W, Feng Z, Tang MS. Nickel (II) enhances benzo[a]pyrene diol epoxide-induced mutagenesis through inhibition of nucleotide excision repair in human cells: a possible mechanism for nickel (II)-induced carcinogenesis. Carcinogenesis. 2004;25:455–462. doi: 10.1093/carcin/bgh012. [DOI] [PubMed] [Google Scholar]