Abstract
Background
Kinase inhibition is an increasingly popular strategy for pharmacotherapy of human diseases. Although many of these agents have been described as “targeted therapy”, they will typically inhibit multiple kinases with varying potency. Pre-clinical model testing has not predicted the numerous significant toxicities identified during clinical development. The purpose of this study was to develop a bioinformatics-based method to predict specific adverse events (AEs) in humans associated with the inhibition of particular kinase targets (KTs).
Methods
The AE frequencies of protein kinase inhibitors (PKIs) were curated from three sources (PubMed, Thompson Physician Desk Reference and PharmGKB), and affinities of 38 PKIs for 317 kinases, representing > 50% of the predicted human kinome, were collected from published in vitro assay results. A novel quantitative computational method was developed to predict associations between KTs and AEs that included a whole panel of 71 AEs and 20 PKIs targeting 266 distinct kinases with Kd < 10uM. The method calculated an unbiased, kinome-wide association score via linear algebra on (i) the normalized frequencies of AEs associated with 20 PKIs and (ii) the negative log-transformed dissociation constant of kinases targeted by these PKIs. Finally, a reference standard was calculated by applying Fisher’s exact test to the co-occurrence of indexed Pubmed terms (p≤0.05, and manually verified) for AE and associated kinase targets (AE-KT) pairs from standard literature search techniques. We also evaluated the enrichment of predictions between the quantitative method and the literature search by Fisher’s Exact testing.
Results
We identified significant associations among already empirically well established pairs of AEs (e.g. diarrhea and rash) and KTs (e.g. EGFR). The following less well recognized AE-KT pairs had similar association scores: diarrhea-(DDR1; ERBB4), rash-ERBB4, and fatigue-(CSF1R; KIT). With no filtering, the association score identified 41 prioritized associations involving 7 AEs and 19 KTs. Among them, 8 associations were reported in the literature review. There were only 78 out of a total of 4,522 AE-KT pairs meeting the evaluation threshold, indicating a strong association between the predicted and the text mined AE-KT pairs (p= 3×10−7). As many of these drugs remain in development, a larger volume of more detailed data on AE-PKI associations is accessible only through non-public databases. These prediction models will be refined with these data and validated through dedicated prospective human studies.
Conclusion and future directions
Our in silico method can predict associations between kinase targets and AE frequencies in human patients. Refining this method should lead to improved clinical development of protein kinase inhibitors, a large new class of therapeutics.
Keywords: Adverse event, toxicity, kinome, kinase inhibitor, computational modeling, translational bioinformatics
1. INTRODUCTION
Motivation
Protein kinase inhibitors (PKIs) are a new class of drugs. Directly or indirectly, kinases regulate nearly every process of cells and tissues. Scientists first described the structure and function of protein kinases through their studies of the molecular basis of cancer. Many of the first discovered “oncogenes” proved to be mutant forms of the genes encoding protein kinases that resulted in autonomous or dysregulated kinase activity. Based on these concepts, PKIs were initially developed to treat cancer, and the first such drug to be approved for marketing by the U.S. Food and Drug Administration was imatinib (Gleevec™) for treatment of chronic myelogenous leukemia (CML). Touted as “targeted therapy”, imatinib revolutionized treatment and prognosis for this disease[1]. But characteristic of PKIs, imatinib not only inhibits the Bcr-Abl oncogene specific to CML, but also other kinase targets (KTs) such as c-Kit, and platelet derived growth factor receptor alpha (PDGFRA). As imatinib was used in increasing numbers of patients, unexpected adverse events (AEs) such as hypophosphatemia[2] and cardiac dysfunction[3] were identified. This pattern of inhibition of both intended and unintended KTs with clear therapeutic benefit limited by unexpected AEs has been reproduced with every PKI approved for widespread use. Many similar agents have failed or been stalled during clinical development because of insufficient benefit and unexpected toxicities. Better understanding of the physiologic consequences of inhibiting specific KTs should lead to development of safer cancer therapeutics, more effective combination treatments, and support the cross-purposing of these drugs from cancer care for use in other diseases[4]. Specifically, confirming the absence of an adverse event is challenging in conventional pre-clinical trials because these trials may not be conducted in animal species exhibiting the AE. For example, rodents may be used to defined efficacy; however, this species is highly resistant to nausea and exotic animal species are required to determine AEs that could occur in human. Consequently, the development of a high throughput computational method over in vitro assays may offer the opportunity to specific the requirements of specific pre-clinical assays to rule out putative AE identified in silico, accelerating the drug development process and reducing costs by avoiding discovery of unexpected AEs of a compound in clinical trials.
Biological Background
Protein kinases constitute much of the signaling pathways and interactive cellular signaling networks. More than 500 protein kinases are encoded in the human genome. The collection of these evolutionarily conserved, modularly structured enzymes is referred to as the kinome[5]. Given the evolutionary history of these proteins reflected in shared sequences, structures, and functions, it is not surprising that drugs screened for capacity to inhibit specific kinases inevitably seem also to inhibit some other kinases unintentionally. Specificity has typically been assessed in in vitro cellular assays. These assays are typically not standardized among laboratories and cross-reacting kinases might not be relevant to cellular function or even be expressed at all in the particular tested cells. Furthermore, the non-conserved sequences of kinase genes contribute to the expression and functional differences in cellular signaling that produce the metabolic and physiologic differences among species, so animal toxicity findings do not consistently predict effects in humans. Consequently, the introduction of PKIs to human subjects has led to novel observations not only for clinical therapeutics but for better understanding human molecular physiology[4].
Although many methods have been developed[6, 7] to predict chemical structure/kinase inhibition relationships, there has been no dedicated effort to associate inhibition of specific kinases with physiologic consequences through systems analysis approaches. Two major obstacles have been observed 1) the limited, labor-intensive assessment of PKI specificity through cellular assays without central laboratory standardization, and 2) limited concurrent comparisons of PKIs in clinical use or development on the same platform. Recently, Zarrinkar and colleagues presented the largest comparative analysis of PKI selectivity (including 38 PKIs) with an unbiased in vitro kinome-wide binding assay[8]. Although with acknowledged limitations, their data provide the opportunity to cross-compare effects of many PKIs in clinical use and development. The structured reporting of AEs in the clinical trials that have tested these agents provides a database with which to infer AE-KT relationships.
Bioinformatics Background
Although there exist many computational approaches to predict individual drug targets, few studies pertain to many targets. We summarized in Table 1 those studies that access multiple-target predictions (high throughput). Some excellent studies focused on computationally predicting the target of drugs (bottom part of the table), while others focused on their toxicities (middle part). On the first row, the present study, focuses on kinases, uses the physical kinome map and provides an evaluation of adverse event relationships, elements that contrast with the majority of other studies.
Table 1.
The comparison with related studies
| Adverse Event- Drug |
Drug Target Prediction |
Protein target- Toxicity |
PKI-Specific Input Data Size |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Literature | Physical Kinome |
Literature | Chemical Structure |
Compt. Prediction |
Evaluation | Kinome-wide Analysis |
Literature | Compt. Prediction |
Evaluation | Adverse Events(#) |
Kinase Inhibitors(#) |
Kinase targets(#) |
|
| Studies Focusing on kinaes Toxicities | |||||||||||||
| Present study | ▪ | ▪ | ▪ | ▪ | C,M | 71 | 20 | 266 | |||||
| General Studies predicting target Toxicities (not focused on kinases) | |||||||||||||
| Hansen 2009[9] | ▪ | ▪ | B | ▪ | 3 | 1* | - | ||||||
| Bender A, et al. 2007[10] | ▪ | ▪ | ▪ | ▪ | 166 | - | 0* | ||||||
| DART 2003[11] | ▪ | ▪ | ▪ | 187 | - | - | |||||||
| Apic 2005 [12] | ▪ | ▪ | - | - | - | ||||||||
| Studies predicting drug targets, but not their Toxicities | Not Applicable | ||||||||||||
| Fliri 2007 [13] | ▪ | ▪ | C,B | 5,923 | 2* | 168* | |||||||
| Garten 2009 [14] | ▪ | 395*(a) | 21*(b) | 121*(c) | |||||||||
| Liebovitch 2007 [15] | ▪ | - | - | - | |||||||||
| Campillos M, et al. 2008[16] | ▪ | ▪ | ▪ | B | 727 | 3* | - | ||||||
C=computational evaluation, B=biological validation, M= manual curation,
count,
: subset of gene targets with protein kinase activity (GO:0004672) or subset of drugs that are kinase inhibitors;
-: no specific detail about the input data
by searching the key words “side effect” in the Pharmspresso on line database (http://pharmspresso.stanford.edu/ygarten/Pharmspresso/html/index.html)
by searching the key words “human”, “kinase” and “Inhibitor” in the Pharmspresso on line database
by searching the key words “human” and “kinase” in the Pharmspresso on line database
Our methodological approach also differs from that of these other publications. First, we used text mining technology to evaluate our computationally predicted results. A few others undertaking such large-scale approaches have incorporated text-mining and natural language processing to unlock relevant data from disparate and heterogeneous sources. For example, Kuhn et al. have created a search tool for interactions of chemicals and proteins, coined “STITCH”, that consolidates chemicals and draws relationships between the chemicals and their activity data in cell lines, MeSH assignments, and literature[17]. Textpresso and other text mining approaches were used to curate the relationships between drugs and genes[11, 14]. However, simple text-mining would not provide enough statistical power for a group of new drugs under active clinical development, since there is insufficient openly accessible literature. For example, searching Pharmspresso by “EGFR side effect” results in no match[14]. Second, we developed computational prediction analyses of toxicity of kinase inhibitors based on kinome-wide association between kinase inhibitors and their targets. Prior studies of adverse effects have mostly generated or used molecular pathway atlases to assess structure-function correlations in complex systems. For example, by understanding the physiological pathways and potential binding partners of a candidate drug, it is theoretically possible to anticipate adverse events of the candidate if the consequences of binding to these non-target partners are known. One commercial provider, Cambridge Cell Networks (CCNet) has created the “PathTox” tool that offers a sequence search in their pathway atlas that reports the probable secondary effects of candidates binding to specific partners[12]. Further, a research group at Pfizer has been able to forecast the effects of drugs on a large scale using computational techniques to establish relationships between structure and function to predict the probability of two drugs exhibiting similar system-wide effects[13]. A recent study based on integration of data on gene-drug interactions, gene-interaction and drug-drug similarity predicted novel candidate genes that might affect inter-individual differences in metabolism, effectiveness, or adverse events for four drugs including one PKI gefitinib[9]. Third, though the feasibility of using adverse events to reveal the molecular and genetic interactions with drugs in humans has already been established, we are the first to develop computational methods focused on the complex, often overlapping binding/inhibition profiles of PKIs specifically. A relevant recent study uncovered 12 drug-target relationships by means of predicting relationships using phenotypic adverse event similarities; they further validated all of these relationships and successfully confirmed 9 of the relationships using in vitro binding assays[16]. Recent computational studies predicted receptor binding based on network features[18], or chemical similarities[19]. In contrast, this study uses in vitro kinome and clinical trials reports to predict mechanisms of toxicities attributable to the off-target binding of kinase inhibitors. . Others predicted combination of drugs with desired effects based on chemical structures of drugs[15]. A pilot study predicted toxicity related targets by combining drug off-targets binding and adverse reactions using Bayesian methods[10], however, their resulting 70 targets contain no kinase targets.
Rationale
To predict computationally the toxicities resulting from inhibition of specific KTs we developed a novel quantitative method which was designed to be comprehensive and unbiased, including all available information on AEs and PKIs. It provided a proof-of-concept that AE-KT relationships can be predicted by analyzing the specific AEs induced by multiple inhibitors and the propensity of these inhibitors to bind specific KTs on a physical kinome map. We then evaluated the prediction method by comparing the results to evidence from literature mining.
Contribution
To our knowledge, we have performed the first computational prediction of toxicity of kinase inhibition using kinome-wide physical mapping. Our work contributed a table of PKIs with reported adverse events and their prevalence.
2. METHODS
In this paper, we aim to offer a proof-of-concept by presenting a significant correlation between frequencies of adverse events and kinome inhibition patterns through a survey of emerging kinase inhibitors in 3 modules: 1) Literature mining to generate quantity mapping between PKIs and adverse events; 2) Computationally predicting the significant association between targets and adverse events using two different methods; 3) Evaluating the predicted results by independent literature searching of co-occurrence and significance estimation. Figure 1 introduces the data and their relationship and Figure 2 demonstrates the three modules.
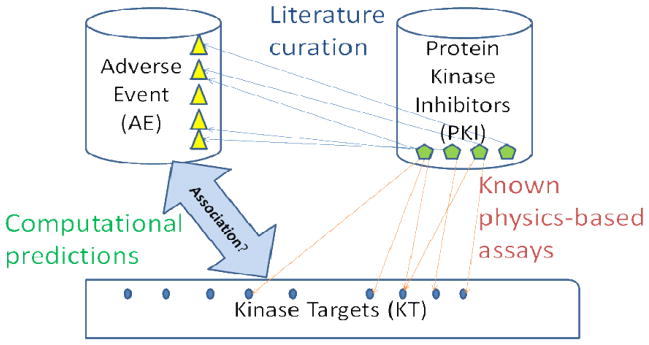
Figure 1. Illustration of raw data and their relationship in this study.
The frequencies of adverse events (AEs) associated with kinase inhibitors were curated from three sources (Thompson Physician Desk Reference, PharmGKB and PubMed). The dissociation constants (Kd) of kinases were summarized from physico-chemical assays[8]. The computational method in this study considers all of these data to predict the association between AEs and targets of kinase inhibitors.
Figure 2. The study consists of three modules: Data organization, computational prediction and computational evaluation.
In the module of Data organization, two steps generated two different associations used as the inputs to the “prediction of kinase toxicity” module. In step ✠, a dissociation constant is found in a physical kinome map, while in step ✠, drug adverse events were curated from multiple sources. In the modules of prediction of kinase toxicity (step ✠), a quantitative method was developed to identify the prioritized AE-KT association scores (PAS). In the evaluation (step ✠), Pubmed was mined to identify enriched AE-KT pairs from a total of 4,522 putative pairs among 17 AEs that have more than five PKI repeats and 266 observed kinases. Then the findings of the predictive method were evaluated against the text mining results.
2.1 Data Organization
2.1.1 Kinome Map Collection and Procession
The physical kinome map was summarized from novel PKI-KT dissociation constant (Kd) data for 38 kinase inhibitors against a panel of 317 kinases[8]. Three steps were performed before statistic computation: First, mutated targets were excluded, except for those without assay for their non-mutation targets which were GCN2, JAK1, JAK2 and JAK3, since the homologous gene targets (mutated kinase) are repeated measures of the related targets comparing to those of more independent genes. Second, 20 PKIs having curated AEs with frequency of incidence in Supplemental Table 1 were included. For example, albeit Flavopiridol is reported to cause diarrhea in some patients, the accurate frequency could not be assessed and enrolled into the Supplemental Table 1, and thus were not included. Finally, we took a negative log-transformation of all Kd values, because logarithm transformation reduces detection noise to an additive level and tends to convert exponential distribution trends to normal distribution trends (Suppl. Figure 1).
2.1.2 Curation for Relationship between PKIs and Adverse Events from various Knowledge Sources (Suppl. Table 1)
Adverse Events Source Selection
To locate the adverse events data for the collection of the 38 kinase inhibitors[8] with kinase binding information, we reviewed a clinical pharmaceutical resource - Thomson Physician’s Desk Reference 2008[20] (PDR), a biomedical informatics resource - PharmGKB, as well as Medline-indexed primary research articles via PubMed. Given the heterogeneity of the PKI data reported in our sources, data found in PDR and PharmGKB was given priority over any journal-derived clinical data. When we failed to find an agent in either PDR or PharmGKB, the most recent journal articles with the most explicitly quantified adverse event data were used. To cross-check the validity of the collected adverse event frequency values in PDR and PharmGKB, we also compared the adverse event frequency data for PKIs found in the electronic PKI database Facts & Comparisons (Version 4.0) for consistency.
Adverse Event Normalization
We normalized both the frequency values and the symptom/adverse event terms of the adverse event data extracted from PharmGKB, PDR, and recent literature. We reported AE frequencies as percentage values. First, to normalize AE frequencies reported in the literature as a range or as different frequencies from various sources, we took the mean value of the range. Additionally, for frequencies presented in a series of comparative doses (e.g. 10mg vs. 100mg vs. 200mg) we used the AE frequencies associated with the highest dose. Second, to normalize the AE terms we created logical AE terms to consolidate related AEs (e.g. “Rash” encompasses “skin Rash”, “vesiculobullous rash”, “acneiform rash”, and “maculopapular rash”). For simplification, we hereafter refer to these normalized AE as AE.
Expected Adverse Event Frequency in Population
In practice, a universal threshold indicating a frequency above that of placebo for all adverse events was inappropriate. Indeed, certain adverse events like headache or back pain occur at much higher incidence in the population than that of neutropenia. Therefore, we reviewed the literature to find out the expected frequency, and then decided a little severer threshold (generally by 50–100% above expected frequency) for every adverse event. The literature sources comprise of: 1) wrong diagnosis (http://www.wrongdiagnosis.com/p/pain/prevalence-types.htm); 2) emedicine.com (http://www.emedicine.com/emerg/topic233.htm); 3) population health metrics (http://www.pophealthmetrics.com); 4) PKIs.com (http://ww.PKIs.com) 5) DailyMed (http://dailymed.nlm.nih.gov); 6) Merck manual online; 7) Google “adverse event” prevalence.
2.2 Computational Prediction on Associations Between Adverse Events and the Kinome
The method was designed to take into account every piece of evidence from the adverse events, kinases and kinome map. The inputs were two quantitative association matrices: the curated AE-PKI frequency matrix and the PKI-KT binding affinity matrix, respectively (Figure 2).
We introduce definitions as follows: (dx), x=1,…,p, denotes a set of protein kinase inhibitor s(PKIs); (ty), y=1,…,q, denotes a set of kinase targets (KTs); and (ez), z=1,…,r, is a set of adverse events (AEs).
Definition 1
The AE-KT association matrix A=(azy) consists of an association score that reflects the association between the kinase target ty and the adverse event ez which is a (r × q) dimension matrix.
Definition 2
The AE-PKI association matrix F=(fzx), a (r × p) dimension matrix, consists of the frequency of normalized adverse event ez for a PKI dx which were curated from three sources (PubMed, Thompson Physician Desk Reference and PharmGKB).
Definition 3
The PKI-KT relationship matrix K=(kxy), which is a (p × q) dimension matrix, consists of an experimental dissociation constant value (Kd value) for a PKI dx targeting ty that was reported in literature[8].
Prioritized Association Scores (PAS)
The PAS method assumes that the toxicity of a single kinase can be repeatedly observed by multiple PKIs targeting this kinase with respects to both their combining affinity and their AE frequencies. By an association operation, the matrix multiplication, we scored the AE-KT associations (Figure 3) as:
| (1) |
where F and K were the normalized frequency of AEs and the dissociation constant of PKIs, respectively. The higher the score, the more prioritized the association of the kinase with the corresponding adverse event would be. Following the study of the false discovery rate, a discovery threshold was set as the top (0.2% quantile) of all observed scores in this study. Moreover, we traced back to identify the PKIs that associated both to the kinase and the adverse event of finding. These PKIs are much of interests because they should be avoided of together using due to the higher possibility of inducing the same adverse event. False discovery rates were computed on each score by permutation resampling of K. Multiple comparisons which were adjusted using the q-values[21, 22]. R language source of the Prioritized Association Scores method is available at http://www.lussierlab.org/publication/PAS/.
Figure 3. The association scores identified prioritized AE-KT pairs were included to generate the output bipartite network.
The AE-KT prioritized association scores in matrix A were constructed by applying a matrix multiplication on the AE-PKI frequency matrix F and the PKI-KT binding affinity matrix K. A prioritized AE-KT pair has an association score higher than the top (0.2% quantile) of all observed scores (q-value < 21%).
2.3 Evaluation
Text Mining of PubMed
To predict the adverse event of kinase by significant co-occurrence in abstracts indexed in PubMed, we used Pubmatrix[23], a multiplex literature mining tool. Let N be the totally records in PubMed, we created a contingency table (Table 2) for each putative pair of AE-KT: n2 is the number of abstracts containing a kinase t, n3 is the number of abstracts about an adverse event e, and n1 is the number of abstracts of both. Then one-sided Fisher’s exact test was applied to evaluate the significance of association between each pair of kinase target and adverse event[7, 24]; and a gold standard of unadjusted p ≤ 0.05 was set as significant for a co-occurrence. We further manually verified the computationally corroborated pairs and rejected targets whose symbols appearing in abstracts refer to other objects instead of genes, e.g. CIT is the abbreviation for “city”, YES refers to “yes”, MET refers to “Methods” or “metabolic equivalents”, Gak as institute name, and TEC is the abbreviation for “toxigenic Escherichia coli” or “total eosinophil counts” in many abstracts.
Table 2.
Contingency table for each AE-KT pair using text mining of Pubmed
| #ref. include kinase t | #ref. not include kinase t | Σ | |
|---|---|---|---|
| #ref. include AE e | n1 | n3−n1 | n3 |
| #ref. not include AE e | n2−n1 | N−n2−n3+n1 | N−n3 |
| Σ | n2 | N−n2 | N |
Using a Fisher’s Exact Test, we further evaluated the enrichment (overlap) between the AE-and-KT co-occurrences in pubmed identified by computational text mining and the AE-KT pairs predicted by the association score method conducted over the kinome map.
3. RESULTS
3.1 Mapping PKIs and adverse events using literature curation
Karaman MA et al. successfully mapped an interaction for 38 PKIs (drugs) across a panel of 317 kinases[8]. Our literature curation resulted in 71 normalized AEs induced by 20 of these 38 PKIs from 181 literature-reported adverse drug activities (Figure 2). The primary analysis was performed on the minimal panel of the 266 distinct kinases targeted by 20 PKIs, whereas the 20 PKIs had 71 reported adverse events. Suppl. Table 1 gives the details of normalized adverse events, their prevalence and the estimated threshold of frequency. In the table, the adverse event “Asthenia” is merged with “Fatigue” because of the similarity of these two categories; conversely, the adverse event “Pain” is further subdivided into “Back Pain”, “Chest Pain” and “Pain” because of their different ratios of incidences. A smaller panel of inputs for evaluation, the 17 AEs with more than 5 repeats of PKIs that target 266 distinct kinases is described in Suppl. Table 2.
3.2 Computational prediction of significant associations between AEs and kinase targets (Methods, Formula 1)
We identified 41 prioritized AE-KT associations among 19 targets and 7 adverse events using a threshold of the top of 0.2% quantile of all 18,866 (71×266) scores (Suppl. Figure 2 and Suppl. Table 3). Eight prioritized associations by PAS were also independently related to significant co-occurrences identified by text mining: nausea to ERBB2, fatigue to VEGFR2, diarrhea to both KIT and EGFR, rash to both EGFR and ERBB2, and asthenia to both ERGF and ERBB2 (red lines in Suppl. Figure 2 and green lines in Suppl. Figure 3). Figure 4 is a bipartite network of the identified associations summarized from the associations between KTs and AEs after merging the vertices of similar AEs into one vertex, which are nausea and vomiting, asthenia and fatigue.
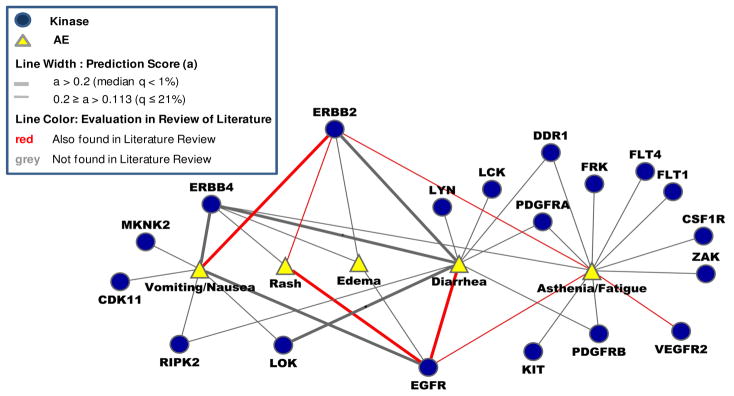
Figure 4. The predicted network of adverse events of kinase targets using association score.
Bipartite network of 41 prioritized AE-KT pairs with the top PAS association scores (a score > 0.2; the best 1/5 of 1% of all calculated scores) consisting of 7 distinct AEs and 19 distinct kinase targets. As shown in the plot, a wider line indicates a higher priority of association. In additional, the predicted AE-KT associations by text mining of PubMed (Fisher’s exact test p≤0.05) are resented in red color. In our evaluation using text mining, a Fisher’s Exact test showed a significant (p=3×10−7) association between the literature review among these 41 predicted AE-KT pairs. Pairs of similar vertices (Nausea and Vomiting, Asthenia and Fatigue) were each merged into a single vertex, respectively (For more details, see Suppl. Figure 2).
3.3 Evaluation
Independently, we searched PubMed using the on line tool Pubmatrix[23] for every putative pair of kinase targets and adverse events. For simplification, we used a smaller panel of inputs, the 17 AEs with more than 5 repeats of PKIs that target 266 distinct kinases. This panel evaluates the literature co-occurrence of 4,522 (266×17) putative AE-KT pairs, which excludes only 3 captured AE-KT pairs from a total of 5,320 (266×20) putative pairs in PAS. As of Oct 21, 2008, there were a total of 19,028,626 (N) abstracts indexed in PubMed. Among them, only 105 AE-KT pairs meet the text mining gold standard (the unadjusted Fisher’s exact tested p-value ≤ 0.05). A stringent correction of p-values turned out to be so conservative that many known associations between target of PKI and its side event were rejected. For example, the known association between Nausea and ERBB2[25], and between EGFR and mucositis[26] will be rejected at a 5% level of adjusted p-values (Suppl. Table 4). Because our main goal is to generate hypotheses, we chose to use the unadjusted FET p-value for a primary evaluation of the computational prediction. After manual verification, 27 pairs involving five targets, which were CIT, YES, MET, Gak and TEC, were rejected due to their searched symbol representing other objects instead of gene in the abstracts indexed in PubMed (For details, see Suppl. Table 4). This kind of rejection keeps the evaluation unbiased but removes a significant number of putative associations between side events and targets. Improved MeSH term qualifiers or text mining approaches may improve the accuracy of the results. Thereafter, 78 predicted AE-KT pairs were presented in as the predicted network of side effects and kinase targets using computational text mining of literature indexed in PubMed (Suppl. Figure 3). Eight out of 41 AE-KT pairs predicted using association score were also predicted by literature review. A Fisher’s Exact test showed a strongly significant association between the discovery of our quantitatively computational method and the literature review: p-value=3×10−7
4. DISCUSSION
Our study recapitulated well recognized associations between AEs and widely used PKIs. The two strongest relationships captured are diarrhea, a recognized, mechanism-based adverse event of epidermal growth factor receptor (EGFR) inhibition, and the other common EGFR inhibitor-induced AE, rash. The rash caused by EGFR kinase inhibitors has been associated with clinical response for multiple cancers, and this serves as evidence that intended KTs have both salutary and adverse effects where the relationship was confirmed by the PAS method[27–29]. Examination of these strongest relationships is useful for assessing our methods development, making modifications for future analyses, and generating hypotheses that might be easily validated on existing clinical specimens or data.
The cumulative adverse event data in this study are collected from phase I, phase II, phase III, and other clinical investigations of numerous kinase inhibitors, both those FDA approved and those not yet completing clinical development; a larger sample than any individual trial. These studies describe the full spectrum of adverse events both drug-related and disease-related. In any single trial (especially if this trial is not placebo controlled- the typical case for oncology phase I trials) it is frequently not apparent which toxicities are reproducibly attributable to the drug, and it is certainly not clear if the adverse events are due to the mechanism of action of the drug or an “off-target” effect. The comparison of drugs with overlapping and non-overlapping molecular binding properties provides a rational approach to determining which adverse events, dose-limiting or not, might be attributed to the inhibition of which targets. Indeed, many of the so-called targeted kinase inhibitors have been shown to target a wider spectrum of kinases than originally planned[30].
An unexpected and exciting finding is the identification of the CSF1R/fatigue and KIT/fatigue associations. As depicted on the kinome dendrogram[31], CSF1R, KIT and VEGFR2 are three kinases with similar sequence and physical structure but with biological roles important to different cell subsets. VEGFR2 is almost exclusively expressed on endothelial cells and is activated in tumor angiogenesis.
Therefore this has been the primary target for several new cancer therapeutics such as sunitinib and AMG-706 (now known as motesanib). CSF1R plays a major role in granulocyte/monocyte/macrophage development, signaling, and regulation of cytokine release and response. KIT signaling has been reported to mediate various roles in different tissues, but is most prominently involved in proliferation and differentiation of various hematopoietic lineages and in mature mast cell and eosinophil signaling. The agents developed to inhibit the structurally similar VEGFR2 tend to have relatively high potency for CSF1R and KIT and fatigue is a common, sometimes dose-limiting adverse effect of these drugs. Although we have identified a strong association between these kinases and fatigue, it remains unclear whether one, two, or all three of these KTs mediates the fatigue. If any of the three is differentially associated, there will be the opportunity in either development of new drugs or in administration of combinations of PKIs to decrease the associated fatigue by either selectively eliminating the offending KT affinity or by selecting 2 PKIs that when added together will maximize inhibition of the intended therapeutic KT, while reducing the inhibition of the unintended, fatigue-inducing target. Given the unbiased identification of this AE-KT association and its strength relative to the panel of relationships tested, our clinical program has commenced studies of human circulating protein biomarkers for disruption of CSF1R signaling and validation studies of the relationship between inhibition of CSF1R and fatigue in cancer patients treated with PKI’s that target VEGFR2.
Mined facts from Pubmed used in the evaluation are specific for each drug and, to our knowledge, only in very few specific cases have been generalized to a group of drugs. While this is a limitation of the evaluation, we believe this may be the first high throughput evaluation of PKI toxicities. While the gold standard of drug target prediction, an older field, is the biological validation in vitro, drug toxicities predictions require a much more complex design for two reasons: in vitro cellular studies cannot reflect more than a cellular toxicity (animal models would be required for most clinical toxicities such as rash, vomiting, etc), and because of interspecies differences in kinase structures and functions animals may frequently not manifest the toxicities seen in humans. Finally, a specific drug target prediction is not generalized to a class of drug, while an adverse event associated to a kinase (target) is a generalized “systems” property and requires a more comprehensive design for validation.
Future studies and limitation
Adverse events occur in clinical studies where the tissue perfusion of kinase inhibitor depends on its specific pharmacokinetics, while the in vitro binding affinity of a kinase receptor to a kinase is measured as dissociation constant Kd. Our approach provides calculations of AE and Kd jointly to create a compound score (PAS) and control for multiplicity. Thus significant PAS scores are associated to a specific kinase, and may imply different underlying Kd values for this kinase under different drug AE. It is a known fact that each drug may have a specific pharmacokinetic profile, and thus the corresponding relevant in vitro Kd remains data driven in our study. This kind of limitation in our study is also shared with previous published efforts to use incidence of adverse events as a means to infer previously unrecognized and untested drug/target relationships[9–11, 13, 14, 16]. An important advance upon the current method will be to integrate data on physiologically relevant pharmacokinetic and pharmacodynamic properties of different drugs and targets. This will be most readily accomplished by first performing clinical validation of some of the associations identified in this study.
We plan to conduct future studies to validate and extend the method with toxicity data from clinical trials of novel kinase inhibitors sponsored by the National Cancer Institute. Better quantitative gold standard data for adverse events might have been obtained from the US adverse event report system (AERS) and the Canadian adverse event reporting and learning system (CAERLS). Additionally, a standardized ontology for adverse event is needed which might be taken from Health Canada’s Canadian Adverse Drug Reaction Monitoring Program (CADRMP, http://www.hc-sc.gc.ca/dhp-mps/medeff/databasdon/index-eng.php). Biological validation of any specific target cannot be conducted using this method and would be a complex pursuit. However, for strong candidates with multiple independent, but consistent data sources, we would consider a human clinical trial the best, gold standard approach to biological validation. Additionally, we will explore additional models involving pathways, multi-kinase effects, and other models of drug/kinase inhibition beyond log-linear ones. However, by design, the physical kinome map limits the breadth of our predictions to kinase toxicities. Arguably, other non-kinasetargets may exist and are beyond our calculations; however it will be challenging to distinguish non-target specific effects from those of collective kinase inhibition. There is increasing evidence that many PKIs aren’t as specific as the concept of “targeted therapy” implies[30].
5. CONCLUSION
Predicting toxicities of kinase inhibitors is an important problem for clinical development of new drugs. Current methods for predicting toxicities mainly rely on pre-clinical cell and animal studies[32]. Our in silico method can predict associations between kinase targets and AE frequencies in human patients. This method was controlled by false discovery, literature and expert review. Expert review further confirmed that the method recapitulates existing empirical and mechanistic knowledge and provides novel and clinically relevant insights (e.g. CSF1R/fatigue and KIT/fatigue). To our knowledge, this is the first study using kinome binding patterns for predictions that proposes a novel Prioritized Association Score and permutation resampling for increased sensitivity of results in spite of the scarcity of information about adverse effects of kinase inhibitors.
Refining the method should lead to improved clinical development of protein kinase inhibitors, a large new class of therapeutics[4]. 1) As some AE-KT relationships will only be apparent at the system physiology level, comprehensive screening for AE-KT relationships in patients exposed to these drugs provides the most readily available method for detecting these relationships. 2) This approach bypasses the species differences in drug pharmacokinetics/pharmacodynamics that impair translation from animal models. 3) Knowing AE-KT relationships can improve the safety and speed the completion of early trials of new kinase inhibitors. For example if inhibition of a particular kinase is associated with hypertriglyceridemia, then determining the active dose range of a novel PKI that blocks that KT could be accelerated by measuring serum triglyceride elevations whether or not that KT is the intended target. 4) This approach also can inform selection from among the thousands of possible combinations of currently available PKI’s, those that have the greatest likelihood for non-overlapping toxicity and hence a better therapeutic index. 5) Finally, uncovering AE-KT relationships with novel PKI’s can reveal new insight into the molecular physiology of the human body, leading to the identification of risk factors for bad outcomes for some drugs and new clinical indications for others[4].
Supplementary Material
Acknowledgments
This work was supported in part by the NIH/NLM/NCI National Center for Multiscale Analyses of Genomic and Cellular Networks (MAGNET, 1U54CA121852), the NIH/NCRR Clinical & Translational Science Awards (1U54 RR023560-01A1), The Cancer Research Foundation, The University of Chicago Cancer Research Center, and The Ludwig Center for Metastasis Research. MLM is supported by Mentored Career Development award K23CA124802. XY is partly supported by the Natural Science Foundation of China 60971099.
Abbreviation
- PKI
Protein Kinase Inhibitor
- KT
Kinase Target
- AE
adverse event
- Kd
dissociation constant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Berman E, et al. Altered bone and mineral metabolism in patients receiving imatinib mesylate. N Engl J Med. 2006;354(19):2006–13. doi: 10.1056/NEJMoa051140. [DOI] [PubMed] [Google Scholar]
- 3.Kerkela R, et al. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12(8):908–16. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 4.Maitland ML, Ratain MJ. Terminal ballistics of kinase inhibitors: there are no magic bullets. Ann Intern Med. 2006;145(9):702–3. doi: 10.7326/0003-4819-145-9-200611070-00015. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Muster W, et al. Computational toxicology in drug development. Drug Discov Today. 2008;13(7–8):303–10. doi: 10.1016/j.drudis.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal P, Searls DB. Literature mining in support of drug discovery. Brief Bioinform. 2008 doi: 10.1093/bib/bbn035. [DOI] [PubMed] [Google Scholar]
- 8.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 9.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther. 2009;86(2):183–9. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender A, et al. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. Chem Med Chem. 2007;2(6):861–73. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 11.Ji ZL, et al. Drug Adverse Reaction Target Database (DART): proteins related to adverse drug reactions. Drug Saf. 2003;26(10):685–90. doi: 10.2165/00002018-200326100-00002. [DOI] [PubMed] [Google Scholar]
- 12.Apic G, et al. Illuminating drug discovery with biological pathways. FEBS Lett. 2005;579(8):1872–7. doi: 10.1016/j.febslet.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Fliri AF, Loging WT, Volkmann RA. Analysis of system structure-function relationships. Chem Med Chem. 2007;2(12):1774–82. doi: 10.1002/cmdc.200700153. [DOI] [PubMed] [Google Scholar]
- 14.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebovitch LS, Tsinoremas N, Pandya A. Developing combinatorial multi-component therapies (CMCT) of drugs that are more specific and have fewer side effects than traditional one drug therapies. Nonlinear Biomed Phys. 2007;1(1):11. doi: 10.1186/1753-4631-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campillos M, et al. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–6. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–8. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim MA, et al. Drug-target network. Nat Biotechnol. 2007;25(10):1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 19.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–81. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson Healthcare Staff. Physicians’ Desk Reference 2008. Matthews Medical Books; 2008. [Google Scholar]
- 21.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B. 2002;64:479–498. [Google Scholar]
- 22.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- 23.Becker KG, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003;4:61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher RA. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. Journal of the Royal Statistical Society. 1922;85(1):87–94. [Google Scholar]
- 25.Warm M, et al. Side-effects of pre-operative epirubicin-paclitaxel therapy in primary breast cancer associated with tumor biology. Anticancer Res. 2009;29(7):2675–80. [PubMed] [Google Scholar]
- 26.Lacouture ME, et al. A proposed EGFR inhibitor dermatologic adverse event-specific grading scale from the MASCC skin toxicity study group. Support Care Cancer. 2010;18(4):509–22. doi: 10.1007/s00520-009-0744-x. [DOI] [PubMed] [Google Scholar]
- 27.Cohen EE, et al. Factors associated with clinical benefit from epidermal growth factor receptor inhibitors in recurrent and metastatic squamous cell carcinoma of the head and neck. Oral Oncol. 2009 doi: 10.1016/j.oraloncology.2009.05.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Soler R, Saltz L. Cutaneous adverse effects with HER1/EGFR-targeted agents: is there a silver lining? J Clin Oncol. 2005;23(22):5235–46. doi: 10.1200/JCO.2005.00.6916. [DOI] [PubMed] [Google Scholar]
- 29.Wacker B, et al. Correlation between development of rash and efficacy in patients treated with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in two large phase III studies. Clin Cancer Res. 2007;13(13):3913–21. doi: 10.1158/1078-0432.CCR-06-2610. [DOI] [PubMed] [Google Scholar]
- 30.Fojo T. Commentary: Novel therapies for cancer: why dirty might be better. Oncologist. 2008;13(3):277–83. doi: 10.1634/theoncologist.2007-0090. [DOI] [PubMed] [Google Scholar]
- 31.Miller ML, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1(35):ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuler ML. Animal Surrogate Systems for Toxicity Testing. In: McAuliffe GJ, Tatosian DA, editors. Encyclopedia of Biomaterials and Biomedical Engineering. 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.