Abstract
Recent studies in Drosophila identified pygopus, which encodes a PHD finger protein, as an additional nuclear component of the canonical Wingless(Wg)/Wnt signaling pathway. In this study, we describe the molecular cloning and expression analysis of a mouse pygopus gene, mpygo2. mpygo2 transcripts were detected in almost all adult mouse tissues examined, whereas transcripts of another mouse pygopus gene, mpygo1, were detected only in heart tissue. Abundant mpygo2 transcripts were observed during embryogenesis in multiple developmental sites. Consistent with the demonstrated role of the Wnt-β-catenin–LEF/TCF signaling pathway in mammalian skin development, mpygo2 expression was detected in the developing epidermis and hair follicles, which suggests that mpygo2 might mediate the effect of this signaling pathway in mouse skin.
Keywords: Wg/Wnt signaling, β-Catenin, LEF/TCF, pygopus, mpygo2, mpygo1, Skin, Hair follicles
Introduction
Wnt signaling is crucial for normal development and tumorigenesis [1–3]. The developmental processes regulated by this signaling event are diverse, ranging from patterning the body axis and the central nervous system to organogenesis that involves reciprocal epithelial-mesenchymal interactions [1,2,4]. Furthermore, Wnt signaling is implicated in the maintenance and differentiation of postnatal stem cells in epithelial tissues such as skin and intestine [1,2,5]. An activated Wnt signal triggers several distinct intracellular signal transduction pathways [6]. The so-called canonical pathway, which is the most extensively characterized, involves a complex series of downstream events culminating in the stabilization of β-catenin, a protein with dual functions in cell–cell adhesion and signaling [3,6]. β-Catenin accumulates in the cytoplasm and enters the nucleus, where it binds to the LEF/TCF family of transcription factors and regulates gene expression.
The Wnt signaling pathway intersects with a complex network of molecular events both outside and inside the nucleus. Studies have shown that several nuclear factors, including Sox, Smad, CBP, CtBP, and Groucho, can associate with either β-catenin or LEF/TCF proteins to modulate the transcription regulatory activity of the β-catenin–LEF/TCF complex [7–14]. Distinct from these nuclear factors that display context-dependent modulatory activities, a recently identified gene in Drosophila, named pygopus (pygo), appears to encode an obligatory and highly specific nuclear component of the β-catenin–LEF/TCF transcription regulatory complex [15–18]. Mutations in pygo specifically disrupt Wg (the fly Wnt counterpart) signaling throughout Drosophila development [15–18]. The Pygo protein contains a PHD domain, often found in chromatin remodeling factors that are thought to alter chromatin structure and thereby allow activation or repression of specific genes [19]. Pygo is recruited to β-catenin–LEF/TCF via an adaptor protein, Lgs/BCL9, and existing evidence suggests that Pygo is required for maximum activation of β-catenin–LEF/TCF-dependent promoters/genes in cell culture [15–18, 20]. Pygo homologs have also been identified in Xenopus, and functional studies indicate that xPygo proteins are required for Wg/Wnt signaling during Xenopus embryogenesis, particularly in embryonic brain patterning [18, 21].
Although previous studies have implicated the existence of two mouse pygopus genes, mpygo1 and mpygo2 [15,18], a full-length sequence of mpygo2 was not reported. Here we describe the cloning of a full-length mpygo2 cDNA and the structure of the mpygo2 gene. Our evidence shows that mpygo2 is expressed during embryonic development and in multiple adult tissues and suggests widespread involvement of mpygo2 in canonical Wnt signaling.
Results
Cloning and molecular structure of mpygo2
BLAST searches of the GenBank database using the sequences of Drosophila pygo, human pygo1 (hpygo1), and human pygo 2 (hpygo2) revealed two mouse cDNA sequences representing mpygo1 and mpygo2, respectively. Although mpygo1 encodes a full-length protein of 417 amino acids, the mpygo2 cDNA thus identified encodes only what may be a truncated open reading frame (ORF) of 338 amino acids. We therefore cloned a full-length mpygo2 cDNA by compiling PCR-amplified cDNA fragments, and the sequence of this cDNA is shown in Fig. 1A. This full-length mpygo2 cDNA encodes an ORF of 405 amino acids. The putative mPygo2 protein shares 81% amino acid sequence identity with hPygo2, whereas mPygo1 shares 56% overall amino acid sequence identity with hPygo1. Like other Pygo proteins, mPygo2 contains a predicted nuclear localization signal (NLS) sequence in its N-terminal region and a Cys4-His-Cys3 PHD finger domain in its C-terminal region (Fig. 1A). The amino acid sequence alignment of all Pygo proteins identified thus far reveals two well-conserved domains across species: a domain rich in proline and charged amino acids at the N-terminus, previously referred to as the N-terminal homology domain (NHD [17]), and the PHD finger domain at the C-terminus (Fig. 1B).
Fig. 1.
Sequence and structural analysis of mpygo2 and its gene products. (A) mpygo2 cDNA sequence and the deduced amino acid sequence. The putative NLS sequence is boxed. The PHD finger domain is underlined. (B) Amino acid sequence alignment of the N-terminal (top) and C-terminal (bottom) domains conserved in Pygo proteins from mouse and human Drosophila and Xenopus. Identical residues are highlighted. NLS residues are marked by black dots under the sequence. Structural residues of the PHD domain are marked by black stars under the sequence. (C) Exon-intron structure of the mpygo2 gene. Rectangular boxes indicate exons (solid boxes, coding sequences; open boxes, noncoding sequences). Horizontal lines indicate introns. Sizes of exons and introns are indicated. D3Mit49 and D3Mit341 are markers on chromosome 3 that flank the mpygo2 gene, and chromosomal locations are indicated in cM. GenBank accession numbers of sequences used in alignment follow: Pygo, NM_143615; xPygo, AF521655; mPygo1, AK011208; hPygo1, AF457207; hPygo2, BC032099.
The structure of the mpygo2 gene was revealed by a comparison between the mpygo2 genomic and cDNA sequences. The gene consists of three exons that span approximately 4.9 kb (Fig. 1C). The predicted physical location of mpygo2 is on the F2 band of mouse chromosome 3, between D3Mit49 and D3Mit341 and approximately 90.5 cM from the centromere. Searches against various available mouse genomic sequence databases consistently identified only one hit on chromosome 3. Southern blot analysis using an mpygo2 genomic fragment also revealed a single band (data not shown). Taken together, these results indicate that mpygo2 is a single gene in the mouse genome.
mpygo2 displays a broader expression spectrum than mpygo1 during development and in adult tissues
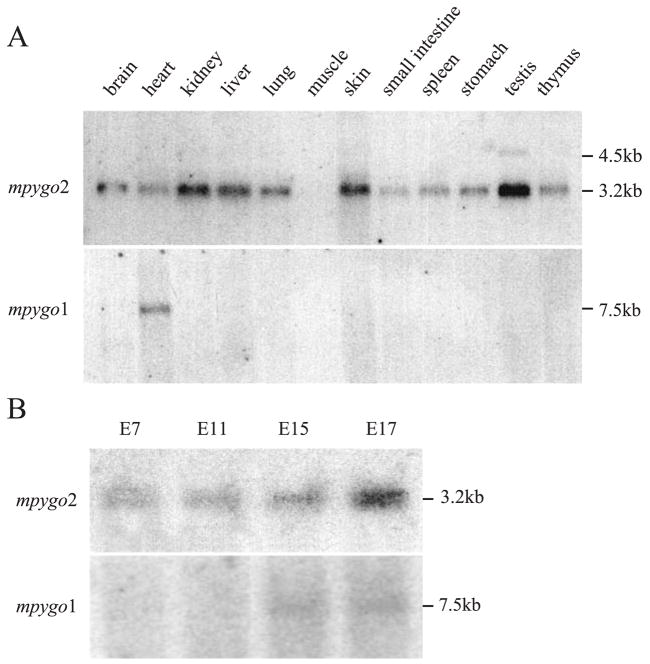
The tissue distribution of mpygo2 and mpygo1 expression was determined. Northern blot analysis identified mpygo2 transcripts in almost all adult mouse tissues examined, including brain, heart, kidney, liver, lung, skin, small intestine, spleen, stomach, testis tissue, and thymus (Fig. 2A). All expressing tissues contained a major mpygo2 transcript of ~3.2 kb, confirming that the cDNA we cloned is indeed full-length, whereas testis contained an additional transcript of 4.5 kb. In contrast, a mpygo1 transcript of 7.5 kb was detected only in heart tissue even after prolonged exposure.
Fig. 2.
Tissue distribution and embryonic expression of mpygo2 and mpygo1. (A) Northern blot analysis of adult mouse tissue poly(A)+ RNA. (B) Northern blot analysis of poly(A)+ RNA from developing embryos at the indicated ages.
The expression of mpygo2 and mpygo1 was also examined during development using a blot containing poly(A)+ RNA prepared from developing mouse embryos. mpygo2 transcripts were detected in embryos of all ages examined, including E7, E11, E15, and E17 (Fig. 2B), with the expression level peaking at E17. In contrast, weak mpygo1 expression was detected in E15 and E17 embryos. Therefore, mpygo2 appears to be expressed more broadly in postnatal tissues and at a higher level in developing embryos than mpygo1.
mpygo2 is expressed in multiple embryonic tissues
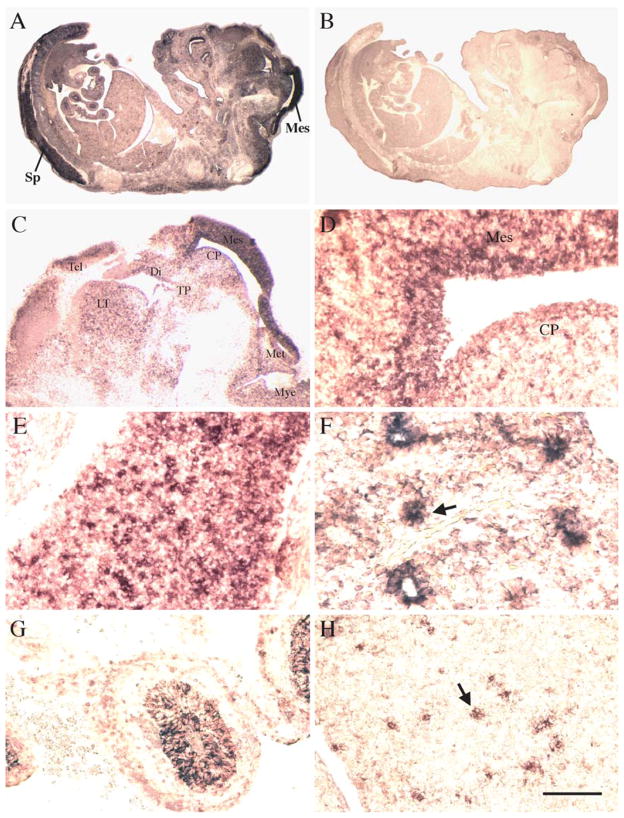
To determine the tissue distribution of mpygo2 during development, we performed in situ hybridization on developing embryos. At E12.5, mpygo2 expression was detected nearly throughout the embryo but was strongest in the mesencephalon of the brain and in the spinal cord (Fig. 3A). A similarly broad pattern of expression was also observed in embryos at E13.5, E15.5, and E16.5 (data not shown). These results are in agreement with the widespread expression of mpygo2 in adult tissues.
Fig. 3.
Developmental expression of mpygo2. Results are from in situ hybridization of E12.5 embryos using antisense (A, C–H) and sense (B) mpygo2 probes. (C–H) High-magnification images of selected developing tissues including brain (C and D), spinal cord (E), lung (F), intestine (G), and liver (H). Sp, spinal cord; Mes, mesencephalon; Tel, telencephalon; Di, diencephalon; Met, metencephalon; Mye, myelencephalon; CP, cerebral peduncle; TP, tuberculum posterius; LT, lamina terminalis. Arrow in F indicates bronchus epithelium of the developing lung. Arrow in H indicates mpygo2 expression in cells scattered throughout fetal liver. Bar, 2 mm in A and B; 800 μm in C; 120 μm in D, E, G, and H; 60 μm in F.
Higher magnification images revealed additional details of mpygo2 expression in selected tissues such as brain (Figs. 3C and 3D), spinal cord (Fig. 3E), lung (Fig. 3F), intestine (Fig. 3G), and liver (Fig. 3H). In the developing brain, mpygo2 hybridization signals were observed in multiple regions including the telencephalon, diencephalon, mesencephalon, metencephalon, myelencephalon, cerebral peduncle, and lamina terminalis, but expression was considerably stronger in the mesencephalon (Figs. 3C and 3D). In the developing lung, mpygo2 transcripts were detected in the bronchus epithelium (arrow in Fig. 3F). In the developing intestine, mpygo2 transcription was observed in the epithelium of the future colon (Fig. 3G).
Expression of mpygo2 in developing and adult hair follicles
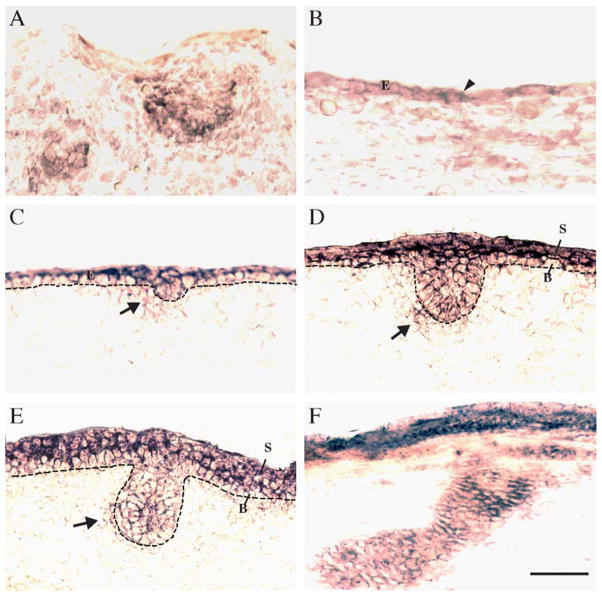
Hair follicle development in mice represents a well-studied system in which Wnt signaling components are functionally required [22]. We therefore examined mpygo2 expression in skin within the developmental window (E12.5–E16.5) during which hair follicle development initiates and proceeds [23]. At E12.5, hair follicle placodes are normally only present in the whisker pad region, and mpygo2 expression was detected in these developing vibrissae placodes (Fig. 4A). Furthermore, mpygo2 expression was seen in cells scattered throughout the surface ectoderm (arrowhead in Fig. 4B). By E13.5, mpygo2 transcripts appeared throughout the ectoderm, which is still single-layered at this stage, and in emerging pelage hair placodes (Fig. 4C). As development proceeds, mpygo2 expression expanded to include the future suprabasal layers of the epidermis and the entire epithelial compartment of the hair germ (Figs. 4D and 4E). Expression was also observed in the dermal condensates around the developing pelage placodes that will form the future dermal papilla of adult hair follicles (arrows in Figs. 4C–4E).
Fig. 4.
Expression of mpygo2 during skin development. Results are from in situ hybridization of developing skin at E12.5 (A and B), E13.5 (C), E15.5 (D), E16.5 (E), and newborn (F) using a mpygo2 cRNA antisense probe. Sense probes showed no specific hybridization signal (not shown). (A) mpygo2 expression in the developing vibrissae. (B–F) mpygo2 expression in developing pelage hair follicles. E, ectoderm; B, basal layer; S, suprabasal layers. Dotted line denotes the basement membrane. Arrows indicate hybridization signals in the dermal condensates. Bar, 50 μm.
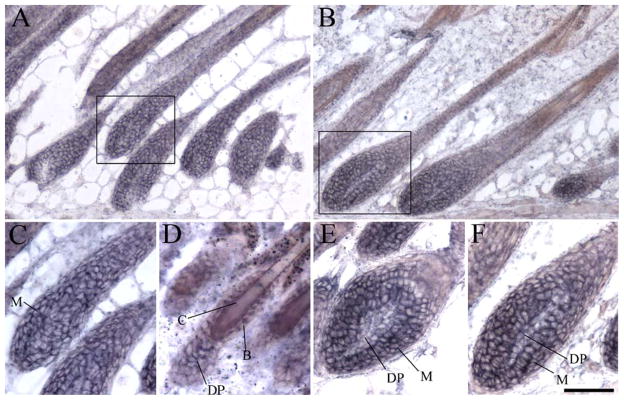
The widespread expression of mpygo2 throughout the epithelial cells of the epidermis and hair follicles persisted in postnatal skin (Figs. 4F, 5A, and 5B and data not shown). Although comparable levels of expression were detected between the hair bulb and the upper portion of the mature hair follicles at P9 (Figs. 5A and 5C), hybridization signals appeared particularly prominent in the matrix of the early-(P23; Fig. 5E) and mid-anagen (P28; Fig. 5F) hair follicles of the new hair cycle. Weaker expression was also observed in the dermal papilla of these follicles. During telogen, mpygo2 transcripts were detected in the ball-shaped dermal papilla at the base of the resting hair follicles and in the epithelial cells including the stem cell-containing bulge that lines the club hair (Fig. 5D).
Fig. 5.
Expression of mpygo2 in mature and cycling hair follicles. Results are from hybridization of postnatal skin at the age of P9 (A and C), P21 (D), P23 (E), and P28 (B and F) using a mpygo2 cRNA antisense probe. (C and F) High-magnification images of the boxed areas in A and B, respectively. M, matrix; D, dermal papilla; B, bulge; C, club hair. Bar, 100 μm in A and B; 60 μm in C, E, and F; 40 μm in D.
Discussion
It is not uncommon that multiple homologs of a single gene in Drosophila exist in higher organisms such as mouse and human. In the case of Drosophila pygo, a newly identified nuclear component of the canonical Wnt signaling pathway, two mouse and human pygopus genes, namely mpygo1/hpygo1 and mpygo2/hpygo2, have been identified [15,17,18]. Are both paralogs required for mediating Wnt signaling in mammalian development? Results of our expression studies revealed a broad transcription pattern for mpygo2 but a restricted pattern of distribution for mpygo1 transcripts. mpygo2 expression occurs in multiple adult and developing tissues, many of which have been shown to require Wnt signaling activity for proper development, morphogenesis, and maintenance [6]. Overall, the embryonic and adult expression pattern of mpygo2 is more consistent than that of mpygo1 with the demonstrated widespread involvement of Wnt signaling in development and morphogenesis. Similarly, in the NCBI-UniGene database, hpygo2 (Hs.172084) cDNAs have been detected in a much broader range of embryonic and adult tissues, including various types of tumor tissues and cells, than hpygo1 (Hs.256587). The strongest embryonic mpygo2 expression was observed in the developing nervous system of the embryo. A functional involvement of Wnt signaling components in the developing and mature central nervous system is well-documented Conventional Wnt1 and conditional β-catenin knockout mice both show brain malformation [24–27]. Wnt1/Wnt3a double mutant mice show additional spinal cord defects [28]. The abundant expression of mpygo2 in the developing brain and spinal cord, together with the finding that a depletion of xpygo2 gene products in Xenopus led to brain defects [21], suggests that mpygo2 is an essential component of the Wnt-β-catenin–LEF/TCF signaling pathway in the development of the central nervous system in mice.
As a putative central component of the Wnt signaling pathway, mpygo2 is expected to be expressed during skin development, particularly during development of the hair follicle. The actual sites in skin where active Wnt signaling occurs have been elegantly mapped using transgenic mice expressing TOP-GAL, a LEF/TCF-responsive β-gal reporter [29,30]. As hair follicle morphogenesis initiates from the pluripotent ectoderm, Wnt signaling is activated in the forming placodes and in the underlying dermal condensates [30]. In postnatal hair follicles, LEF1 transcripts were detected primarily in the hair matrix and precortex cells, whereas TOP-GAL is activated in the precortex [30]. We observed active mpygo2 transcription in all of these locations, placing mpygo2 in the right place and at the right time to mediate Wnt signaling in these developmental sites. Interestingly, mpygo2 transcripts were also detected in the developing interfollicular epidermis, where canonical Wnt signaling does not appear to be required. Overall, mpygo2 appears to be expressed more ubiquitously than the actual sites of active Wnt signaling. This is also true for Drosophila pygo [16], yet pygo action appears to be highly specific to Wg signaling [15–18]. Studies are ongoing to examine the function of mpygo2 in the development and differentiation of epidermis and its appendages and to address whether it is involved in both Wnt-dependent and Wnt-independent pathways.
Materials and methods
Cloning and sequence analysis
mpygo2 genomic sequence was obtained from the Ensembl database and can be found in the GenBank database (accession number NT_078386). A full-length mpygo2 cDNA of 3198 bp was obtained by RT-PCR on RNAs isolated from adult mouse skin using the following primers: 5′ TGTCGAGTGGTGCGTTCCACTCT 3′ (5′PY2–4) and 5′ GGATTTTCAAGTCATTTTATTAATC-CAACCG 3′ (PY2–3′). The design was based on mpygo2 genomic sequence and an existing mpygo2 EST sequence (GenBank accession number. BC025531). Sequencing was performed using this cDNA as well as cDNA fragments obtained by RT-PCR using several internal primer pairs, and a full-length sequence was assembled from that of overlapping fragments. mPygo2 protein sequence analysis was conducted using Pfam, Prosite, and Predict NLS server.
Northern blot analysis
Northern blot containing poly(A)+ RNA from various adult mouse tissues was purchased from Origene (Rockville, MD). Northern blot containing poly(A)+ RNA from four embryonic stages was purchased from Clontech (Palo Alto, CA). These blots were hybridized with a random primed, 32P-labeled 531-bp probe containing mpygo1 cDNA sequence (GenBank accession number AK011208; probe corresponds to positions 1054–1584 of the sequence) or a 894-bp probe containing the 3′ UTR sequence of mpygo2 (corresponds to positions 1927–2820 of the mpygo2 cDNA sequence in Fig. 1A) using standard procedures.
In situ hybridization
Digoxigenin-labeled cRNA sense and antisense probes were synthesized from either a 894-bp mpygo2 fragment corresponding to the 3′ UTR region or a 1215-bp ORF-encoding mpygo2 fragment corresponding to positions 357–1571 of the mpygo2 cDNA sequence in Fig. 1A. Identical results were obtained using these two different probes. In situ hybridizations with these cRNA probes were performed using a procedure adapted from Deng and Lin [31]. Specifically, freshly dissected embryos were fixed in 4% paraformaldehyde/PBS overnight at 4 °C and subsequently passed through a series of sucrose/PBS solutions of increasing sucrose concentration. After an overnight incubation in 30% sucrose/PBS:OCT (1:1), the samples were frozen in 30% sucrose/PBS:OCT (1:3); 10-μm sections were then cut and dried at 50 °C for 2 hours, followed by drying at room temperature overnight. Sections were treated with Proteinase K (30 μg/ml in PBS) for 5 min at room temperature, rinsed briefly in PBS, and refixed in 4% paraformaldehyde/PBS for 7 min. For postnatal skin, 10- to 15-μm frozen sections were fixed in 4% paraformaldehyde/PBS for 10 min at room temperature. Postfixation washes, probe hybridizations, posthybridization washes, and immunological detection were as described in reference 31, followed by a colorimetric reaction using the NBT/BCIP substrates.
Acknowledgments
We thank Virginia Bilanchone for critical reading of the manuscript and Ming Hu for technical assistance. This work was in part supported by an NIH Research Grant R01 AR47320 and a U.S. Army Grant W81XWH-04-1-0516 awarded to X.D.
Footnotes
Sequence data from this article have been deposited with the EMBL/GenBank Data Library under accession number AY486147 (mouse mpygo2 full-length cDNA).
References
- 1.Huelsken J, et al. Beta-catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 2.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 3.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 4.Andl T, et al. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 5.Alonso L, Fuchs E. Stem cells in the skin: waste not, Wnt not. Genes Dev. 2003;17:1189–1200. doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- 6.Huelsken J, Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 7.Zorn AM. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 8.Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishita M, et al. Interaction between Wnt and TGF-beta signaling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 10.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signaling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 11.Cavallo RA, et al. Drosophila Tcf and Groucho interact to repress Wingless signaling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 12.Gradl D, Kuhl M, Wedlich D. The Wnt/Wg signal transducer beta-catenin controls fibronectin expression. Mol Cell Biol. 1999;19:5576–5587. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valenta T, Lukas J, Korinek V. HMG box transcription factor TCF-4’s interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31:2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson B, et al. A new nuclear component of the Wnt signaling pathway. Nat Cell Biol. 2002;4:367–373. doi: 10.1038/ncb786. [DOI] [PubMed] [Google Scholar]
- 16.Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- 17.Kramps T, et al. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 18.Belenkaya TY, et al. Pygopus encodes a nuclear protein essential for wingless/Wnt signaling. Development. 2002;129:4089–4101. doi: 10.1242/dev.129.17.4089. [DOI] [PubMed] [Google Scholar]
- 19.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 20.Townsley FM, Thompson B, Bienz M. Pygopus residues required for its binding to Legless are critical for transcription and development. J Biol Chem. 2004;279:5177–5183. doi: 10.1074/jbc.M309722200. [DOI] [PubMed] [Google Scholar]
- 21.Lake BB, Kao KR. Pygopus is required for embryonic brain patterning in Xenopus. Dev Biol. 2003;261:132–148. doi: 10.1016/s0012-1606(03)00305-1. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 23.Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- 24.Brault V, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;198:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 25.McMahon AP, et al. The midbrain-hindbrain phenotype of Wnt-1−/Wnt-1− mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- 26.Mastick GS, et al. Early deletion of neuromeres in Wnt-1−/− mutant mice: evaluation by morphological and molecular markers. J Comp Neurol. 1996;374:246–258. doi: 10.1002/(SICI)1096-9861(19961014)374:2<246::AID-CNE7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Serbedzija GN, Dickinson M, McMahon AP. Cell death in the CNS of the Wnt-1 mutant mouse. J Neurobiol. 1996;31:275–282. doi: 10.1002/(SICI)1097-4695(199611)31:3<275::AID-NEU1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Ikeya M, et al. Wnt signaling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 29.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 31.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]