Abstract
The geno- and phenotypic diversity of commercial Saccharomyces cerevisiae wine yeast strains provides an opportunity to apply the system-wide approaches that are reasonably well established for laboratory strains to generate insight into the functioning of complex cellular networks in industrial environments. We have previously analyzed the transcriptomes of five industrial wine yeast strains at three time points during alcoholic fermentation. Here, we extend the comparative approach to include an isobaric tag for relative and absolute quantitation (iTRAQ)-based proteomic analysis of two of the previously analyzed wine yeast strains at the same three time points during fermentation in synthetic wine must. The data show that differences in the transcriptomes of the two strains at a given time point rather accurately reflect differences in the corresponding proteomes independently of the gene ontology (GO) category, providing strong support for the biological relevance of comparative transcriptomic data sets in yeast. In line with previous observations, the alignment proves to be less accurate when assessing intrastrain changes at different time points. In this case, differences between the transcriptome and proteome appear to be strongly dependent on the GO category of the corresponding genes. The data in particular suggest that metabolic enzymes and the corresponding genes appear to be strongly correlated over time and between strains, suggesting a strong transcriptional control of such enzymes. The data also allow the generation of hypotheses regarding the molecular origin of significant differences in phenotypic traits between the two strains.
Saccharomyces cerevisiae has long been a model organism to investigate the biology of the eukaryotic cell. The yeast genome, which is compact and contains only around 6,000 protein-encoding genes, was completely sequenced in 1996 (18), but nearly 10% of putative proteins remain without predicted functions. The majority, if not all of these remaining gene products, are nonessential, and the deletion of these genes in most cases does not lead to a detectable phenotype.
A major limitation of most current approaches in this regard is that research is conducted using a limited number of laboratory yeast strains which, while displaying characteristics that are useful for genetic and molecular analyses, represent limited genetic and phenotypic diversity. These laboratory strains are furthermore significantly different from the strains that are used for industrial and commercial purposes. Industrial environments, however, constitute much of the evolutionary framework of the species S. cerevisiae in the past centuries, and many genes that appear not to be associated with a specific function in laboratory strains may be responsible for specific phenotypes in industrial strains. Such strains will therefore be better suited for the analysis of complex genetic and molecular networks and of their phenotypic relevance or biological meaning. The recent sequencing of wine yeast strains (9, 31) showed that a significant number of genes that are not found in the standard S288c laboratory strain were present in these strains and that a large number of other significant differences exist between these genomes. Furthermore, different wine yeast strains exhibit great variation in chromosome size and number, as well as ploidy, and cover a wide range of phenotypic traits, many of which are absent in laboratory yeast (6).
Large-scale gene expression analysis with microarrays is one of the most powerful and best-developed functional genomics methodologies that can be applied to yeast (5). Transcriptome analysis of wine yeast strains has already proven useful to analyze the broad genetic regulation of fermentative growth in wine environments and has allowed identification of stress response mechanisms that are active under these conditions (3, 16, 29). Rossouw et al. (37) showed that a comparative analysis of the transcriptome and exometabolome could be used to identify genes that are involved in aroma metabolism and to predict some of the impact of changed gene expression levels. While of great usefulness, transcription data alone are of limited value, since they cannot be directly correlated with protein levels and, a fortiori, with in vivo metabolic fluxes (13, 19, 36, 48). All omics data sets would indeed be significantly strengthened in combination with other layers of the biological information transfer system (36, 44, 47).
A current bottleneck of such approaches is that most “omics” tools are not developed to the same degree as transcriptomics. In particular, genome-scale protein quantification faces significant challenges, but methods for determining relative levels of protein between samples have been developed (42). Two-dimensional (2-D) gel electrophoresis has been and continues to be employed to separate complex protein mixtures and is frequently combined with in-gel tryptic digestion and mass spectrometry for the identification of proteins (27, 32). In general, most yeast proteomic studies to date have been conducted using this 2-D gel electrophoresis technology (10, 25, 36, 46). While over 1,400 soluble proteins of yeast have been identified using 2-D analyses, this approach has not addressed the issue of quantification in a satisfactory manner and also suffers from the relatively low number of proteins which are identified in a single analysis, combined with an underrepresentation of low-abundance and hydrophobic proteins (17, 35, 36). In wine yeast, the 2-D gel approach coupled to mass spectrometry has been used to study postinoculation changes in protein levels (39) and the proteomic response of fermenting yeast to glucose exhaustion (45). Rossignol et al. (36) used this approach to identify 59 proteins and compare the transcriptome and proteome of a single wine yeast strain during various stages of fermentation. Based on this analysis, those authors found limited alignment between these two layers of the biological information transfer system.
To overcome some of these limitations, whole-proteome analysis can also be implemented by a high-throughput chromatography approach in combination with mass spectrometry (28). The separation of peptides from complex protein digests is usually achieved by two-dimensional nano-liquid chromatography-mass spectrometry (LC/MS) (30). A total of 1,504 yeast proteins have been unambiguously identified in a single analysis using this 2-D chromatography approach coupled with tandem mass spectrometry (MS/MS) (34). Advances in LC/MS-based proteome analysis, in combination with advances in computational methods, have led to a more comprehensive identification and accurate quantification of endogenous yeast proteins (14, 26). Yet most of the above-mentioned studies were carried out with laboratory yeast strains, mostly under confined experimental conditions limited to steady, exponential growth rates. No such studies have been conducted using different wine yeast strains at different stages of the industrial growth cycle.
In our study we made use of such a chromatography-coupled mass spectrometry approach for the comparative analysis of wine yeast strains. To enable relative quantification between samples, we employed the 8-plex isobaric tag for relative and absolute quantitation (iTRAQ) labeling strategy. The strategy enables relative quantification of up to eight complex protein samples in a single analysis using isobaric tags (11). In short, unlabeled protein samples are trypsin digested, then labeled using isobaric tags (the eight reporter ions), and subsequently separated by liquid chromatography, followed by MS/MS. The covalently bound isobaric tags have the same charge and overall mass but produce different low-mass signatures upon MS/MS, thus enabling relative quantification between different samples in a single analysis (2).
In this paper, we extend the comparative omics approach by aligning the transcriptomes and proteomes of two industrial wine yeast strains. The transcriptomes of these strains, generated at the same time points under the same conditions, have been partially analyzed in a previous paper (37). Our data show that the differences in transcript levels of the two strains at a given time point are a reasonably accurate reflection of the differences in the corresponding protein levels independently of the gene ontology (GO) category. This provides strong support for the biological relevance of comparative transcriptomic data sets in yeast, showing that intrinsic differences between strains may form a more reliable platform for analyses of biologically relevant and meaningful genetic features of a system. Interstrain comparative transcriptome and proteome analyses (as opposed to single-strain analyses) appear to substantially increase our ability to provide a biologically relevant interpretation of omics data sets and to understand metabolic and physiological changes that occur during wine fermentation. Such combinatorial comparative approaches should ultimately enable accurate model building for industrial wine yeast and facilitate the generation of intelligent yeast improvement strategies.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Two yeast strains were used in this study, namely, VIN13 (Anchor Yeast, South Africa) and BM45 (Lallemand Inc., Canada). All are diploid Saccharomyces cerevisiae strains used in industrial wine fermentations. Yeast cells were cultivated at 30°C in yeast extract-peptone-dextrose (YPD) synthetic media, with 1% yeast extract (BioLab, South Africa), 2% peptone (Fluka, Germany), and 2% glucose (Sigma, Germany). Solid medium was supplemented with 2% agar (BioLab, South Africa).
Fermentation media.
Fermentation experiments were carried out with synthetic must MS300, which approximates to a natural must as previously described (7). The medium contained 125 g/liter glucose and 125 g/liter fructose, and the pH was buffered at 3.3 with NaOH.
Fermentation conditions.
All fermentations were carried out under microaerobic conditions in 100-ml glass bottles (containing 80 ml of the medium) sealed with rubber stoppers with a CO2 outlet. The fermentation temperature was approximately 22°C, and no continuous stirring was performed during the course of the fermentation. Fermentation bottles were inoculated with YPD cultures in logarithmic growth phase (around an optical density at 600 nm [OD600] of 1) to an OD600 of 0.1 (i.e., a final cell density of approximately 106 CFU·ml−1). The cells from the YPD precultures were briefly centrifuged and resuspended in MS300 to avoid carryover of YPD to the fermentation media. The fermentations followed a time course of 14 days, and the bottles were weighed daily to assess the progress of fermentation. Samples of the fermentation media and cells were taken at days 2, 5, and 14 as representative of exponential, early stationary, and late stationary growth phases, respectively.
Microarray analysis.
Transcriptome data were generated (using the Affymetrix platform) at three time points during fermentation, namely, day 2 (exponential growth phase), day 5 (early stationary phase), and day 14 (late stationary phase) at the end of fermentation. These data were evaluated in part for a previous publication (38). Sampling of cells from fermentation and total RNA extraction were performed as described by Abbott et al. (1). For a complete description of the hybridization conditions, normalization, and statistical analysis, refer to the work of Rossouw et al. (37). Transcript data can be downloaded from the Gene Expression Omnibus (GEO) repository under accession number GSE11651.
Protein extraction.
General chemicals for sample preparation were acquired from Merck. Samples of the cells were taken from the fermentations (at days 2, 5, and 14) by centrifugation and weighed after being washed with double-distilled water (ddH2O). The pellets were sonicated using a Soniprep 150 probe sonicator on ice in 30-s bursts and then spun at 16,000× g, and the supernatants were collected. Protein content was assayed by the EZQ method (Invitrogen), and aliquots containing 50 μg of total protein underwent reduction (incubation with 10 mM dithiothreitol [DTT] at 56°C for 1 h) and alkylation (incubation with 30 mM iodoacetamide at pH 8.0 in the dark for 1 h) and were then quenched with further DTT. Samples were subsequently digested by incubation with 2 μg of trypsin (Promega, Madison, WI) at 37°C overnight. The resulting peptides were desalted on 10-mg Oasis SPE cartridges (Waters Corporation, MA) and completely dried down using a speed vacuum concentrator (Thermo Savant, Holbrook, NY).
iTRAQ labeling.
Dried protein digests were reconstituted with 30 μl of dissolution buffer from the iTRAQ reagents multiplex kit (Applied Biosystems, Foster City, CA) and labeled with 8-plex iTRAQ reagents, according to the manufacturer's instructions. Labeled material from six different samples were then combined, acidified, desalted as described above, concentrated to approximately 50 μl, and finally diluted to 250 μl in 0.1% formic acid.
Chromatographic method.
Pooled samples were fractionated in an on-line fashion on a BioSCX II 0.3- by 35-mm column (Agilent Technologies, Santa Clara, CA) using the following 10 salt steps: 10, 20, 40, 60, 80, 100, 140, 200, 260, and 500 mM KCl. Peptides were captured on a 0.3- by 5-mm PepMap cartridge (LC Packings, Dionex Corporation, Sunnyvale, CA) before being separated on a 0.3- by 100-mm Zorbax 300SB-C18 column (Agilent). The high-pressure liquid chromatography (HPLC) gradient between buffer A (0.1% formic acid in water) and buffer B (0.1% formic acid in acetonitrile) was formed at 6 μl/min as follows: 10% buffer B for the first 3 min, increasing to 35% buffer B by 80 min, increasing to 95% buffer B by 84 min, held at 95% until 91 min, back to 10% buffer B at 91.5 min, and held there until 100 min.
MS conditions.
The LC effluent was directed into the IonSpray source of the QStar XL hybrid quadrupole time-of-flight mass spectrometer (Applied Biosystems), scanning from 300 to 1,600 m/z. The top three most abundant multiply charged peptides were selected for MS/MS analysis (55 to 1,600 m/z). The mass spectrometer and HPLC system were under the control of the Analyst QS software package (Applied Biosystems).
Data analysis.
All of the data files from each 2-D liquid chromatography-MS/MS experiment were searched as a set by ProteinPilot 2.0.1 (Applied Biosystems) against a yeast protein database from Stanford University's Saccharomyces Genome Database (5,884 sequences, downloaded November 2008). The data were also searched against the same set of sequences in reverse to estimate the false discovery rate for each run, which was below 0.3% for all three runs. The proteomic data set is available in the supplemental material.
Network analysis.
Microarray data were normalized with the GCRMA method (50). Ratios of the RNA levels for each gene at each time point comparing BM45 to VIN13 were subsequently created by the means of technical replicates performed for each strain. If the resulting ratio was less than 1, it was transformed by taking its negative inverse in order to express relative expression levels on the same scale. Ratios for protein levels between BM45 and VIN13 were similarly created. Ratios for the RNA and protein levels were also created to show the differences between time points within each strain.
XML files for the KEGG pathway database (21, 22, 23) were downloaded, parsed, and used to create an undirected graph consisting of nodes representing pathways and nodes representing gene products which participate in said pathways. Edges between the gene product nodes and each of the pathway nodes in which they are thought to participate were created. A neighborhood walking algorithm was implemented in order to extract subgraphs corresponding to all of the gene products and their associated pathways for which we had ratios for both protein and RNA levels. Given that the proteins identified by iTRAQ varied across time points (within and between each strain), this subgraph extraction was done separately for each time point.
The resulting subgraphs were visualized with Cytoscape 2.6.1 (12, 41). Pathways representing differences between strains as well as reasonable concordance in the regulation of RNA and protein levels were subsequently selected. An unweighted force-directed layout algorithm was applied to the selected subgraphs, and finally, the order of gene product nodes around pathway nodes was manually adjusted to be consistent across time points. Manual node order adjustment was necessary due to the variation in protein data identified by iTRAQ from time point to time point.
The resulting visually mapped subgraphs provide an effective visualization method with which to observe the ratios of RNA and proteins involved in specific pathways simultaneously and, as such, give further insight into the differences in metabolic regulation between strains and time points for both types of molecules.
All programming required for ratio creation, data parsing, graph creation, and neighborhood walking was implemented in Perl.
RESULTS AND DISCUSSION
Interstrain alignment of transcriptomes and proteomes.
Protein abundance data for the BM45 and VIN13 strains were generated at three time points during fermentation, namely, day 2 (exponential growth phase), day 5 (early stationary phase), and day 14 (late stationary phase). Three repeats each for both of the strains were combined for each time point in a single 8-plex iTRAQ analysis. In other words, the repeats for BM45 and VIN13 were grouped for comparative analyses into three sets according to time points (i.e., all day 2 samples were grouped together, all day 5 samples were grouped together, and all day 14 samples were grouped together). A total of 436 proteins were unambiguously identified. Not all of these proteins were identified for both strains across all three time points, but for each time point, at least 250 common proteins were quantified for the three BM45 samples and the three VIN13 samples.
To get an impression of the general data structure and overall alignment of transcript and protein data when comparing the two strains at each time point, we first calculated the ratios of the concentrations of identified proteins and the ratios of the corresponding gene expression values between the two strains (i.e., for BM45 versus VIN13 at each of the three time points). As a broad measure of alignment, we used the log ratios of these protein and transcript comparisons (Fig. 1). In these representations, values above 1 and below −1 represent cases for which the fold change differences in protein concentration diverges by a factor of more than 2 from the fold change in transcription levels between the two strains. In other words, the changes in transcript levels are not aligned with the observed changes in protein levels outside these 1 and −1 value cutoffs.
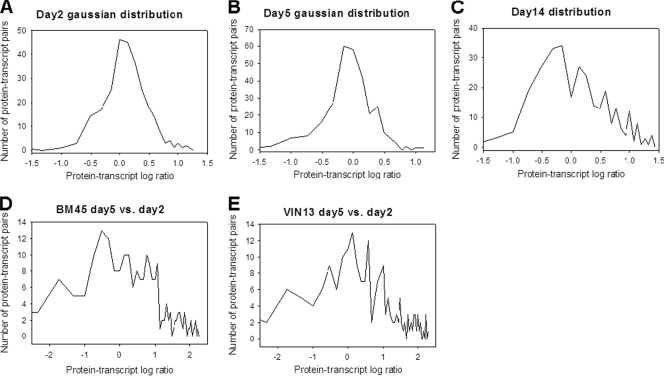
FIG. 1.
Distribution of protein/transcript ratios. The distribution of the different protein-transcript pairs across the spectrum of ratios was determined for days 2 (A), 5 (B), and 14 (C) of the BM45 versus VIN13 comparative analysis. For the intrastrain analysis, the distribution of protein/transcript ratios for day 5 compared to those for day 2 is shown for BM45 (D) and VIN13 (E).
Figure 1 shows the general alignment that the log2-transformed protein/mRNA ratios represented as a distribution curve. Log2-transformed ratios close to zero indicate very strong agreement between the protein levels and gene expression levels for comparisons between strains (for protein and mRNA levels). Hence, the steeper the gradient of the slopes of the Gaussian-shaped curves, the closer the alignment of transcript and protein data sets as a whole. For the interstrain analysis at specific time points, there is clearly a significant peak for days 2 and 5 around the optimal alignment point of zero, with sharply declining slopes in the direction of the 2-fold change indicators (namely, values of 1 and −1). The narrow peaks for these 2 days are a clear indicator of the close alignment of the protein and transcript data sets. The opposite is clearly true for day 14 (Fig. 1C), where no clear Gaussian distribution is evident, but rather, a segmented pattern of increase and decrease across the wide range of protein/transcript ratios is shown.
For a more-detailed analysis of individual protein-transcript pairs, standard t tests were applied to the three repeats of BM45 and VIN13 to determine significant differences in gene or protein levels. The interstrain ratios for transcripts or proteins are set to 1 in cases where no statistically significant differences exist for either the mRNA or protein levels between these two strains. Where interstrain differences are significant, the fold changes are reported for BM45 versus VIN13. This enables comparisons of transcript and corresponding protein fold changes to be made. Examples of the interstrain alignments of mRNA-protein pairs involved in general metabolism (Table 1) and cell rescue and defense (Table 2) for BM45 versus VIN13 are shown in the tables.
TABLE 1.
GO category of energy and metabolism for protein-mRNA pairs at days 2, 5, and 14
| Gene name | ORFa | Functional descriptionb | Fold change in energy and metabolism at dayc: |
|||||
|---|---|---|---|---|---|---|---|---|
| 2 |
5 |
14 |
||||||
| BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | |||
| ACC1 | YNR016C | Acetyl-CoA carboxylase | 1.22 | 1.46 | 1 | 1 | 1.27 | 1.25 |
| ACO1 | YLR304C | Aconitate hydratase | M | M | 1 | 1 | 1 | 1 |
| ACS2 | YLR153C | Acetyl-CoA synthetase | 1.38 | 1.54 | 1 | 1.64 | 1 | 1.52 |
| ADE12 | YNL220W | Adenylosuccinate lyase | 1 | 1 | 1 | 1 | M | M |
| ADE13 | YLR359W | Adenylosuccinate lyase | −1.44 | 1 | M | M | −1.59 | 1 |
| ADE17 | YMR120C | 5-Aminoimidazole-4-carboxamide ribotide transformylase | −1.30 | 1 | 1 | 1 | −1.49 | 1.11 |
| ADE2 | YOR128C | Phosphoribosylaminoimidazole carboxylase | M | M | 1 | 1 | M | M |
| ADE3 | YGR204W | C1-tetrahydrofolate synthase (trifunctional enzyme) | −1.19 | 1 | 1 | −1.19 | 1.25 | 1 |
| ADE4 | YMR300C | Amidophosphoribosyltransferase | −1.86 | 1 | M | M | M | M |
| ADE5,7 | YGL234W | 7-Phosphoribosylamine-glycine ligase | −1.25 | 1 | 1 | 1 | 1 | 1 |
| ADH1 | YOL086C | Alcohol dehydrogenase I | 1 | 1 | 1 | 1 | 1 | 1 |
| ADH3 | YMR083W | Alcohol dehydrogenase III | 1 | 1.14 | M | M | M | M |
| ADK1 | YDR226W | Adenylate kinase, cytosolic | −1.41 | 1 | 1 | 1 | 1 | 1 |
| ALD6 | YPL061W | Aldehyde dehydrogenase, cytosolic | 1.51 | 1.44 | M | M | 1 | 2.62 |
| ADO1 | YJR105W | Strong similarity to human adenosine kinase | M | M | 1 | 1 | M | M |
| APE2 | YKL157W | Aminopeptidase yscII | M | M | 1 | 1 | M | M |
| ARG1 | YOL058W | Argininosuccinate synthetase | M | M | 1 | −1.19 | 1 | −1.18 |
| ARG4 | YHR018C | Arginosuccinate lyase | M | M | 1 | −1.37 | M | M |
| ARO1 | YDR127W | Arom pentafunctional enzyme | −1.43 | −1.28 | M | M | M | M |
| ARO2 | YGL148W | Chorismate synthase | 1 | 1 | M | M | M | M |
| ARO3 | YDR035W | 2-Dehydro-3-deoxyphosphoheptonate aldolase | 1 | 1 | 1 | 1 | −1.39 | 1 |
| ARO4 | YBR249C | 2-Dehydro-3-deoxyphosphoheptonate aldolase | 1 | 1 | 1 | 1 | 1 | 1 |
| ARO8 | YGL202W | Aromatic amino acid aminotransferase I | −1.07 | 1 | M | M | 1 | 1 |
| ASN1 | YPR145W | Asparagine synthetase | 1 | 1 | M | M | 1 | 1 |
| ATP1 | YBL099W | F1F0-ATPase complex, F1 alpha subunit | −1.42 | 1.11 | 1 | 1 | 1 | 1 |
| ATP16 | YDL004W | YDL004W | M | M | 1 | 1 | −1.18 | −1.50 |
| ATP2 | YJR121W | F1F0-ATPase complex, F1 beta subunit | 1 | 1 | 1 | 1 | −1.66 | 1 |
| ATP4 | YPL078C | F1F0-ATPase complex, F0 subunit B | M | M | 1 | 1 | M | M |
| BAT1 | YHR208W | Branched-chain amino acid aminotransferase | 1 | 1 | 1 | −1.29 | 1 | −1.25 |
| BGL2 | YGR282C | Endo-beta-1,3-glucanase of the cell wall | 1 | 1 | 1 | 1 | 1 | 1 |
| CDC19 | YAL038W | Pyruvate kinase | 1 | −1.06 | 1 | −1.10 | 1 | 1 |
| CIT1 | YNR001C | Citrate (si)-synthase, mitochondrial | M | M | 1 | 1 | 2.00 | 1 |
| COR1 | YBL045C | Ubiquinol-cytochrome c reductase 44K core protein | M | M | 1 | 1 | 1 | 1 |
| COX4 | YGL187C | Cytochrome c oxidase chain IV | 1 | 1 | M | M | 1 | 1 |
| CYS3 | YAL012W | Cystathionine gamma-lyase | 1.22 | 1.53 | 1 | 1.38 | 1.85 | 1.51 |
| CYS4 | YGR155W | Cystathionine beta-synthase | 1.08 | 1 | 1 | 1.20 | 1 | 1.26 |
| DAK1 | YML070W | Dihydroxyacetone kinase, induced in high salt | 1.11 | 1 | M | M | M | M |
| DPM1 | YPR183W | Dolichyl-phosphate beta-d-mannosyltransferase | −1.57 | 1 | M | M | M | M |
| ECM17 | YJR137C | Involved in cell wall biogenesis and architecture | 1 | 1 | 1 | 1 | 1.92 | −1.23 |
| EGD1 | YPL037C | GAL4 DNA-binding enhancer protein | −1.23 | 1 | 1 | 1 | M | M |
| EGD2 | YHR193C | Alpha subunit of the nascent polypeptide-associated complex | −1.41 | 1 | 1 | 1 | −1.54 | 1 |
| ENO1 | YGR254W | Enolase I (2-phosphoglycerate dehydratase) | 1 | −1.25 | 1 | −1.23 | 1 | −1.11 |
| ENO2 | YHR174W | Enolase II (2-phosphoglycerate dehydratase) | 1 | 1 | 1 | 1 | 1 | 1 |
| ERG1 | YGR175C | Squalene monooxygenase | 1 | 1.23 | 1 | 1 | 1 | 1 |
| ERG10 | YPL028W | Acetyl-CoA C-acetyltransferase, cytosolic | 1.33 | 1.26 | 1 | 1 | M | M |
| ERG13 | YML126C | 3-Hydroxy-3-methylglutaryl CoA synthase | 1.23 | 1 | M | M | −3.45 | 1 |
| ERG20 | YJL167W | Farnesyl-pyrophosphate synthetase | 1 | 1.19 | M | M | −2.03 | 1 |
| ERG6 | YML008C | S-Adenosyl-methionine delta-24-sterol-C-methyltransferase | 1.12 | 1 | 1 | 1 | 1 | 1 |
| EXG1 | YLR300W | Exo-beta-1,3-glucanase (I/II), major isoform | 1.33 | 1 | 1 | 1 | 1 | 1 |
| FAS1 | YKL182W | Fatty-acyl-CoA synthase, beta chain | 1.13 | 1.28 | 1 | 1 | 1 | 1.17 |
| FAS2 | YPL231W | Fatty-acyl-CoA synthase, alpha chain | 1 | 1.17 | 1 | 1.12 | 1.19 | 1 |
| FBA1 | YKL060C | Fructose-bisphosphate aldolase | 1 | 1 | 1 | 1 | 1 | 1 |
| FUR1 | YHR128W | Uracil phosphoribosyltransferase | M | M | 1 | 1.71 | M | M |
| GAD1 | YMR250W | Similarity to glutamate decarboxylases | M | M | −1.63 | 1 | M | M |
| GDH1 | YOR375C | Glutamate dehydrogenase (NADP+) | −1.26 | 1.07 | 1 | 1 | M | M |
| GFA1 | YKL104C | Glucosamine-fructose-6-phosphate transaminase | 1 | 1 | 1 | −1.75 | M | M |
| GLK1 | YCL040W | Aldohexose specific glucokinase | 1 | −1.07 | 1 | 1 | 1 | 1 |
| GND1 | YHR183W | 6-Phosphogluconate dehydrogenase | 1 | 1.22 | 1 | 1.17 | 1 | 1.22 |
| GPD1 | YDL022W | Glycerol-3-phosphate dehydrogenase (NAD+) | 1 | 1.05 | 1 | 1 | 1 | 1 |
| GPD2 | YOL059W | Glycerol-3-phosphate dehydrogenase (NAD+) | 4.23 | 1.43 | M | M | 1 | 1.44 |
| GPH1 | YPR160W | Glycogen phosphorylase | 1 | 1 | M | M | M | M |
| GPM1 | YKL152C | Phosphoglycerate mutase | 1 | 1 | 1 | 1 | 1 | −1.08 |
| GRE3 | YHR104W | Aldose reductase | 1.65 | 1.59 | M | M | 1 | 1.44 |
| HEM13 | YDR044W | Coproporphyrinogen III oxidase | −1.97 | −1.35 | M | M | 1 | −1.29 |
| HIS1 | YER055C | ATP phosphoribosyltransferase | 1 | 1 | M | M | 1 | 1 |
| HIS3 | YOR202W | Imidazoleglycerol-phosphate dehydratase | M | M | 1 | 1 | M | M |
| HIS4 | YCL030C | Phosphoribosyl-ATP pyrophosphatase | 1.16 | 1 | 1 | 1 | 1 | 1 |
| HOM2 | YDR158W | Aspartate-semialdehyde dehydrogenase | 1 | 1 | 1 | 1 | −1.55 | 1 |
| HOM6 | YJR139C | Homoserine dehydrogenase | 1 | 1 | M | M | 1.28 | 1 |
| HOR2 | YER062C | dl-Glycerol phosphatase | M | M | 1 | 1 | M | M |
| HXK1 | YFR053C | Hexokinase I | −1.39 | 1 | 1 | 1 | 1.45 | 1 |
| HXK2 | YGL253W | Hexokinase II | −1.39 | −1.16 | 1 | 1.22 | 1 | 1.28 |
| HXT3 | YDR345C | Low-affinity hexose transporter | 1.31 | 1 | 1 | −1.11 | M | M |
| HYP2 | YEL034W | Translation initiation factor eIF5A.1 | 1 | 1.41 | 1 | 1.41 | 1 | 1 |
| ILV1 | YER086W | Anabolic serine and threonine dehydratase precursor | 1 | 1 | M | M | −1.64 | 1 |
| ILV2 | YMR108W | Acetolactate synthase | 1 | 1 | M | M | 1 | −1.15 |
| ILV3 | YJR016C | Dihydroxy acid dehydratase | −1.53 | −1.58 | 1 | −1.65 | −1.86 | −1.55 |
| ILV5 | YLR355C | Ketol-acid reductoisomerase | −1.15 | 1 | 1 | 1 | 1 | 1 |
| ILV6 | YCL009C | Acetolactate synthase, regulatory subunit | M | M | 1 | 1 | 1 | 1 |
| IMD2 | YHR216W | IMP dehydrogenase | 1 | 2.10 | 1 | 1.70 | 1 | 1.89 |
| IMD3 | YLR432W | Strong similarity to IMP dehydrogenases | M | M | 1 | 1 | M | M |
| IMD4 | YML056C | Strong similarity to IMP dehydrogenases | −1.87 | 1 | M | M | 1 | 1 |
| IPP1 | YBR011C | Inorganic pyrophosphatase, cytoplasmic | −1.10 | −1.19 | 1 | 1 | 1 | −1.17 |
| LEU2 | YCL018W | Beta-isopropyl-malate dehydrogenase | 1 | 1 | 1 | −1.22 | 1 | −1.13 |
| LEU4 | YNL104C | 2-Isopropylmalalate synthase | −1.09 | 1 | M | M | −1.38 | 1 |
| LPD1 | YFL018C | Dihydrolipoamide dehydrogenase precursor | M | M | 1 | 1.15 | M | M |
| LYS1 | YIR034C | Saccharopine dehydrogenase | 1.27 | 1 | 1 | 1 | −1.88 | 1 |
| LYS12 | YIL094C | Homoisocitrate dehydrogenase | −1.24 | 1 | 1 | 1 | 1 | 1.18 |
| LYS20 | YDL131W | Homocitrate synthase | 1 | 1 | M | M | M | M |
| LYS4 | YDR234W | Homoaconitase | −1.83 | 1 | 1 | 1 | −4.42 | 1 |
| LYS9 | YNR050C | Saccharopine dehydrogenase | 1 | 1 | 1 | 1 | 1 | 1 |
| MAE1 | YKL029C | Malic enzyme | −1.75 | 1 | 1 | 1 | 1 | 1 |
| MCR1 | YKL150W | Cytochrome b5 reductase | M | M | 1 | 1 | 1 | 1 |
| MDH1 | YKL085W | Malate dehydrogenase precursor | M | M | 1 | 1 | 1 | 1 |
| MET10 | YFR030W | Sulfite reductase flavin-binding subunit | M | M | 1 | 1.28 | M | M |
| MET17 | YLR303W | O-Acetylhomoserine sulfhydrylase | 1 | 1.33 | 1 | 1.48 | 1.86 | 1.53 |
| MET22 | YOL064C | Protein Ser/Thr phosphatase | 1.18 | 1 | M | M | M | M |
| MET3 | YJR010W | Sulfate adenylyltransferase | 1 | 1 | 1 | 1 | 1 | 1.27 |
| MET6 | YER091C | 5-Methyltetrahydropteroyltriglutamate methyltransferase | 1 | 1.43 | 1 | 1.45 | 1 | 1.52 |
| MIR1 | YJR077C | Phosphate transport protein | 1 | 1 | 1 | 1 | M | M |
| NCP1 | YHR042W | NADPH-cytochrome P450 reductase | 1.15 | 1 | M | M | M | M |
| OYE2 | YHR179W | NADPH dehydrogenase | 1 | 1 | 1 | 1 | 2.00 | 1 |
| PDA1 | YER178W | Pyruvate dehydrogenase alpha chain precursor | 1 | 1 | M | M | −1.27 | 1 |
| PDC1 | YLR044C | Pyruvate decarboxylase, isozyme 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| PDC5 | YLR134W | Pyruvate decarboxylase, isozyme 2 | M | M | 1 | −2.32 | −1.32 | −2.00 |
| PDI1 | YCL043C | Protein disulfide-isomerase precursor | 1 | 1 | 1 | −1.16 | 1 | −1.18 |
| PDX3 | YBR035C | Pyridoxamine-phosphate oxidase | 1 | 1 | M | M | 1 | 1 |
| PFK1 | YGR240C | 6-Phosphofructokinase, alpha subunit | 1 | 1.15 | 1 | 1.14 | 1.63 | 1.25 |
| PFK2 | YMR205C | 6-Phosphofructokinase, beta subunit | 1 | 1 | 1 | 1 | M | M |
| PGI1 | YBR196C | Glucose-6-phosphate isomerase | 1 | 1.18 | 1 | 1.18 | 1.88 | 1.27 |
| PGK1 | YCR012W | Phosphoglycerate kinase | 1 | 1 | 1 | 1 | 1 | −1.04 |
| PGM2 | YMR105C | Phosphoglucomutase, major isoform | 1.82 | 1.27 | 1 | 1 | 1.50 | 1 |
| PSA1 | YDL055C | Mannose-1-phosphate guanyltransferase | 1 | −1.19 | 1.66 | 1 | 1.93 | −1.17 |
| PYC2 | YBR218C | Pyruvate carboxylase 2 | M | M | −2.74 | −1.26 | M | M |
| QCR7 | YDR529C | Ubiquinol-cytochrome c reductase subunit 7 | −1.52 | 1 | 1 | 1 | 1 | −1.25 |
| RHR2 | YIL053W | dl-Glycerol phosphatase | −1.70 | −1.25 | M | M | 1 | 1 |
| RIB3 | YDR487C | 3,4-Dihydroxy-2-butanone 4-phosphate synthase | 1 | 1 | M | M | 1 | 1 |
| RNR2 | YJL026W | Ribonucleoside-diphosphate reductase, small subunit | −1.82 | 1 | −1.86 | −1.19 | −1.51 | −1.17 |
| RNR4 | YGR180C | Ribonucleotide reductase, small subunit | −1.66 | −1.22 | −1.80 | 1 | 1 | −1.22 |
| RPP1B | YDL130W | F1 ATPase stabilizing factor, 10 kDa | −1.14 | 1 | 1 | 1 | 1 | 1 |
| SAH1 | YER043C | S-Adenosyl-l-homocysteine hydrolase | 1 | 1.15 | 1 | 1.15 | −2.07 | 1.17 |
| SAM1 | YLR180W | S-Adenosylmethionine synthetase 1 | M | M | 1 | 1.44 | M | M |
| SAM2 | YDR502C | S-Adenosylmethionine synthetase 2 | 1 | 1 | 1 | 1 | 1.72 | 1 |
| SEC53 | YFL045C | Phosphomannomutase | 1 | 1 | M | M | −1.45 | 1.13 |
| SER1 | YOR184W | Phosphoserine transaminase | M | M | 1 | 1 | 1 | 1 |
| SER33 | YIL074C | 3-Phosphoglycerate dehydrogenase | M | M | 1 | 1.16 | 1 | 1.23 |
| SHM2 | YLR058C | Serine hydroxymethyltransferase | −1.27 | 1.18 | 1 | 1 | 1 | 1 |
| STM1 | YLR150W | Specific affinity for guanine-rich quadruplex nucleic acids | M | M | 1 | 1 | 1 | 1 |
| TAL1 | YLR354C | Transaldolase | 1 | 1.22 | 1 | 1.14 | 1 | 1.19 |
| TDH1 | YJL052W | Glyceraldehyde-3-phosphate dehydrogenase 1 | 1 | −1.34 | 1 | −1.40 | 1 | −1.34 |
| TDH3 | YGR192C | Glyceraldehyde-3-phosphate dehydrogenase 3 | 1 | 1 | 1 | 1 | 1.25 | 1 |
| THR1 | YHR025W | Homoserine kinase | −1.99 | 1 | 1 | 1 | 1 | 1 |
| THR4 | YCR053W | Threonine synthase (o-p-homoserine p-lyase) | 1 | 1 | M | M | 1 | 1 |
| THS1 | YIL078W | Threonyl-tRNA synthetase, cytosolic | M | M | 1 | 1.21 | M | M |
| TKL1 | YPR074C | Transketolase 1 | 1 | 1 | M | M | M | M |
| TPI1 | YDR050C | Triose-phosphate isomerase | 1 | 1.20 | 1 | 1.20 | 1 | 1 |
| TPS1 | YBR126C | Alpha,alpha-trehalose-phosphate synthase | 1 | 1.26 | 1 | 1.08 | 1 | 1.19 |
| TRP5 | YGL026C | Tryptophan synthase | 1 | 1 | 1 | 1 | 1 | 1 |
| TRR1 | YDR353W | Thioredoxin reductase (NADPH) | M | M | 1 | 1 | M | M |
| TSL1 | YML100W | Alpha,alpha-trehalose-phosphate synthase | 3.55 | 1 | 1 | 1 | M | M |
| URA2 | YJL130C | Multifunctional pyrimidine biosynthesis protein | −1.26 | 1.30 | 1 | 1.18 | 1 | 1.31 |
| YDL124W | YDL124W | Similarity to aldose reductases | M | M | M | M | 1 | 1.27 |
| YEL047C | YEL047C | Soluble fumarate reductase | 1 | 1 | 1 | 1 | 1 | 1 |
| YPR1 | YDR368W | Strong similarity to aldo/keto reductase | −1.15 | 1 | 1 | 1 | 1 | 1 |
ORF, open reading frame.
CoA, coenzyme A.
Transcript fold changes are indicated by the letter G, and protein fold changes are indicated by the letter P. Values are the averages of three repeats. Where protein data are unavailable for a particular time point, the letter M is used to indicate missing values. Where no statistically significant differences for gene or protein values in BM45 compared to those in VIN13 exist, the ratio is set to a default value of 1.
TABLE 2.
GO category of cell rescue and defense for protein-mRNA pairs at days 2, 5, and 14
| Gene name | ORF | Functional description | Fold change in cell rescue and defense at daya: |
|||||
|---|---|---|---|---|---|---|---|---|
| 2 |
5 |
14 |
||||||
| BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | BM45 vs VIN13 (G) | BM45 vs VIN13 (P) | |||
| AHP1 | YLR109W | Alkyl hydroperoxide reductase | 1 | 1 | 1 | 1.21 | 1 | 1 |
| CCS1 | YMR038C | Copper chaperone for superoxide dismutase SOD1P | 1 | 1 | 1 | 1 | M | M |
| CPR1 | YDR155C | Cyclophilin (peptidylprolyl isomerase) | 1 | 1 | 1 | 1 | 1 | 1 |
| DAK1 | YML070W | Dihydroxyacetone kinase, induced in high salt | 1.11 | 1 | M | M | M | M |
| DDR48 | YMR173W | Heat shock protein | 1.19 | 1 | 1 | −1.37 | −1.33 | −1.52 |
| GPD1 | YDL022W | Glycerol-3-phosphate dehydrogenase (NAD+) | 1 | 1.05 | 1 | 1 | 1 | 1 |
| GRE3 | YHR104W | Aldose reductase | 1.65 | 1.59 | M | M | 1 | 1.44 |
| GRX1 | YCL035C | Glutaredoxin | M | M | 1 | −1.43 | 1 | −1.75 |
| GRX5 | YPL059W | Member of the subfamily of yeast glutaredoxins | m | M | 1 | −1.44 | 1 | 1 |
| HMF1 | YER057C | Heat shock inducible inhibiter of cell growth | 1 | 1 | M | M | 1 | −1.29 |
| HOR2 | YER062C | dl-Glycerol phosphatase | m | M | 1 | 1 | M | M |
| HSP104 | YLL026W | Heat shock protein | 1 | 1 | 1 | 1 | 1.30 | 1.14 |
| HSP12 | YFL014W | Heat shock protein | 1 | 5.51 | 1 | 2.41 | 1 | 1.98 |
| HSP26 | YBR072W | Heat shock protein | 2.65 | 1.72 | 1 | 1 | 1 | 1 |
| HSP30 | YCR021C | Heat shock protein | m | M | 1 | 1 | 1 | 1.66 |
| HSP60 | YLR259C | Heat shock protein | −1.50 | −1.24 | 1 | −1.21 | 1 | −1.26 |
| HSP78 | YDR258C | Heat shock protein | 1 | 1 | 1 | −1.30 | 1.51 | 1 |
| HSP82 | YPL240C | Heat shock protein | 1 | −1.23 | M | M | M | M |
| LAP3 | YNL239W | Member of the GAL regulon | m | M | 1 | 1 | M | M |
| MET22 | YOL064C | Protein Ser/Thr phosphatase | 1.18 | 1 | M | M | M | M |
| MRH1 | YDR033W | Membrane protein related to HSP30P | 1.13 | 1.27 | 1 | 1 | 1 | 1.32 |
| NCP1 | YHR042W | NADPH-cytochrome P450 reductase | 1.15 | 1 | M | M | M | M |
| PRX1 | YBL064C | Similarity to thiol-specific antioxidant enzyme | 1 | 1 | 1 | −1.11 | M | M |
| SOD1 | YJR104C | Copper-zinc superoxide dismutase | 1 | 1 | 1 | 1 | 1 | 1 |
| SSA1 | YAL005C | Heat shock protein of HSP70 family | 1 | 1 | M | M | 1 | 1 |
| SSC1 | YJR045C | Mitochondrial heat shock protein | −1.59 | −1.19 | 1 | −1.17 | 1.49 | −1.23 |
| SSE1 | YPL106C | Heat shock protein of HSP70 family | 1 | 1.16 | 1 | 1.11 | M | M |
| SSZ1 | YHR064C | Protein involved in pleiotropic drug resistance | 1 | 1 | 1 | 1 | −2.15 | 1 |
| STI1 | YOR027W | Stress-induced protein | 1 | 1 | M | M | 1.36 | −1.34 |
| TPS1 | YBR126C | Alpha,alpha-trehalose-phosphate synthase | 1 | 1.26 | 1 | 1.08 | 1 | 1.19 |
| TRX2 | YGR209C | Thioredoxin II | m | M | 1 | 1 | 1.43 | −1.32 |
| TSA1 | YML028W | Thiol-specific antioxidant | 1 | 1 | 1 | 1 | 1 | 1 |
| YDJ1 | YNL064C | Mitochondrial and ER import protein | −1.84 | 1 | M | M | M | M |
| YHB1 | YGR234W | Flavohemoglobin | −1.29 | −2.37 | 1 | −2.38 | 1 | −2.37 |
Transcript fold changes are indicated by the letter G, and protein fold changes are indicated by the letter P. Values are the averages of three repeats. Where protein data are unavailable for a particular time point, the letter M is used to indicate missing values. Where no statistically significant differences for gene or protein values in BM45 compared to those VIN13 exist, the ratio is set to a default value of 1. ER, endoplasmic reticulum.
For the day 2 analysis, only 9 of the 248 protein/mRNA ratios (for the entire set of identified proteins) differed significantly by a fold change of more than 2. This means that comparisons between strains at a given time point are surprisingly reliable, as fold changes in gene expression and in protein abundance data align with close to 95% overlap within the 2-fold threshold. The same observation holds for the day 5 analysis, where once again only ±4% (8 out of 260) of the protein/mRNA pair ratios differed by a fold change of 2 or greater. These data clearly suggest that comparisons of transcript levels are surprisingly reliable in predicting differences in protein levels between two strains. This appears to hold true for all GO categories and is in stark contrast with previous data (36) which suggest that similar predictions are not reliable when analyzing the evolution of transcriptomes and proteomes during fermentation across time points for a given strain.
By day 14 of fermentation, the close alignment of transcript and protein ratios between strains breaks down slightly. Here, 32 of the 277 protein-mRNA pairs show significant discrepancies in the comparative ratios between BM45 and VIN13. The poorer alignment at this stage of fermentation can probably be explained by the fact that active fermentation has stopped and that cells are exposed to severe stress in the form of high ethanol levels and nutrient depletion. At this stage, active transcription is at a minimum, except for those genes related to the mobilization of reserve nutrients or tolerance of the severe stress conditions faced as the cells slow down metabolically. The levels of accumulated proteins still present at this point may thus bear limited correlation to the levels of mRNA in the cells.
Intrastrain comparison of the evolution of transcriptomes and proteomes.
In order to compare peptide signal areas between different runs (i.e., for comparisons between different time points for either VIN13 and BM45), the data were normalized as follows: all of the iTRAQ signals for peptides that are not shared among multiple detected proteins and that have a confidence score of at least 1.00 were selected. The area for each label in these peptides was calculated as a percentage of the total iTRAQ signal for each of the labels. This final transformed value is more conducive for comparisons across multiple iTRAQ experiments. The agreement among the replicates when expressed as a percentage of the total signal, as per our calculations, was very good and enabled intrastrain comparisons across time points to be made.
When the analysis of transcript versus protein ratios was applied to the intrastrain data sets established at different time points, the results indicated a largely random distribution of protein/transcript ratios (Fig. 1D and E). The intrastrain comparisons clearly do not conform to the distribution curve seen for interstrain alignments. It must be kept in mind that in this analysis, a large positive or negative change in the expression of a particular gene or protein, along with a moderate or large change in the corresponding protein levels (in the same direction), would fall outside the threshold applied here for a good alignment. However, such an alignment would in many cases be considered a good fit from a biological perspective.
To overcome the inherent stringency of this form of analysis, and considering the breakdown of correlation between transcripts and protein levels observed for the intrastrain analysis, we decided to use trends in transcript and protein levels as a second criterion. This assessment is much less stringent since it queries only whether up or down changes in transcript levels over time points would generally correlate with similar trends in protein levels. In this case, ratios in which both transcripts and proteins were less than 1 or greater than 1 were considered aligned. Inverse ratios (i.e., one ratio was less than 1 and the other was greater than 1) constituted a negative result (nonaligned).
Using this approach, the alignment of protein versus transcript data for the VIN13 and BM45 strains between time points (i.e., day 5 versus day 2 and day 14 versus day 5) was only around 60% for all three comparisons. Considering that a random sample would yield 50%, this value is surprisingly low but in line with previous reports. Even when protein-transcript pairs for only the top 50 genes in terms of the magnitude of the increase/decrease in mRNA levels were evaluated, the trend analysis did not improve in any noteworthy manner. For day 5 versus day 2 for both of the strains, the alignment value increased slightly from 65 to 68%, but for day 14 versus day 5, there was a decrease to close to 50%, much lower than the 60% value calculated for the entire gene set. This is surprising, since the transcript levels of these genes were changed by at least 1.8-fold (and up to 32-fold), and such relatively significant changes would generally be expected to be reflected on the proteome level.
There are several possible explanations for this discordant alignment of transcript and protein levels for the intrastrain comparisons. First, our transcriptome and proteome data were generated at the same stage of fermentation. However, the proteome at a specific time point is a reflection of previous rather than concomitant transcript levels. In other words, it would be expected that a particular transcriptomic data set should be more closely aligned with proteomic data that are generated at a later time point, i.e., after the translation and posttranslational modification workflow has responded to the earlier changes in transcription levels. Second, the time points assessed here represent very different environmental conditions within a dynamically changing system, whereas the comparison of different strains at the same time points de facto normalizes for the environmental background. Another point to consider involves the half-lives of proteins and protein turnover. Differences in the turnover rate of mRNA versus the half-lives of encoded proteins would also lead to a discrepancy in the correlation of the mRNA and protein, particularly during stationary phase when the half-lives of certain proteins are extended.
These findings help to explain our observation that the predictive capacity of the omics matrix that was derived from the alignment of transcriptome and exometabolome data sets (37) was statistically reliant mainly on the comparative analysis of several strains and much less reliant on intrastrain comparisons.
Our data set also confirms previous observations (14, 15, 36) that transcriptomic and proteomic data sets are frequently difficult to align across different time points and that transcriptome data need to be interpreted with caution. This is particularly the case when only a single strain is analyzed, as any changes at the transcript level might be specific to the strain in question and not represent a generally relevant response. In this sense, transcriptome comparisons of different strains under the same experimental conditions (regarding time point and medium composition, etc.) represent a more reliable system for inferring biological meaning, since only the genetic background will provide the basis for differences in physiological or phenotypic changes. Using different strains in comparative transcriptome analyses represents an inherent control system that is self-standardized to limit “noisy” outputs.
Transcript-protein pairs showing discordant regulation between time points (i.e., opposite trends in protein and mRNA levels) were investigated more closely in the two strains. Interestingly, the nonaligned gene-protein pairs followed similar trends in both of the strains, suggesting that these trends are not due to experimental error or noise but rather to a consistent feature of the system. To clarify, for the total of 95 gene-protein pairs showing opposite trends in expression levels for the day 5 analysis versus the day 2 analysis in either of the two strains, only 21 of these gene-protein pairs did not overlap between strains. For the day 14 analysis versus the day 5 analysis, only 37 of the total of 124 nonaligned mRNA-protein pairs did not overlap between the BM45 and VIN13 strains. Thus, discordant alignment between transcriptomes and proteomes between time points is relatively consistent for the different strains, which is helpful for elucidating the regulation of expression/translation of these consistently nonaligned genes. Without the use of multiple strains, this feature of the transcriptome/proteome would have been overlooked. The set of overlapping, yet discordant, transcript-protein pairs were classified according to functional activity and translation, cysteine metabolism, and biopolymer biosynthesis were strongly represented categories.
Functional categorization.
For comparisons within a single experiment, the ratios of BM45 and VIN13 for both expressed genes and proteins were determined and compared. To facilitate evaluation of the data, the protein-mRNA pairs were categorized according to GO classification terms. The proteins identified in our analysis can reasonably be considered representative of the entire proteome, as all functional categories are well represented (i.e., approximately 160 proteins are involved in energy and metabolism, 25 in cell cycle regulation, 35 in cellular transport, 35 in cell rescue and defense, 80 in protein synthesis, and 25 in transcription). Furthermore, no bias toward any generic protein feature, such as concentration or hydrophobicity profiles, was obvious in the data. In this section, the following two relevant categories are further discussed as examples: energy and metabolism as well as cell rescue and defense (Tables 1 and 2).
As can be seen in Tables 1 and 2, and as would be expected when considering the overall good alignment presented for the interstrain comparisons at similar time points, the relative over- or underexpression of genes generally coincides with a similar trend in the protein abundance data (particularly for the first two time points during fermentation).
The same functional categories were also analyzed for the intrastrain data. Surprisingly, when considering the rather poor general alignment of changes in transcript and protein levels in this case, gene expression and protein levels also aligned well for the specific functional categories of amino acid metabolism and fermentative metabolism, suggesting a strong transcriptional control of such metabolic enzymes (Table 3). This is in contrast to the results reported by Rossignol et al. (36), in which most of the glycolytic and amino acid metabolic proteins identified showed opposite correlations between mRNA and proteins between the two fermentative stages considered (exponential phase versus stationary phase) during alcoholic fermentation in synthetic must (MS300).
TABLE 3.
Relative protein and transcript ratios for day 5 versus day 2 analyses of VIN13 and BM45 for genes involved in fermentation and amino acid metabolisma
| Gene name | ORF | Ratio |
Trend |
||||
|---|---|---|---|---|---|---|---|
| BM45 (P) | BM45 (G) | VIN13 (P) | VIN13 (G) | BM45 | VIN13 | ||
| ACS2 | YLR153C | 0.29 | 0.41 | 0.27 | 0.47 | + | + |
| ARO3 | YDR035W | 0.39 | 0.90 | 0.48 | 1.53 | + | − |
| ARO4 | YBR249C | 0.41 | 0.24 | 0.44 | 0.24 | + | + |
| ASN1 | YPR145W | 0.65 | 0.39 | 0.65 | 0.23 | + | + |
| BAT1 | YHR208W | 0.75 | 0.44 | 0.98 | 0.63 | + | + |
| GDH1 | YOR375C | 0.60 | 0.18 | 0.69 | 0.08 | + | + |
| GPD1 | YDL022W | 1.53 | 1.49 | 1.63 | 1.43 | + | + |
| GPH1 | YPR160W | 2.41 | 0.93 | 3.07 | 1.32 | − | + |
| ILV3 | YJR016C | 0.76 | 0.39 | 0.78 | 0.42 | + | + |
| ILV5 | YLR355C | 0.70 | 0.62 | 0.69 | 0.52 | + | + |
| LEU2 | YCL018W | 1.88 | 0.26 | 2.32 | 0.49 | − | − |
| LYS12 | YIL094C | 0.22 | 0.34 | 0.24 | 0.24 | + | + |
| LYS21 | YDL131W | 0.25 | 0.28 | 0.31 | 0.27 | + | + |
| LYS4 | YDR234W | 0.22 | 0.68 | 0.28 | 0.42 | + | + |
| LYS9 | YNR050C | 0.66 | 0.11 | 0.61 | 0.07 | + | + |
| PDC1 | YLR044C | 0.60 | 0.91 | 0.62 | 0.87 | + | + |
| PFK1 | YGR240C | 0.60 | 0.90 | 0.59 | 0.82 | + | + |
| PFK2 | YMR205C | 0.67 | 0.69 | 0.64 | 0.50 | + | + |
| PGM2 | YMR105C | 1.27 | 1.31 | 1.59 | 2.91 | + | + |
| SAM2 | YDR502C | 0.26 | 0.86 | 0.27 | 0.74 | + | + |
| SHM2 | YLR058C | 0.56 | 0.30 | 0.61 | 0.30 | + | + |
| TDH1 | YJL052W | 1.34 | 0.95 | 1.38 | 0.98 | − | + |
| TDH3 | YGR192C | 0.45 | 0.68 | 0.41 | 0.79 | + | + |
| THR1 | YHR025W | 0.22 | 0.35 | 0.35 | 0.18 | + | + |
| TPI1 | YDR050C | 0.68 | 0.87 | 0.68 | 0.79 | + | + |
| TPS1 | YBR126C | 0.72 | 1.93 | 0.83 | 3.01 | − | − |
| TRP5 | YGL026C | 0.37 | 0.48 | 0.37 | 0.48 | + | + |
| TSL1 | YML100W | 3.55 | 2.13 | 2.71 | 7.83 | + | + |
Transcript ratios are indicated by the letter G, and protein ratios are indicated by the letter P. Matching trend alignments are indicated by a plus sign, while opposite trends in transcript and protein levels are indicated by a minus sign. Values are the averages of three repeats.
Other categories showed almost no relationship between changes in transcript and protein levels. As an example, Table 4 shows data from the GO category of transcription and cell cycle control. The difference in the alignment of protein and transcript data between different functional categories becomes quite apparent when contrasting it with the results depicted in Tables 3 and 4. Transcriptomic data thus appears to be reasonably representative of protein levels for metabolic enzymes but not for most other GO categories such as general cell maintenance and growth.
TABLE 4.
Relative protein and transcript ratios for day 5 versus day 2 analyses of VIN13 and BM45 for the GO categories of transcription and cell cycle controla
| Category/gene name | ORF | Ratio |
Trend |
||||
|---|---|---|---|---|---|---|---|
| BM45 (P) | BM45 (G) | VIN13 (P) | VIN13 (G) | BM45 | VIN13 | ||
| Transcription | |||||||
| NOP1 | YDL014W | 0.74 | 0.39 | 0.69 | 0.19 | + | + |
| SUB2 | YDL084W | 1.36 | 0.72 | 1.34 | 0.52 | − | − |
| HTA1 | YDR225W | 0.47 | 1.09 | 0.43 | 0.97 | − | + |
| NPL3 | YDR432W | 0.83 | 1.53 | 0.85 | 1.00 | − | + |
| SNU13 | YEL026W | 1.45 | 0.85 | 1.73 | 0.65 | − | − |
| PAB1 | YER165W | 0.90 | 0.55 | 1.06 | 0.55 | + | − |
| ARC1 | YGL105W | 1.26 | 0.32 | 1.86 | 0.22 | − | − |
| ADE3 | YGR204W | 0.22 | 0.81 | 0.24 | 0.64 | + | + |
| EGD2 | YHR193C | 1.41 | 0.39 | 1.61 | 0.27 | − | − |
| DED1 | YOR204W | 0.29 | 2.72 | 0.29 | 3.33 | − | − |
| NOP58 | YOR310C | 0.02 | 0.50 | 0.01 | 0.32 | + | + |
| EGD1 | YPL037C | 0.62 | 0.30 | 1.03 | 0.23 | + | − |
| RPO26 | YPR187W | 0.60 | 0.76 | 0.70 | 0.37 | + | + |
| Cell cycle | |||||||
| CMD1 | YBR109C | 7.75 | 0.83 | 9.03 | 0.81 | − | − |
| CDC48 | YDL126C | 4.40 | 1.47 | 4.44 | 1.18 | + | + |
| HSP12 | YFL014W | 5.63 | 1.77 | 12.68 | 2.06 | + | + |
| ACT1 | YFL039C | 1.37 | 0.85 | 1.40 | 0.90 | − | − |
| MLC1 | YGL106W | 1.93 | 0.71 | 2.30 | 0.37 | − | − |
| RNR4 | YGR180C | 0.45 | 0.39 | 0.44 | 0.42 | + | + |
| RNR2 | YJL026W | 0.87 | 0.25 | 0.91 | 0.26 | + | + |
| RPL10 | YLR075W | 0.95 | 0.69 | 1.10 | 0.74 | + | − |
| HSP82 | YPL240C | 0.50 | 5.52 | 0.45 | 4.97 | − | − |
Transcript ratios are indicated by the letter G, and protein ratios are indicated by the letter P. Positive trend alignments are indicated by a plus sign, while opposite trends in transcript and protein levels are indicated by a minus sign. Values are the averages of three repeats.
Correlations between protein levels and phenotype.
In a related work, the strains VIN13 and BM45 were phenotypically profiled (38), and some differences in protein abundance between the two strains can tentatively be correlated to specific phenotypic differences. For instance, the significantly lower levels of several heat shock proteins, such as Hsp60, Hsp82, and Ddr48 in BM45 in comparison to those in VIN13 (Table 2), could account for the generally lower tolerance of this strain under various stress conditions, including heat stress. This hypothesis is strongly supported by data that show that individual overexpression of these gene results in higher stress resistance, and lifting the expression level of these genes in BM45 to the level observed in VIN13 should therefore result in a recognizable phenotypic change (8, 40). Similarly, lower levels of antioxidant proteins such as Tsa1 and Yhb1 (Table 2) could also explain the increased susceptibility of BM45 to oxidative stress in comparison to the susceptibility of VIN13 (49). Lower protein abundances of Erg13, Erg20, and Erg6 (Table 1) in BM45 versus those in VIN13 could also account for the lower ethanol and osmotic shock tolerance of BM45, given that these proteins are involved in the production of a variety of sterols with roles in cell membrane stabilization (33, 51).
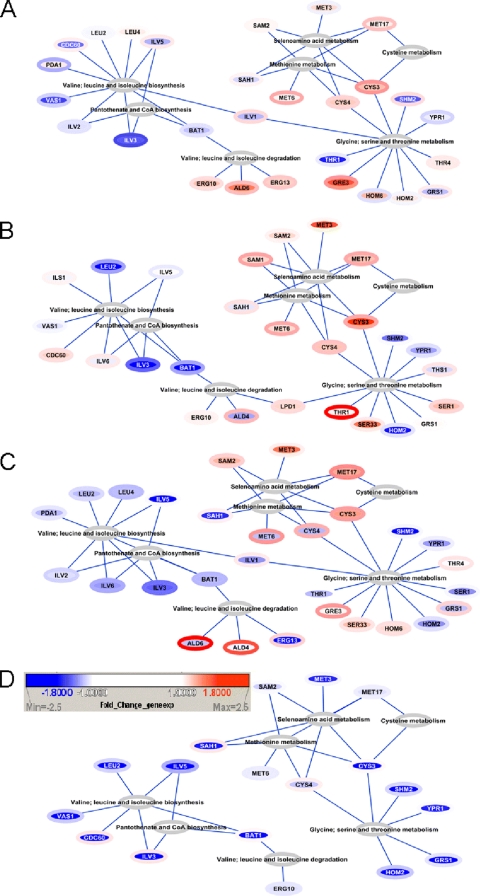
Regarding metabolism, the data indicate why the alignment of exometabolome and transcriptome data have previously proven successful (37). Indeed, differences in the ratios of several proteins involved in the synthesis of the aromatic amino acids (namely, Aro1, Aro3, Aro4, and Aro8) (Table 1) are reflected by differences in the concentrations of the end products of these pathways. Likewise, Bat1 is involved in catalyzing the first transamination step of the catabolic formation of fusel alcohols via the Ehrlich pathway (24). Differences in Bat1 expression (Fig. 2; Table 1) have proven to effect large changes in higher alcohol production by wine yeast strains (37). BAT1 gene expression and Bat1 protein levels are quite notably concordant (Fig. 2), and the decrease in expression of BM45 relative to that of VIN13 agrees with metabolite data showing significantly lower propanol, butanol, and methanol production by BM45 in comparison to that by VIN13 (37). In fact, this close alignment between transcript and protein levels appears to be the case for almost all of the gene-protein pairs linked to the metabolism of the amino acids shown in Fig. 2, at both days 2 and 5 and even at day 14. In Fig. 2, it is clear that there is a direct correlation between transcript and protein abundance in central metabolic pathways, (such as those pathways related to amino acid metabolism in this example).
FIG. 2.
Network visualization of protein and gene expression ratios in metabolic hubs linked to amino acid metabolism. The pathway networks for BM45 versus VIN13 at days 2 (A), 5 (B), and 14 (C) are presented. (D) Changes in gene and protein levels for day 5 versus day 2 in VIN13. Visual mapping was used to represent the ratios of RNA and proteins as follows. RNA ratios are represented by a linear color scale assigned to the interior of each node, and protein ratios are represented by a linear color scale assigned to the border of each node. Both of the linear color scales are constructed such that the maximum intensity is set to correspond to ratios equal to or above a positive or negative 2-fold difference between strains or between time points within each strain. The blue scale represents negative ratios, while the red scale represents positive ratios. White indicates a ratio of 1, i.e., no difference for that molecule.
Amino acid metabolism is of particular interest from a wine-making perspective, as amino acids serve as the precursors of important volatile aroma compounds. For instance, sulfur-containing amino acids such as methionine (and cysteine to a lesser extent) are the precursors for the volatile thiols that are significant aroma compounds in wine (43). The branched-chain amino acids such as valine, leucine, and isoleucine, on the other hand, serve as the precursors for various higher alcohols. Of the enzymes involved in branched-chain amino acid metabolism, BAT1 was been discussed above. Other genes that encode enzymes in this pathway and that were identified in our previous study (37) for their strong statistical link between expression levels and the production of specific aroma compounds include LEU2, encoding a beta-isopropylmalate dehydrogenase that catalyzes the third step in the leucine biosynthesis pathway (4). Expression of this gene showed a significant statistical correlation with compounds such as isobutanol (37), and as can be seen in Fig. 2, the relative transcript and protein abundance ratios align well for this gene. Of the genes involved in the metabolism of isoleucine and valine (precursors for higher alcohol synthesis), the ILV gene family (ILV1, ILV2, ILV3, ILV5, and ILV6) encode isoforms of acetohydroxyacid reductoisomerases involved in branched-chain amino acid biosynthesis (20). Expression of the ILV gene isoforms showed strong positive correlations with many higher alcohols analyzed in a previous study, and expression differences between BM45 and VIN13 once again align with differences in the exometabolite profiles of these two strains, as reported by Rossouw et al. (37). The ILV gene/protein ratio is also well-aligned, again confirming the tight, concordant regulation of transcript levels and enzyme abundance in key metabolic pathways.
In terms of intrastrain comparisons between time points, the alignment of changes in transcription and protein abundance is also good when considering metabolic pathways such as those of amino acid metabolism (Fig. 2D). Although the intensity of the fold change differs for mRNA and proteins, the overall trends match up well. Figure 2D shows that there is a general downregulation of transcripts (and their corresponding proteins) involved in amino acid metabolism as fermentation proceeds from exponential growth phase (day 2) to early stationary phase (day 5). This is in line with yeast growth behavior, as day 5 represents a fermentative phase characterized by continued high rates of fermentative metabolism associated with a significant reduction in growth and biomass formation.
Concluding remarks.
Although our coverage of the yeast proteome was only around 5%, the identified proteins were distributed over all functional categories. This coverage is also significantly higher than that obtained in previous studies (36) and appears sufficient to assess the biological relevance and reliability of the transcriptome data. In our study, the alignment of relative protein abundance ratios with gene expression data was accurate for data generated in the early stages of fermentation (days 2 and 5), when active cell growth and metabolism is occurring. In the case of data comparisons across time points, the quality of gene expression to protein correlations deteriorates substantially, due to the lag time between the expressed transcriptome and later changes in the protein profile. In the intrastrain analysis, only the alignment of protein and transcript levels within metabolic pathways specifically proved to be extremely reliable. This confirms the observations by Rossignol et al. (36).
Clearly, transcriptomic studies involving analyses across different time points are fraught with significant complication and therefore may be more difficult to interpret in a biologically meaningful manner. On the other hand, comparison of transcription patterns in the context of different genetic backgrounds appears to provide a reliable indication of underlying genetic differences and phenotypes. This means that many of the molecular causes of phenotypical differences between strains can most probably be directly derived from transcriptomic data sets.
Most notably, the concordance of gene and protein levels of enzymes involved in metabolism confirms transcriptional control of at least some of the important metabolic pathways in yeast. This implies that transcriptomic data can theoretically be applied to evaluate and model certain aspects of yeast metabolism with relative confidence. The agreement of protein abundance ratios between strains with the phenotypic characteristics of these strains further strengthens our belief that the “omics” data sets we have generated provide valuable and reliable insights into the fundamental molecular mechanisms at work in industrial wine yeast strains during alcoholic fermentation.
Supplementary Material
Acknowledgments
Funding for the research presented in this paper was provided by the NRF and Winetech, and personal sponsorship was provided by the Wilhelm Frank Trust.
Proteomic analysis was performed by Martin Middleditch at the Centre for Genomics and Proteomics at the University of Auckland. We thank Jo McBride and the Cape Town Centre for Proteomic and Genomic Research for the microarray hybridization and subsequent signal detection and the staff and students at the IWBT for their support and assistance in numerous areas.
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbott, D. A., T. A. Knijnenburg, L. M. de Poorter, M. J. Reinders, J. T. Pronk, and A. J. van Maris. 2007. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 7:819-833. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal, K., L. H. Choe, and K. H. Lee. 2005. Quantitative analysis of protein expression using amine-specific isobaric tags in Escherichia coli cells expressing rhsA elements. Proteomics 5:2297-2308. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 4.Andreadis, A., Y. P. Hsu, M. Hermodson, G. Kohlhaw, and P. Schimmel. 1984. Yeast LEU2. Repression of mRNA by leucine and primary structure of the gene product. J. Biol. Chem. 259:8059-8062. [PubMed] [Google Scholar]
- 5.Ashby, M., and J. Rine. October 2006. Methods for drug screening. U.S. patent 5,569,588.
- 6.Bakalinsky, A. T., and R. Snow. 1990. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6:367-382. [DOI] [PubMed] [Google Scholar]
- 7.Bely, L., J. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Viticult. 40:319-324. [Google Scholar]
- 8.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman, A. R., A. Forgan, I. S. Pretorius, and P. J. Chambers. 2008. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 8:1185-1195. [DOI] [PubMed] [Google Scholar]
- 10.Brejning, J., N. Arneborg, and L. Jespersen. 2005. Identification of genes and proteins induced during the lag and early exponential phase of lager brewing yeasts. J. Appl. Microbiol. 98:261-271. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X., L. W. Sun, Y. P. Yu, Y. Xue, and P. Yang. 2007. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics 4:25-37. [DOI] [PubMed] [Google Scholar]
- 12.Cline, M. S., M. Smoot, E. Cerami, A. Kuchinsky, N. Landys, C. Workman, R. Christmas, I. Avila-Campilo, M. Creech, B. Gross, K. Hanspers, R. Isserlin, R. Kelley, S. Killcoyne, S. Lotia, S. Maere, J. Morris, K. Ono, V. Pavlovic, A. R. Pico, A. Vailaya, P. L. Wang, A. Adler, B. R. Conklin, L. Hood, M. Kuiper, C. Sander, I. Schmulevich, B. Schwikowski, G. J. Warner, T. Ideker, and G. D. Bader. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2:2366-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daran-Lapujade, P., M. L. Jansen, J. M. Daran, W. van Gulik, J. H. de Winde, and J. T. Pronk. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J. Biol. Chem. 279:9125-9138. [DOI] [PubMed] [Google Scholar]
- 14.de Godoy, L. M. F., J. V. Olsen, J. Cox, M. L. Nielsen, N. C. Hubner, F. Fröhlich, T. C. Walther, and M. Mann. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251-1254. [DOI] [PubMed] [Google Scholar]
- 15.De Groot, M. J. L., P. Daran-Lapujade, B. van Breukelen, T. A. Knijnenburg, E. A. F. de Hulster, M. J. T. Reinders, J. T. Pronk, A. J. R. Heck, and M. Slijper. 2007. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153:3864-3878. [DOI] [PubMed] [Google Scholar]
- 16.Erasmus, D. J., G. K. van der Merwe, and H. J. J. van Vuuren. 2003. Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 3:375-399. [DOI] [PubMed] [Google Scholar]
- 17.Fey, S. J., and P. M. Larsen. 2001. 2D or not 2D. Two-dimensional gel electrophoresis. Curr. Opin. Chem. Biol. 5:26-33. [DOI] [PubMed] [Google Scholar]
- 18.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldman, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 19.Griffin, T. J., S. P. Gygi, T. Ideker, B. Rist, J. Eng, L. Hood, and R. Aebersold. 2002. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1:323-333. [DOI] [PubMed] [Google Scholar]
- 20.Holmberg, S., and J. G. Petersen. 1988. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 13:207-217. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, and Y. Yamanishi. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480-D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kispal, G., H. Steiner, D. A. Court, B. Rolinski, and R. Lill. 1996. Mitochondrial and cytosolic branched chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J. Biol. Chem. 271:24458-24464. [DOI] [PubMed] [Google Scholar]
- 25.Kobi, D., S. Zugmeyer, S. Potier, and L. Jaquet-Gutfreund. 2004. Two-dimensional map of an “ale”-brewing yeast strain: proteome dynamics during fermentation. FEMS Yeast Res. 5:213-230. [DOI] [PubMed] [Google Scholar]
- 26.Kolkman, A., P. Daran-Lapujade, A. Fullaondo, M. M. A. Olsthoorn, J. T. Pronk, M. Slijper, and A. J. R. Heck. 2006. Proteome analysis of yeast response to various nutrient limitations. Mol. Sys. Biol. 2:0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maillet, I., G. Lagniel, M. Perrot, H. Boucherie, and J. Labarre. 1996. Rapid identification of yeast proteins on two-dimensional gels. J. Biol. Chem. 271:10263-10270. [DOI] [PubMed] [Google Scholar]
- 28.Mann, M., R. C. Hendricksen, and A. Pandey. 2001. Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem. 70:437-473. [DOI] [PubMed] [Google Scholar]
- 29.Marks, V. D., S. J. Ho Sui, D. Erasmus, G. K. van den Merwe, J. Brumm, W. W. Wasserman, J. Bryan, and H. J. van Vuuren. 2008. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 8:35-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nägele, E., M. Vollmer, and P. Hörth. 2004. Improved 2D nano-LC/MS for proteomics applications: a comparative analysis using yeast proteome. J. Biomol. Tech. 15:134-143. [PMC free article] [PubMed] [Google Scholar]
- 31.Novo, M., F. Bigey, E. Beyne, V. Galeote, F. Gavory, S. Mallet, B. Cambon, J.-L. Legras, P. Wincker, S. Casaregola, and S. Dequin. 2009. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U. S. A. 106:16333-16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 33.Parks, L. W., S. J. Smith, and J. H. Crowley. 1995. Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids 30:227-230. [DOI] [PubMed] [Google Scholar]
- 34.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2002. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43-50. [DOI] [PubMed] [Google Scholar]
- 35.Rabilloud, T. 2002. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs the mountains. Proteomics 2:3-10. [PubMed] [Google Scholar]
- 36.Rossignol, T., D. Kobi, L. Jacquet-Gutfreund, and B. Blondin. 2009. The proteome of a wine yeast strain during fermentation, correlation with the transcriptome. J. Appl. Microbiol. 107:47-55. [DOI] [PubMed] [Google Scholar]
- 37.Rossouw, D., T. Naes, and F. F. Bauer. 2008. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9:530-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossouw, D., R. Olivares-Hernandes, J. Nielsen, and F. F. Bauer. 2009. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl. Environ. Microbiol. 75:6600-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvadó, Z., R. Chiva, S. Rodríguez-Vargas, F. Rández-Gil, A. Mas, and J. M. Guillamón. 2008. Proteomic evolution of a wine yeast during the first hours of fermentation. FEMS Yeast Res. 8:1137-1146. [DOI] [PubMed] [Google Scholar]
- 40.Sanyal, A., A. Harington, C. J. Herbert, O. Groudinsky, P. P. Slonimsky, B. Tung, and G. S. Getz. 1995. Heat shock protein HSP60 can alleviate the phenotype of mitochondrial RNA-deficient temperature-sensitive mna2 pet mutants. Mol. Gen. Genet. 246:56-64. [DOI] [PubMed] [Google Scholar]
- 41.Shannon, P., A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, and T. Ideker. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 11:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smolka, M., H. Zhou, and R. Aebersold. 2002. Quantitative protein profiling using two-dimensional gel electrophoresis, isotope-coded affinity tag labeling, and mass spectrometry. Mol. Cell. Proteomics 1:19-29. [DOI] [PubMed] [Google Scholar]
- 43.Swiegers, J. H., D. L. Capone, K. H. Pardon, G. M. Elsey, M. A. Sefton, I. L. Francis, and I. S. Pretorius. 2007. Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast 24:561-574. [DOI] [PubMed] [Google Scholar]
- 44.Tong, A. H., B. Drees, G. Nardelli, G. D. Bader, B. Brannetti, L. Castagnoli, M. Evangelista, S. Ferracuti, B. Nelson, S. Paoluzi, M. Quondam, A. Zucconi, C. W. Hogue, S. Fields, C. Boone, and G. Cesareni. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321-324. [DOI] [PubMed] [Google Scholar]
- 45.Trabalzini, L., A. Paffeti, A. Scaloni, F. Talamo, E. Ferro, G. Coratza, L. Bovalini, P. Lusini, P. Martelli, and A. Santucci. 2003. Proteomic response to physiological fermentation stresses in a wild-type wine strain of Saccharomyces cerevisiae. Biochem. J. 370:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vido, K., D. Spector, G. Lagniel, S. Lopez, M. B. Toledano, and J. Labarre. 2001. A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276:8469-8474. [DOI] [PubMed] [Google Scholar]
- 47.Walhout, A. J., J. Reboul, O. Shtanko, N. Bertin, P. Vaglio, H. Ge, H. Lee, L. Doucette-Stamm, K. C. Gunsalus, A. J. Schetter, D. G. Morton, K. J. Kemphues, V. Reinke, S. K. Kim, F. Piano, and M. Vidal. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12:1952-1958. [DOI] [PubMed] [Google Scholar]
- 48.Washburn, M. P., R. Ulaszek, C. Deciu, D. M. Schielts, and J. R. Yates. 2002. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74:1650-1657. [DOI] [PubMed] [Google Scholar]
- 49.Wong, C. M., Y. Zhou, R. W. Ng, H. F. Kung, and D. Y. Jin. 2002. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 277:5385-5394. [DOI] [PubMed] [Google Scholar]
- 50.Wu, Z., R. Irizarry, R. Gentleman, F. M. Murillo, and F. Spencer. 2004. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909-917. [Google Scholar]
- 51.Yoshikawa, K., T. Tanaka, C. Furusawa, K. Nagahisa, T. Hirasawa, and H. Shimizu. 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9:32-44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.