Abstract
A vast colour palette of monomeric fluorescent proteins has been developed to investigate protein localization, motility and interactions. However, low brightness has remained a problem in far-red variants, which hampers multicolour labelling and whole-body imaging techniques. In the present paper, we report mKate2, a monomeric far-red fluorescent protein that is almost 3-fold brighter than the previously reported mKate and is 10-fold brighter than mPlum. The high-brightness, far-red emission spectrum, excellent pH resistance and photostability, coupled with low toxicity demonstrated in transgenic Xenopus laevis embryos, make mKate2 a superior fluorescent tag for imaging in living tissues. We also report tdKatushka2, a tandem far-red tag that performs well in fusions, provides 4-fold brighter near-IR fluorescence compared with mRaspberry or mCherry, and is 20-fold brighter than mPlum. Together, monomeric mKate2 and pseudo-monomeric tdKatushka2 represent the next generation of extra-bright far-red fluorescent probes offering novel possibilities for fluorescent imaging of proteins in living cells and animals.
Keywords: far-red fluorescence, fluorescent protein, fusion protein, photostability, whole-body imaging
INTRODUCTION
Among the numerous homologues of avGFP (Aequorea victoria green fluorescent protein), naturally occurring orange and red fluorescent proteins isolated from other species are either dimeric or tetrameric. For this reason, they are unsuitable in applications as fusion tags for studying localization, intermolecular binding and motility dynamics of proteins of interest [1–3]. To overcome this limitation, monomeric orange, red and far-red fluorescent proteins have been developed in recent years, including the entire palette of `mFruits' [4–6], mKO [7], TagRFP [8] and mKate [9]. However, brightness remains a problem in the far-red monomeric fluorescent proteins. The brightest variant yet reported, mKate, is still dramatically dimmer than any of the wild-type red fluorescent proteins [9]. At the same time, bright far-red monomers are required both for the multicolour labelling in cells and for the whole-body imaging of fusion proteins in living transgenic organisms. In fact, bright far-red fluorescence is a vital characteristic, since the near-IR optical window is exceptionally advantageous for light penetration into living tissues [9]. The present study focused on developing the photophysical characteristics of mKate and has resulted in a new derivative exhibiting dramatic improvements.
EXPERIMENTAL
Methods concerning cloning and gene construction and characterization of fluorescent proteins in vitro can be found in the Supplementary Online Data at http://www.BiochemJ.org/bj/418/bj4180567add.htm.
Microscopy and live-cell imaging
All filters for fluorescence screening and imaging were purchased from Chroma Technology, Omega Filters and Semrock. HeLa epithelial (CCL-2, A.T.C.C., Manassas, VA, U.S.A.) and grey fox lung fibroblast (CCL-168, A.T.C.C.) cells were grown in a 50:50 mixture of DMEM (Dulbecco's modified Eagle's medium) and Ham's F12 with 12.5 % Cosmic calf serum (Hyclone) and transfected with Effectene (Qiagen). Imaging was performed in Delta-T culture chambers (Bioptechs) under a humidified atmosphere of 5 % CO2 in air. Imaging in widefield mode was performed with a Nikon TE-2000 inverted microscope equipped with Omega QuantaMax™ filters and a Photometrics Cascade II camera or an Olympus IX71 inverted microscope equipped with Semrock BrightLine™ filters and a Hamamatsu ImagEM™ camera. Laser-scanning confocal microscopy was conducted using a Nikon C1Si and an Olympus FV1000, both equipped with argon-ion (457 and 488 nm) and helium–neon or diode (543 and 561 nm respectively) lasers and proprietary filter sets. Spinning disk confocal microscopy was performed on an Olympus DSU-IX81 equipped with a Lumen 200 illuminator (Prior Scientific), a Hamamatsu 9100-13 EMCCD camera, Semrock filters and ten-position filter wheels driven by a Lambda 10-3 controller (Sutter). In some cases, cell cultures expressing fluorescent protein fusions were fixed after imaging in 2 % (w/v) paraformaldehyde (Electron Microscopy Sciences) and washed several times in PBS containing 0.05 M glycine before mounting with a poly(vinyl alcohol)-based medium. Morphological features in all fusion constructs were confirmed by imaging fixed cell preparations on coverslips using a Nikon 80i upright microscope and Semrock Texas Red filter set (4040B) coupled to a Hamamatsu Orca ER or a Photometrics CoolSNAP™ HQ2 camera.
Photobleaching assays
Widefield and laser-scanning confocal microscopy photobleaching experiments were conducted with N-terminal fusions of the appropriate fluorescent protein to human histone H2B (six-residue linker) to confine fluorescence to the nucleus in order to closely approximate the dimensions of aqueous droplets of purified fluorescent proteins used in widefield measurements by the Roger Y. Tsien laboratory [1]. HeLa-S3 cells (A.T.C.C.) of average nucleus diameter 17 μm were transfected with the H2B construct using Effectene and maintained in a 5 % CO2 atmosphere in Bioptechs Delta-T imaging chambers for at least 36 h before imaging. The chambers were transferred to a Bioptechs stage adapter, imaged at low magnification to ensure cell viability, and then photobleached using a 40× oil-immersion objective for confocal [Olympus UPlan Apo, NA (numerical aperture)=1.00] or a 40× dry objective (Nikon Plan Fluorite, NA=0.85) for widefield studies. A metal halide illumination source (EXFO) was used without neutral-density filters for widefield photobleaching assays. Output power at the objective focal plane was measured with a Newport 1918-C meter. Laser lines (543 nm, helium–neon and 488 nm, argon-ion) were adjusted to an output power of 100 μW, measured with a FieldMaxII-TO (Coherent) power meter equipped with a high-sensitivity silicon/germanium optical sensor (Coherent OP-2Vis). The instrument (Olympus FV1000) was set to a zoom of 4×, a region of interest of 341.2 μm2 (108 pixels × 108 pixels), a photomultiplier voltage of 650 V and an offset of 9 % with a scan time of 0.181 s/frame. Nuclei having approximately the same dimensions and intensity under the fixed instrument settings were chosen for photobleaching assays. Fluorescence using the 543 nm laser was recorded with a 570 nm dichromatic mirror and 656 nm longpass barrier filter, whereas emission using the 488 nm laser was reflected directly by a mirror through a 510 nm longpass barrier filter. The photobleaching half-times were calculated as described in [6] as the time required to reduce the scan-averaged emission rate to 50 % from an initial emission rate of 1000 photons/s per fluorescent protein chromophore.
Transgenic Xenopus laevis embryos
To generate a construct expressing mKate2 under the control of Xanf1 promoter, the 2200 bp fragment of this promoter sufficient for specifically targeting the expression to the anterior neural plate [10] was subcloned into VspI (blunted)/AgeI sites of mKate2-N vector, instead of the multiple cloning site region and CMV (cytomegalovirus) promoter. For transgenic experiments, vectors were linearized by SfiI and purified using Qiagen columns. Transgenic embryos were generated by the REMI (restriction enzyme-mediated integration) technique exactly as described previously [10] and were analysed using a Leica MZFLIII fluorescent stereomicroscope with the rhodamine filter set: excitation filter 546/10 nm; suppression filter 565 nm. To visualize a fluorescent signal on tissue sections, transgenic embryos were fixed overnight in 4 % (w/v) formaldehyde in 0.1 % PBS, embedded in 3 % agarose in 0.1 % PBS and sectioned using a HB 650 Vibratome (Microme) in 50 μm sections. The animal work was performed in Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, in accordance with the regulations of the Department of Health and Human Services, National Institutes of Health.
RESULTS AND DISCUSSION
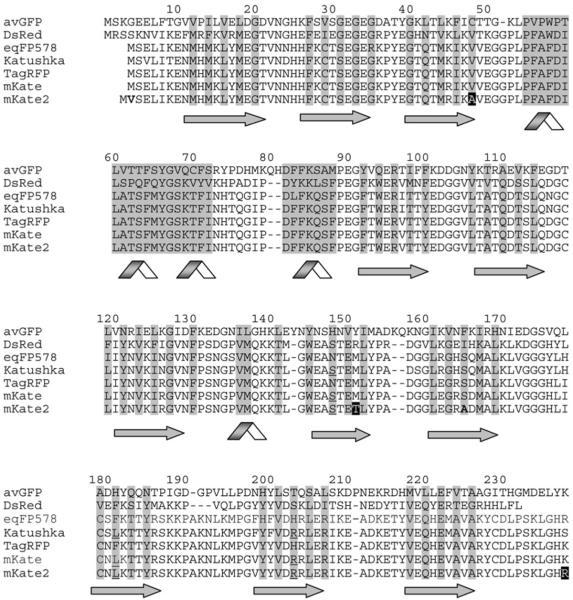
Based on analysis of the mKate crystal structure solved at several pH values (V. Z. Pletnev, unpublished work), we have concluded that the chromophore is capable of forming either a cis or trans conformation. The cis conformation is brightly fluorescent and this increases proportionally with pH, whereas the trans conformation is much dimmer and decreases with increasing pH. These data correlate very well with the kindling-like [11] photoactivation behaviour of mKate [9]. Indeed, at physiological pH, mKate exhibits a 10–20 % increase in fluorescent brightness in response to irradiation by green light (see [9] and below), which corresponds well to a light-induced chromophore trans–cis isomerization. Whereas Ser148 (numbering in accordance with avGFP alignment, Figure 1) stabilizes the fluorescent cis conformation of mKate (similar to most red fluorescent proteins, including DsRed [12,13], and kindling fluorescent protein [11,14]), the trans conformation of the chromophore is stabilized by Ser165 (similar to eqFP611 [15] and chromoproteins [16]). We therefore assumed that mutagenesis at position 165 could disrupt the hydrogen-bonding system that stabilizes the chromophore in trans conformation, and thus produce a new variant with higher brightness and lower pH-dependence.
Figure 1. Alignment of the amino acid sequences for the selected fluorescent proteins.
Structurally important regions are highlighted in grey, β-strands are shown by arrows, and α-helixes are shown by ribbons. Buried residues are shaded. Amino acid residues responsible for the far-red fluorescence of the cis-chromophore of Katushka, mKate and mKate2 are underlined. Random mutations generated in the course of mKate optimization are shown white on black, targeted mutations are shown bold. Overall alignment numbering follows that of avGFP.
Accordingly, we performed saturation mutagenesis (using all 20 amino acids) of Ser165 and screened for brighter variants of mKate. The brightest and fastest maturing variant at 37 °C contained the S165A substitution. This result generally confirmed our assumption: in contrast with the polar hydrophilic residue serine, the non-polar hydrophobic alanine residue is unable to take part in the hydrogen-bonding system that stabilizes the low-fluorescent trans conformation of the chromophore. The S165A mutation should therefore increase the portion of the brightly fluorescent cis chromophore in the physiological and acidic pH range.
We optimized further the new variant using two rounds of random mutagenesis (see the Supplementary Online Data at http://www.BiochemJ.org/bj/418/bj4180567add.htm) and obtained the beneficial mutation V48A, which essentially increases the protein maturation rate, assessed by the relative brightness of the freshly grown Escherichia coli expressing mKate variants (results not shown). Additionally, we uncovered a second mutation, K238R, which is found in the wild-type eqFP578 sequence [8] and also seems to enhance maturation slightly. Valine was inserted as the second amino acid in order to generate the optimal Kozak sequence for eukaryotic translation [17]. Collectively, these major and minor changes resulted in a much brighter, faster maturing, acid-resistant and more photostable monomeric far-red fluorescent protein that we named mKate2.
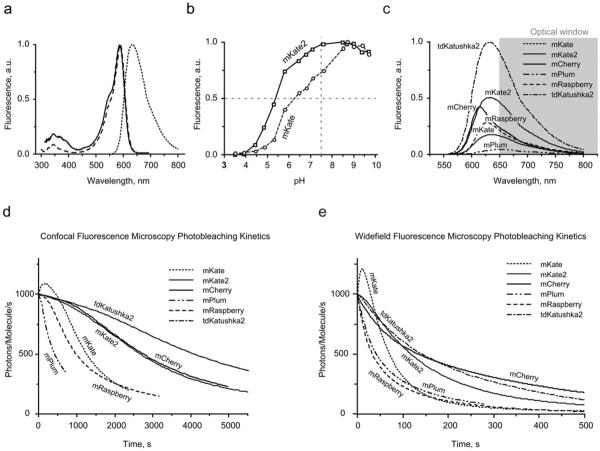
The main characteristics of mKate2 are summarized in Table 1. The spectral profiles are almost identical with those of mKate (Figure 2a), with excitation and emission peaking at 588 and 633 nm respectively. Compared with mKate, the fluorescence of mKate2 is far more pH-stable (Figure 2b), thus increasing its brightness at physiological pH and making mKate2 a more reliable reporter for imaging in acidic organelles, such as late and recycling endosomes and lysosomes. mKate2 is characterized by a ~ 2-fold higher molar absorption coefficient and 1.4-fold higher fluorescence quantum yield compared with mKate, resulting in 2.8-fold brighter fluorescence. The calculated brightness for mKate2 is approx. 74% of EGFP (enhanced green fluorescent protein), which makes the new variant 10-fold brighter than mPlum (Table 1). Within the optical window optimal for light penetration in living tissues, calculated brightness of mKate2 is at least 2-fold higher compared with any monomeric fluorescent protein reported to date (Figure 2c and Table 1).
Table 1. Key characteristics of tdKatushka2 and mKate2 in comparison with other monomeric far-red fluorescent proteins.
Brightness is calculated as a product of molar absorption coefficient and the fluorescence quantum yield, and is presented as a percentage of the brightness of mEGFP. Photostability is given in s as the normalized (from 1000 photons/molecule/s) fluorescent state half-life upon irradiation. Brightness beyond 650 nm (see Figure 2c) is presented as a percentage of the brightness of tdKatushka2.
| Fluorescent protein | λex (nm) | λem (nm) | Quantum yield | Molar absorption coefficient (M−1 .cm−1) | Brightness (% relative to mEGFP) | Brightness beyond 650 nm (% relative to tdKatushka2) | pKa | Photostability, widefield | Photostability, confocal | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| mCherry | 587 | 610 | 0.22 | 72 000 | 47 | 24 | <4.5 | 135 | 2770 | [1] |
| mRaspberry | 598 | 625 | 0.15 | 86 000 | 38 | 26 | - | 35 | 1010 | [5,9] |
| mPlum | 590 | 649 | 0.10 | 41 000* | 12‡ | 11‡ | <4.5 | 77‡ | 770‡ | [5,9] |
| 22 000† | 7§ | 6§ | 42§ | 410§ | ||||||
| mKate | 588 | 635 | 0.28 | 31 500 | 26 | 19 | 6.2 | 71 | 1260 | [9] |
| mKate2 | 588 | 633 | 0.40 | 62 500 | 74 | 52 | 5.4 | 118 | 2690 | - |
| tdKatushka2 | 588 | 633 | 0.37 | 66 250×2 | 146 | 100 | 5.4 | 144 | 4090 | - |
Literature data.
Our data.
Calculated using literature data.
Calculated using our data.
Figure 2. Spectral characteristics of mKate2 in comparison with selected fluorescent proteins.
(a) Normalized mKate2 absorption (solid line), fluorescence excitation (– –) and emission (- - -) spectra. (b) pH stability of mKate2 (solid line) and mKate (broken line) fluorescence. (c) Emission spectra of mCherry, far-red monomeric fluorescent proteins and tdKatushka2 relative to their calculated brightness (Table 1). Scaling was applied to the area of the peak. The favourable `optical window' is shaded grey. (d) Normalized photobleaching curves for mCherry, far-red monomeric fluorescent proteins and tdKatushka2 using laser-scanning confocal microscopy. (e) Normalized photobleaching curves, widefield fluorescence microscopy under metal halide illumination.
Importantly, molar absorption coefficients and fluorescence quantum yields for mKate, mKate2 and tdKatushka2 (see below) were measured in direct comparison with mCherry at the physiological pH of 7.5, to allow relevant comparison of theoretical brightness in cytosol of eukaryotic cells. Previously, we reported higher molar absorption coefficient and quantum yield for mKate, measured at pH of 8.2 [9]. However, since mKate is rather pH-sensitive, the pH change from 8.2 to 7.5 leads to pronounced decrease of the protein brightness. In the present paper, we publish corrected data for mKate at pH 7.5.
Our experiments indicate that mKate2 is characterized by fast chromophore maturation at 37 °C. Standard maturation assay (see the Supplementary Online Data at http://www.BiochemJ.org/bj/418/bj4180567add.htm and [8]) could barely capture the very end of the maturation curve, thus maturation half-time for mKate2 should be <20 min (compared with 40 min for mCherry, 75 min for mKate and 100 min for TagRFP [8,9]). Maturation of the mKate2 red chromophore is complete, as is evident by a single narrow peak in the absorbance spectrum, which is largely identical with the fluorescence excitation spectrum (Figure 2a). We verified that mKate2 remains monomeric by gel filtration (see the Supplementary Online Data at http://www.BiochemJ.org/bj/418/bj4180567add.htm).
A high level of photostability is one of the primary characteristics of fluorescent proteins that is necessary for long-term time-lapse imaging. To determine the photostability of mKate2 in comparison with several well-established fluorescent proteins, we performed photobleaching experiments in living HeLa cells using procedures described previously [6]. Photobleaching assays were conducted with selected fluorescent proteins fused to human histone H2B (Figure 3r) using widefield fluorescence microscopy under metal halide illumination with commercial filter sets and laser-scanning confocal microscopy using the 488 and 543 nm spectral lines from an argon-ion and helium-neon laser respectively (see the Experimental section). The raw photobleaching data were normalized to 1000 photons/molecule/s as described previously [1,6] and are listed in Table 1. Remarkably, mKate2 is more photostable under both widefield and confocal illumination than other monomeric far-red proteins, including mKate, mRaspberry and mPlum (Figures 2d and 2e and Table 1). In contrast with mKate, mKate2 demonstrates almost no kindling (photoactivation) phenomena, consistent with the structural considerations discussed above. We presume that, at physiological pH, the chromophore of mKate2 exists mostly as the cis isomer, with limited potential for additional light-induced trans–cis isomerization of the chromophore to a bright cis conformation.
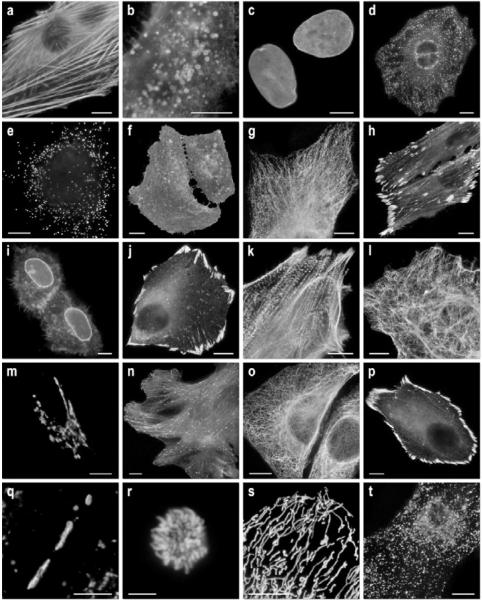
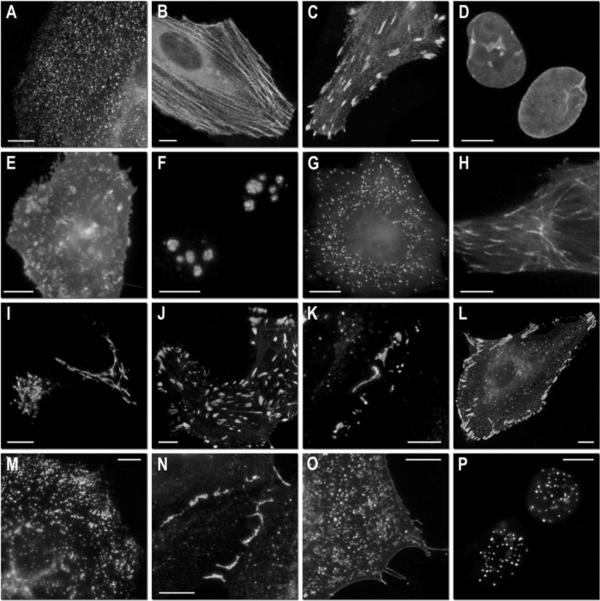
Figure 3. Fluorescence imaging of mKate2 fusion vectors.
(a)–(g) C-terminal mKate2 fusion constructs: (a) mKate2–β-actin-7 (human); (b) mKate2–endosomes-14 (human RhoB GTPase and c-Myc epitope tag); (c) mKate2–lamin B1–10 (human); (d) mKate2–clathrin light chain-15 (human); (e) mKate2–peroxisomes-2 (peroxisomal targeting signal 1; PTS1); (f) mKate2–membrane-5 (20-amino-acid farnesylation signal from c-Ha-Ras); (g) mKate2–α-tubulin-6 (human); (h) mKate2–VASP-5 (mouse vasodilator-stimulated phosphoprotein); (i) mKate2–annexin (A4)-12 (human). (j)–(t) N-terminal fusion constructs; (j) mKate2–paxillin-22 (chicken); (k) mKate2–α-actinin-19 (human non-muscle); (l) mKate2–vimentin-7 (human); (m) mKate2–Golgi-7 (N-terminal 81 amino acids of human β-1,4-galactosyltransferase); (n) mKate2–EB3-7 (human microtubule-associated protein; RP/EB family); (o) mKate2–keratin-17 (human cytokeratin 18); (p) mKate2–zyxin-7 (human); (q) mKate2–Cx43-7 (rat α-1 connexin 43); (r) mKate2–H2B-6 (human; prophase); (s) mKate2–mitochondria-7 (human cytochrome c oxidase subunit VIII); (t) mKate2–lysosomes-20 (human lysosomal membrane glycoprotein 1). The cell line used for expressing mKate2 fusion vectors was grey fox lung fibroblast cells (FoLu) in panels (a), (g) and (n), and human cervical adenocarcinoma cells (HeLa; ATCC, CCL2) in all other panels. A different strain of HeLa (ATCC, S3) was used for mitosis studies in (r). Scale bars, 10 μm. For each fusion protein, the linker amino acid length is indicated after the name of the targeted organelle or fusion protein.
To characterize mKate2 in living cells, we performed transient transfections of HeLa and Phoenix Eco cell lines with a vector driving mKate2 expression without any fusion or localization signals. The fluorescent signal was evenly distributed within the cytosol and nucleus with no visible aggregates or non-specific localization observed within 4 days of transfection in both cell lines. These data indicate that mKate2 should be non-toxic when expressed in stable cell lines, as well as the previously reported mKate (cell lines available from Marinpharm GmbH).
To verify the utility of mKate2 as a fusion tag, we constructed 20 vectors and expressed them in epithelial and fibroblast cell lines to examine performance. mKate2 performed well in most C- and N-terminal fusions (Figure 3), similar to the results observed with the parent proteins, mKate and TagRFP, and demonstrated excellent brightness and photostability that allowed the capture of a long time series (see Supplementary Videos S1–S7 at http://www.BiochemJ.org/bj/418/bj4180567add.htm). A fusion of mKate2 to annexin (A4) translocated as expected from the cytoplasm to the nuclear and plasma membranes upon induction with ionomycin (Figure 3i and Supplementary Video S2). mKate2 also performed well in fusions targeting the Golgi complex (Figure 3m), clathrin vesicles (Figure 3d), peroxisomes (Figure 3e), lysosomes (Figure 3t), mitochondria (Figure 3s), endosomes (Figure 3b) and gap junctions (Figure 3q), as well as assorted nuclear, focal adhesion and cytoskeletal targets (Figures 3c, 3g, 3h, 3k, 3l, 3n, 3o and 3p).
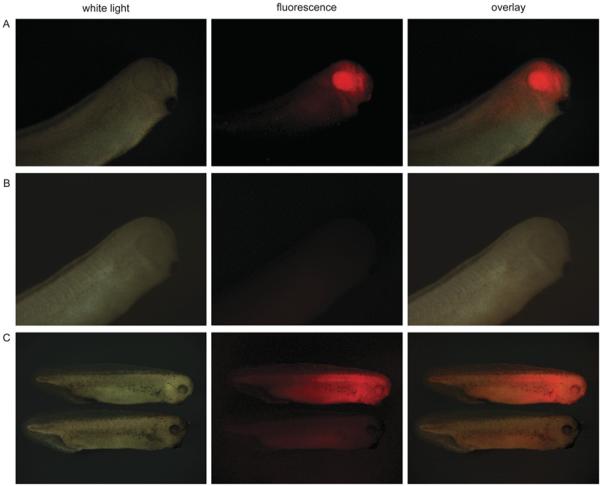
To verify the low toxicity of mKate2 in transgenic animals, we expressed mKate2 under the control of Xanf1 promoter in X. laevis embryos. We have used the 2200 bp fragment of this promoter, which is sufficient for specific targeting of the expression to the anterior neural plate [10]. As expected, we observed bright red fluorescence in the forehead region, including eyes, the forebrain and nasal placodes (Figures 4A and 4B).
Figure 4. Imaging of mKate2 in X. laevis embryos.
(A) Expression of mKate2 under the Xanf1 promoter in the transgenic embryos at stage 28 is specifically localized in the forehead region, including eyes, the forebrain and nasal placodes. The embryo is shown from the right-hand side, with the dorsal side at the top and left. (B) Control images, non-transgenic embryo at the same stage. (C) Upper image: expression of mKate2–zyxin under CMV promoter in the transgenic embryo of stage 35 (2 days). Despite quite intensive and ubiquitous expression of mKate2–zyxin, the embryo appears normal and healthy. Lower image: control embryo of the same stage. The embryos are shown from the right-hand side, with the dorsal side at the top.
To compare brightness and maturation rates of mKate and mKate2 in vivo, X. laevis embryos were microinjected with mKate-N and mKate2-N vectors at the stage of two blastomeres. Living embryos were photographed from the animal pole side at the middle gastrula stages (10.5 h after fertilization). As expected, embryos microinjected with mKate2-N demonstrated superior brightness (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/418/bj4180567add.htm).
In addition, to test in an embryonic model the performance of mKate2 in a targeting protein fusion, we generated transgenic X. laevis embryos bearing a CMV–mKate2–zyxin fusion construct. Despite quite extensive and ubiquitous expression of mKate2–zyxin under the control of the CMV promoter, these embryos appear normal and healthy (Figure 4C and Supplementary Figures S2 and S3 at http://www.BiochemJ.org/bj/418/bj4180567add.htm). Importantly, the tadpoles bearing mKate2 and mKate2–zyxin transgenic constructs have demonstrated roughly comparable viability with tadpoles expressing EGFP transgenes (on average 39% of transgenes selected at stage 46 reached stage 60), which was observed in our previous experiments ([10,18] and A. G. Zaraisky, unpublished work). This indicates that mKate2, as well as EGFP, exerts a low toxic effect on living cells in transgenic organisms.
The dimeric far-red fluorescent protein Katushka is a close relative of mKate and mKate2 [9]. It was therefore intriguing to verify the influence of the key S165A substitution on the spectral properties of Katushka. As could be expected, site-directed mutagenesis S165A resulted in a notable (approx. 20%) increase of fluorescence brightness. Similarly to mKate2, Katushka2 lost the kindling property and mostly preserved the spectral characteristics of the parental Katushka. To generate a Katushka2 variant suitable for fusions, we employed the popular `tandem' approach [4,19] to create a pseudo-monomeric probe. In spite of the larger size, tandem versions of dimeric fluorescent proteins behave similarly to the true monomers and usually perform well in fusions with many proteins of interest. Indeed, the resulting tdKatushka2 performed well in a number of fusions, including those targeting clathrin vesicles, the nuclear lamina, nucleoli, the Golgi, peroxisomes, centrosomes, lysosomes, plasma membrane, focal adhesions and even several fusion proteins that themselves form biopolymers, including actin and gap junctions (Figure 5). Further characterization in vitro and in living cells showed that extra-bright (1.46-fold brighter than EGFP per molecule) far-red tdKatushka2 demonstrates fast maturation, high pH-stability and high photostability (Table 1).
Figure 5. Fluorescence imaging of tdKatushka2 fusion vectors.
(a)–(g) C-terminal tdKatushka2 fusion constructs: (a) tdKatushka2–clathrin light chain-15 (human); (b) tdKatushka2–β-actin-7 (human); (c) tdKatushka2–VASP-5 (mouse vasodilator-stimulated phosphoprotein); (d) tdKatushka2–lamin B1–10 (human); (e) tdKatushka2–membrane-5 (20-amino acid farnesylation signal from c-Ha-Ras); (f) tdKatushka2–fibrillarin-7 (human); (g) tdKatushka2–peroxisomes-2 (peroxisomal targeting signal 1; PTS1). (h)–(p) N-terminal tdKatushka fusion constructs: (h) tdKatushka2–EB3-7 (human microtubule-associated protein; RP/EB family); (i) tdKatushka2–Golgi-7 (N-terminal 81 amino acids of human β-1,4-galactosyltransferase); (j) tdKatushka2–zyxin-7 (human); (k) tdKatushka2–Cx26-7 (rat β2 connexin 26); (l) tdKatushka2–paxillin-22 (chicken); (m) tdKatushka2–lysosomes-20 (human lysosomal membrane glycoprotein 1); (n) tdKatushka2–VE-cadherin (human vascular epithelial cadherin); (o) tdKatushka–VSVG-12 (vesicular stomatitis virus glycoprotein; membrane); (p) tdKatushka–CENPB-22 (human CENP-B DNA-binding domain). The cell line used for expressing tdKatushka2 fusion vectors was grey fox lung fibroblast cells (FoLu) in panel (h) and human cervical adenocarcinoma cells (HeLa; ATCC, CCL2) in all other panels. Scale bars, 10 μm. For each fusion protein, the linker amino acid length is indicated after the name of the targeted organelle or fusion protein.
Advantageous characteristics of mKate2 and tdKatushka2 make them probably the best fluorescent tags yet reported in the far-red range. Bright monomeric fluorescent proteins now span almost the entire visible range, which greatly expands the possibilities of using these probes as monitors of gene expression, multicolour labelling, dynamic interactions, and FRET (fluorescence resonance energy transfer)/FLIM (fluorescence lifetime imaging microscopy) techniques. Importantly, the emission spectra of mKate2 and tdKatushka2 extend into the near-IR optical window that is optimal for light penetration into living tissues [9]. This unique combination of high brightness and far-red emission definitely makes mKate2 and tdKatushka2 the fluorescent proteins of choice for visualizing fusion tags of interest in whole-body imaging of live animals.
Supplementary Material
Acknowledgments
FUNDING This work was supported by the Howard Hughes Medical Institute [grant numbers 55005618 and 55005630], Molecular and Cell Biology Program, Russian Academy of Sciences, RFBR 08-04-01702-a, European Union Framework Programme 6 Integrated Project [grant number LSHG-CT-2003-503259], `State Support of the Leading Scientific Schools' programme [grant number NS-2395.2008.4], National Institutes of Health [grant numbers GM070358 and GM073913], Russian Federal Agency for Science and Innovations [grant number 02.513.12.3013]. D.M.C. is supported by the Grant of the President of Russian Federation [grant number MK-6119.2008.4] and the Russian Science Support Foundation.
Abbreviations used
- avGFP
Aequorea victoria green fluorescent protein
- CMV
cytomegalovirus
- EGFP
enhanced green fluorescent protein
- NA
numerical aperture
REFERENCES
- 1.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J. Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 4.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 9.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 10.Martynova N, Eroshkin F, Ermakova G, Bayramov A, Gray J, Grainger R, Zaraisky A. Patterning the forebrain: FoxA4a/Pintallavis and Xvent2 determine the posterior limit of Xanf1 expression in the neural plate. Development. 2004;131:2329–2338. doi: 10.1242/dev.01133. [DOI] [PubMed] [Google Scholar]
- 11.Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. J. Biol. Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 12.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 13.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38Å resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 15.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0-Å crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J. Biol. Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 16.Beddoe T, Ling M, Dove S, Hoegh-Guldberg O, Devenish RJ, Prescott M, Rossjohn J. The production, purification and crystallization of a pocilloporin pigment from a reef-forming coral. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:597–599. doi: 10.1107/s0907444902023466. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 18.Ermakova GV, Solovieva EA, Martynova NY, Zaraisky AG. The homeodomain factor Xanf represses expression of genes in the presumptive rostral forebrain that specify more caudal brain regions. Dev. Biol. 2007;307:483–497. doi: 10.1016/j.ydbio.2007.03.524. [DOI] [PubMed] [Google Scholar]
- 19.Fradkov AF, Verkhusha VV, Staroverov DB, Bulina ME, Yanushevich YG, Martynov VI, Lukyanov S, Lukyanov KA. Far-red fluorescent tag for protein labelling. Biochem. J. 2002;368:17–21. doi: 10.1042/BJ20021191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.