Abstract
A polyolefinic hydrocarbon was found in nonpolar extracts of Shewanella oneidensis MR-1 and identified as 3,6,9,12,15,19,22,25,28-hentriacontanonaene (compound I) by mass spectrometry, chemical modification, and nuclear magnetic resonance spectroscopy. Compound I was shown to be the product of a head-to-head fatty acid condensation biosynthetic pathway dependent on genes denoted as ole (for olefin biosynthesis). Four ole genes were present in S. oneidensis MR-1. Deletion of the entire oleABCD gene cluster led to the complete absence of nonpolar extractable products. Deletion of the oleC gene alone generated a strain that lacked compound I but produced a structurally analogous ketone. Complementation of the oleC gene eliminated formation of the ketone and restored the biosynthesis of compound I. A recombinant S. oneidensis strain containing oleA from Stenotrophomonas maltophilia strain R551-3 produced at least 17 related long-chain compounds in addition to compound I, 13 of which were identified as ketones. A potential role for OleA in head-to-head condensation was proposed. It was further proposed that long-chain polyunsaturated compounds aid in adapting cells to a rapid drop in temperature, based on three observations. In S. oneidensis wild-type cells, the cellular concentration of polyunsaturated compounds increased significantly with decreasing growth temperature. Second, the oleABCD deletion strain showed a significantly longer lag phase than the wild-type strain when shifted to a lower temperature. Lastly, compound I has been identified in a significant number of bacteria isolated from cold environments.
Currently, there is industrial interest in nongaseous microbial hydrocarbons for specialty chemical applications and, more recently, as high-energy biofuels (20, 27, 34). Microbes produce hydrocarbons of different types, for example, aliphatic isoprenoid compounds (20) and alkanes from fatty aldehyde decarbonylation (10). Fatty aldehyde decarbonylation is not well understood but offers a clean route to diesel fuels from fatty acids.
Certain microbes also make a distinctly different class of long-chain hydrocarbons, generally C25 to C33 in chain length, that contain a double bond near the middle of the chain (1, 3, 5, 15, 30, 31, 33, 34). These long-chain olefinic hydrocarbons are thought to derive from processes different than isoprene condensation and decarbonylation mechanisms. This class of hydrocarbons has been shown by carbon-14-labeling studies (2) to derive from fatty acids. The process, described in 1929 by Channon and Chibnall (9), has become known as head-to-head hydrocarbon biosynthesis. Albro and Ditmar (3) defined the head-to-head condensation as coupling of the head (C1) and the α-carbon (C2) of two fatty acids with decarboxylation, a reaction that should not be confused with an acyloin-like carboxyl carbon-to-carboxyl carbon coupling. Products of the head-to-head mechanism have been identified in Gram-positive bacteria such as Micrococcus luteus (29, 30) and Arthrobacter aurescens (13) and in Gram-negative bacteria such as Stenotrophomonas maltophilia (28). Micrococcus and Arthrobacter strains produce fatty acids that are methyl branched terminally and subterminally (8, 29, 30). The long-chain olefinic hydrocarbons from those strains similarly contain a mixture of terminal and subterminal methyl group branching (2, 13, 31).
Albro and Ditmar (3, 4) acquired direct evidence for the head-to-head mechanism occurring in microbial whole organisms and cell extracts. In cell extracts, it was shown that one of the fatty acid carboxyl groups is lost as carbon dioxide, with the remaining carbon atoms being retained in the resultant hydrocarbon (4). The hydrocarbons contain a double bond at the point of condensation. More recently, Beller et al. described the genes encoding head-to-head fatty acid condensation pathway enzymes from Micrococcus luteus, which are known as ole genes for the olefin products formed (5). Three genes from Micrococcus luteus were shown to confer on Escherichia coli the ability to make long-chain olefinic hydrocarbons. Two recent patent applications by L. Friedman et al. (18 September 2008, WO2008/113041; 4 December 2008, WO2008/147781) also described a three- or four-gene cluster as being involved in head-to-head hydrocarbon biosynthesis to make olefins. The patent applications identified homologs to ole genes in different bacteria, including strains of Shewanella.
Bacteria of the genus Shewanella have been heavily studied over the last decade because they are widespread and have the ability to use a startling variety of electron acceptors for respiration (11). There are more than 20 completed genome sequences for Shewanella strains. The model system for studying Shewanella is S. oneidensis MR-1. The genome sequencing of S. oneidensis MR-1 was reported in 2002 (16), and the organism has been shown to be highly amenable to genetic manipulation (11).
The present study used Shewanella oneidensis strain MR-1 as a model system to investigate hydrocarbon biosynthetic genes and the possible biological function of the proteins they encode. The hydrocarbon produced by the Ole proteins in S. oneidensis MR-1 was found to be very different from hydrocarbons previously identified as deriving from a head-to-head condensation mechanism (28, 29, 32). The product was identified here as 3,6,9,12,15,19,22,25,28-hentriacontanonaene by chemical modification studies, mass spectrometry, and nuclear magnetic resonance (NMR) spectroscopy. Previously, a similar polyolefin had been identified in many Antarctic bacteria (22). Cloning of a heterologous oleA gene into S. oneidensis MR-1 was found to produce a completely different set of products. A hydrocarbon deletion mutant showed a distinctly longer growth lag than wild-type cells when shifted to a lower temperature, suggesting that the ole genes in S. oneidensis MR-1 may aid the cells in adapting to a sudden drop in temperature.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and growth.
A list of Shewanella strains used in this study can be found in Table 1. Cultures of S. oneidensis MR-1 were routinely grown in Luria-Bertani (LB) medium under ideal conditions (aerobic, 30°C) unless stated otherwise. Cultures were grown to early stationary phase at 36°C, 22°C, 15°C, or 4°C for experiments in which the relative amount of hydrocarbon was determined (n = 6). In cold adaption experiments (n = 6), the oleABCD mutant and wild-type strains were first grown to a similar optical density (OD) on LB medium overnight at 30°C and then diluted by the same dilution factor into fresh medium at 4°C with a beginning OD of approximately 0.01. Aerobic growth was continued at 4°C, and optical densities were measured using a Beckman DU 7400 spectrophotometer. For each treatment (six flasks), three OD measurements were made and then averaged.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic(s) | Reference/source |
|---|---|---|
| Strains | ||
| S. oneidensis MR-1 | Wild type | Lab stock |
| S. oneidensis Δole | S. oneidensis MR-1, Δole; does not produce hydrocarbon | This study |
| S. oneidensis ΔoleC | S. oneidensis MR-1, ΔoleC; does not produce hydrocarbon | This study |
| S. oneidensis ΔpfaA | S. oneidensis MR-1, ΔpfaA; does not produce hydrocarbon | This study |
| E. coli UQ950 | E. coli DH5α λ(pir) host for cloning; F− Δ(argF-lac)169 φ80dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96(NalR) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | 26 |
| E. coli WM3064 | Donor strain for conjugation; thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | 26 |
| Plasmids | ||
| pSMV3 | 9.5-kb vector; Kmr-only version of pSMV8; lacZ sacB | 26 |
| pSMV3-Δole | 2.3-kb fusion PCR fragment containing Δole cloned into SpeI/SacI site of pSMV3; used to make S. oneidensis Δole strain | This study |
| pSMV3-ΔoleC | 2.2-kb fusion PCR fragment containing ΔoleC cloned into SpeI/SacI site of pSMV3; used to make S. oneidensis ΔoleC strain | This study |
| pSMV3-ΔpfaA | 2.0-kb fusion PCR fragment containing ΔpfaA cloned into SpeI/ApaI site of pSMV3; used to make S. oneidensis ΔpfaA strain | This study |
| pBBR1MCS-2 | 5.1-kb broad-host-range plasmid; lacZ; Kmr | 19 |
| pOleC | 2.1-kb PCR fragment containing S. oneidensis oleC, cloned into SpeI/SacI site of pBBR1MCS-2 | This study |
| pPfaA | 7.6-kb PCR fragment containing S. oneidensis pfaA, cloned into ApaI/SpeI site of pBBR1MCS-2 | This study |
| pOleA-S.m. | 1.1-kb PCR fragment containing S. maltophilia oleA, cloned into SpeI/SacI site of pBBR1MCS-2 | This study |
For maintenance of plasmids in S. oneidensis strains, 50 μg/ml of kanamycin (Km) was added to the medium. For selection for recombinants (see “Mutagenesis,” below), Km was added to a final concentration of 50 μg/ml while sucrose was added to a final concentration of 5% (wt/vol). Escherichia coli strains and their genotypes are listed in Table 1. All E. coli strains were grown aerobically at 37°C in LB. Where appropriate, Km was added to the growth medium at a final concentration of 50 μg/ml and diaminopimelic acid was added to a final concentration of 0.3 mM.
Hydrocarbon and ketone analysis.
Hydrocarbons and ketones were analyzed by gas chromatography-mass spectrometry (GC-MS) as previously described (13). Early-stationary-phase cultures, cells and medium together, were extracted. The resulting evaporated residue was recovered in 1 ml of methyl-tert-butyl ether and applied to a 4.0-g silica gel column, eluted with 35 ml of hexanes, concentrated, and subjected to molecular distillation using a Bantamware sublimation apparatus. The hydrocarbon distillate was collected between 100 and 115°C (0.02 torr), and the ketone distillate was collected between 120 and 130°C (0.02 torr). The distillates were recovered in 1 ml of pentanes and subjected to GC-MS analysis using an HP6890 gas chromatograph connected to an HP5973 mass spectrometer (Hewlett Packard, Palo Alto, CA). GC conditions consisted of the following: helium gas at 1 ml/min; HP-1ms column (100% dimethylpolysiloxane capillary, 30 m by 0.25 mm by 0.25 μm); temperature ramp, 100 to 320°C, at 10°C/min, with a 5-min hold at 320°C. The mass spectrometer was run in electron impact mode at 70 eV and 35 μA.
The 3,6,9,12,15,19,22,25,28-hentriacontanonaene (compound I) produced by wild-type S. oneidensis MR-1 was purified and identified through GC-MS and NMR analyses. NMR was performed using a Varian INOVA 500 MHz NMR apparatus. Olefin hydrogenation used 5% palladium on carbon as the catalyst under hydrogen at 1 to 2 atm pressure. Chemical characterization: thin-layer chromatography (TLC; hexanes:dichloromethane at 80:20 [vol/vol]), RF = 0.13; (hexanes:dichloromethane, 80:20 [vol/vol], silver nitrate), RF = 0.027; 1H-NMR (500 MHz, CDCl3): 5.28 to 5.45 ppm (17.8 H), 2.76 to 2.92 (14.0 H), 2.14 to 2.22 (3.9 H), 2.00 to 2.12 (4.8 H), 0.94 to 1.02 (5.9 H); UV/vis: λmax 208 nm; medium-resolution MS (m/z): [M]+ calculated for C31H46: 418.7; found: 418.3.
Mutagenesis.
Deletion of the oleABCD cluster and oleC from MR-1 was achieved utilizing homologous recombination between flanking regions of the target gene(s) cloned into a suicide vector (26). Briefly, upstream and downstream regions of the target deletion were cloned into the suicide vector pSMV3 in a compatible E. coli cloning strain UQ950. The suicide vector was transformed into an E. coli mating strain WM3064 and then conjugated into MR-1. The initial recombination event was selected for by resistance to Km. Cells containing the integrated suicide vector were grown in the absence of selection overnight at 30°C and then plated onto LB plates containing 5% sucrose (26). Cells retaining the suicide vector were unable to grow due to the activity of SacB, encoded on the vector, while cells that had undergone a second recombination event formed colonies. Colonies were then screened by PCR to determine strains containing the deletion. For creation of the oleABCD cluster knockout strain, primers oleclusterUF, oleclusterUR, oleclusterDF, and oleclusterDR containing SpeI, BsaI, BsaI, and SacI restriction sites, respectively, were designed for the regions flanking the two ends of the oleABCD cluster (gi numbers 24373309, 24373310, 24373311, and 24373312, respectively; locus tags SO_1742, SO_1743, SO_1744, and SO1745, respectively). For creation of the oleC knockout strain, primers oleCUF, oleCUR, oleCDF, and oleCDR containing SpeI, BsaI, BsaI, and SacI restriction sites, respectively, were designed for the regions flanking the ends of oleC (gi 24373311; locus tag SO_1744). Finally, for the creation of the pfaA knockout strain, primers pfaA1F, pfaA1R, pfaA2F, and pfaA2R containing the SpeI, BamHI, BamHI, and ApaI restriction sites, respectively, were designed for the regions flanking the ends of pfaA (gi 24373171; locus tag SO_1602). Primer names and sequences are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| oleclusterUF | TTACTAGTATCATGCCAACCCTTTTCGC |
| oleclusterUR | TTGGTCTCCATCGGATAATTGATGCC |
| oleclusterDF | TTGGTCTCTCGATAGAAGAGGGGATG |
| oleclusterDR | AAGAGCTCGCACTCGGTGTTGATACAAA |
| oleCUF | TTACTAGTTTTAACGAAGGTGCGCTAAGG |
| oleCUR | AAGGTCTCCTCGAACAGCGCATCATCCA |
| oleCDF | TTGGTCTCATCGAGCTTGATCAATCTTT |
| oleCDR | AAGAGCTCCAGCTTCAGCTTACCTAAAC |
| pfaA1F | ACTAGTGCACTCAAGTCGCAGATATTGTTCGCA |
| pfaA1R | GGATCCACCAACGATGGCAATGGGCAT |
| pfaA2F | GGATCCAGTAAGACGCTTAACCAAGCAT |
| pfaA2R | GGGCCCGGTCAATGAATCAATCAGTTGCAACAAC |
| SO1744Fcomp | ACTAGTGATTACCCATATCAAGCACTTTATGACTGAGA |
| SO1744Rcomp | GAGCTCTTGAATGCAATGGGATAATGTTTCATCCC |
| pfaAcomplementF | GGGCCCATGAGCCATACCCCTTCACAGCCT |
| pfaAcomplementR | ACTAGTTAATGCGGCATGTGCGATTGGGTTGAGTG |
| SmclusterCompF | ACTAGTCCCCCTTTTGCCTGAGCCTTGGCGC |
| SmthiolaseCompR | GAGCTCGAAGATCATCGCTGTCCGTCGCGAGC |
Mutant complementation and heterologous gene expression.
Complementation of the oleC and pfaA mutants was performed using the pBBR1MCS-2 expression vector (19) and the endogenous lac promoter (which is constitutive in MR-1 due to the absence of lacI). Primers SO1744Fcomp and SO1744Rcomp containing SpeI and SacI restriction sites or pfaAcomplementF and pfaAcomplementR containing ApaI and SpeI restriction sites were designed for the regions flanking the ends of oleC (gi 24373311; locus tag SO_1744) or pfaA (gi 24373171; locus tag SO_1602), respectively. The Stenotrophomonas maltophilia oleA (gi 194363945; locus tag Smal_0167) was amplified using primers SmclusterCompF and SmthiolCompR containing the SpeI and SacI restriction sites. Resulting PCR products were ligated into the Strataclone cloning system (Agilent Technologies) followed by ligation of the product into the pBBR1MCS-2 expression vector. Constructs were introduced into E. coli WM3064 and conjugated into the oleC deletion, pfaA deletion, or wild-type S. oneidensis MR-1 strain. Appropriately oriented inserts were verified by PCR analysis. The expression of the cloned genes was verified by detection of product activity using GC-MS analysis.
Sequence analysis.
Sequence comparisons were made using the National Center for Biotechnology Information BLAST (bl2seq) tool. Ole protein sequences from S. oneidnesis MR-1 and M. luteus were compared. The gi numbers and sequences were obtained from the GenBank database.
RESULTS AND DISCUSSION
A long-chain hydrocarbon is present in S. oneidensis cells at all growth phases.
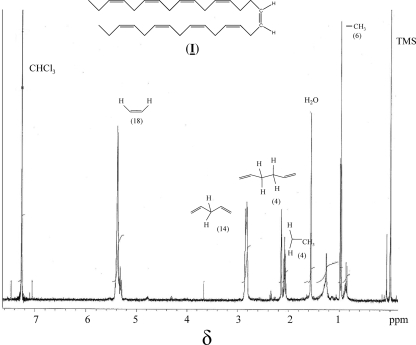
The hydrocarbon was identified in the nonpolar fraction following solvent extraction from the cultures. Gas chromatography-mass spectrometry showed a single sharp peak at 20.2 min that had a parent ion at 418 mass units (Fig. 1 A). Reduction of the product with hydrogen yielded a single product with a slightly longer retention time and a parent ion of 436 mass units (Fig. 1). The reduced product behaved identically to the C31 n-alkane hentriacosane. This indicated that the biological product was a hentriacontanonaene, but the positions of the nine double bonds could not be deduced from mass spectrometry. The compound had no appreciable UV absorbance above 230 nm, suggesting that the double bonds were not in conjugation. The proton NMR was decisive (Fig. 2) and consistent with one nearly centrosymmetric structure only, specifically, 3,6,9,12,15,19,22,25,28-hentriacontanonaene (compound I). The absolute stereochemistry at the double bonds remains to be determined but is shown in the figure as all-cis because of further data on its biosynthetic origin (see below). The structure of compound I was consistent with it being derived from a head-to-head condensation between two fatty acyl chains to produce long-chain olefins containing a double bond between the central and an adjacent carbon atom in the chain.
FIG. 1.
Gas chromatograph of the S. oneidensis hydrocarbon compound I (20.2 min) (A) and the product of its hydrogenation (20.8 min) that comigrates with and has an identical mass spectrum to n-hentriacosane (B).
FIG. 2.
NMR spectrum of the hydrocarbon compound I produced by S. oneidensis strain MR-1 in deuterated chloroform (CHCl3) with tetramethylsilane (TMS) as the reference standard. The fragment representing each resonance and the number of protons on integration are indicated. The structure of the compound represented by the spectrum is shown at the top.
Origin of the fatty acids undergoing head-to-head condensation.
The structure of the hydrocarbon (compound I) produced by S. oneidensis MR-1 would require the condensation of two molecules of hexadeca-4,7,10,13-tetraenoic acid or an acyl equivalent of this, for example, the acyl-coenzyme A (CoA) derivative. This specific acyl derivative is known to be an intermediate in the biosynthesis of long-chain polyunsaturated fatty acids (PUFAs) (21). PUFAs such as eicosapentaenoic acid are known to be produced by various Shewanella species (6). Moreover, PUFA biosynthetic genes from Shewanella have been identified by heterologous expression (18) and in S. oneidensis strain MR-1 via genome annotation (16).
To confirm the involvement of the PUFA pathway genes in the biosynthesis of compound I, a pfaA (annotated as a multidomain β-keto acyl synthase; gi 24373171, locus tag SO_1602) deletion mutant was constructed. When this mutant was tested for hydrocarbon biosynthesis, neither compound I nor any hydrocarbon product could be detected. Hydrocarbon biosynthesis was restored by the presence of the plasmid-encoded pfaA (data not shown).
Genetic analysis of ole gene homologs.
We next sought to study the genes responsible for the condensation of a PUFA intermediate leading to the formation of compound I. A cluster of genes in Shewanella oneidensis MR-1 was observed to be homologous to genes (ole) previously implicated in head-to-head hydrocarbon biosynthesis (Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781). These were Shewanella proteins (gi 24373309, 24373310, 24373311, and 24373312), which were annotated in the GenBank database as a 3-oxoacyl-(acyl carrier protein) synthase III, an α/β-fold family hydrolase, a peptide hydrolase, and a 3-hydroxysteroid dehydrogenase/isomerase family protein, respectively. The first protein (gi 24373309) had 31% sequence identity to the Mlut_13230 protein identified by Beller et al. to be involved in a head-to-head condensation pathway in M. luteus (5). The two proteins gi 243733310 and gi 24373311 from S. oneidensis MR-1 resembled the N terminus and carboxy terminus, respectively, of the protein Mlut_13240 in M. luteus. Protein 4 (gi 24373312) showed 31% sequence identity to the Mlut_13250 protein of M. luteus. The bioinformatics data suggested that S. onidenesis MR-1 proteins gi 24373309 through 24373312 were, like the M. luteus proteins, involved in a head-to-head condensation reaction. This was investigated genetically to both confirm the genes' involvement and to investigate the effect of gene alteration on product formation.
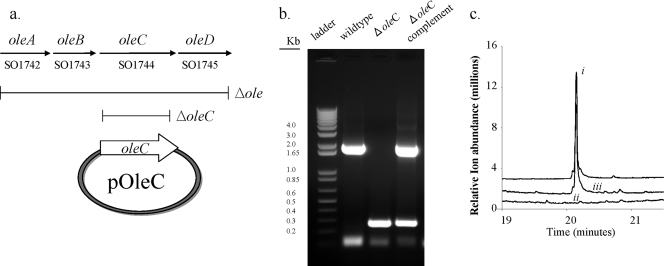
The choice of S. oneidensis strain MR-1 allowed us to use well-established gene deletion methods to test if the oleABCD genes were involved in olefin biosynthesis (Fig. 3a). In-frame deletions of the entire ole cluster, and of oleC individually, were generated. The gene deletion was verified using PCR. A 1.7-kb band corresponding to the oleC-containing gene cluster in the wild type became a 0.3-kb fragment in ΔoleC, resulting from deletion of the 1.5-kb oleC (Fig. 3b). The complement showed both 0.3- and 1.7-kb bands representing the deleted gene region plus the full oleC present on the pOleC plasmid. Figure 3c shows the gas chromatograph of the region where compound I, produced by wild-type S. oneidensis, eluted at approximately 20.2 min. The oleC mutant showed no detectable peak in this region. The complemented strain showed a restoration of the 20.2-min peak. The identity of the compound eluting at 20.2 min was confirmed by mass spectrometry. GC experiments were performed in triplicate. Similarly, the oleABCD deletion strain did not produce compound I (see Fig. S1 in the supplemental material).
FIG. 3.
The oleABCD genes are required for long-chain olefin production by S. oneidensis. (a) Illustration of the oleABCD and oleC regions deleted and plasmid pOleC containing the oleC gene that complemented the oleC deletion. (b) DNA gel confirming gene deletion and complementation (primers used for analysis were SO1744CompF and SO1744CompR). (c) Gas chromatograph of solvent extracts from S. oneidensis wild-type (i), the oleC deletion mutant (ii), and the oleC mutant complemented with the pOleC plasmid (iii).
Formation of ketones and implications for the function of OleA.
The S. oneidensis MR-1 oleC deletion mutant did not produce a hydrocarbon, but it made another compound that was purified from a different distillation fraction than the hydrocarbon. The mass spectrum of the compound, compound III, had a parent ion of m/z 434. These data were consistent with a symmetrical molecule with eight double bonds and having the carbonyl functionality at the center of the hydrocarbon chain. Compound III was hydrogenated to produce a molecule with m/z 450 and showed an ion fragment of m/z 239. This confirmed the structure of compound III to be 3,6,9,12,19,22,25,28-hentriacontaoctaene-16-one. Compound III was not found in the S. oneidensis MR-1 oleABCD mutant.
Ketone products were also observed in an additional experiment involving heterologous oleA gene expression in S. oneidensis MR-1. The oleA gene homolog from S. maltophilia strain R551-3 was cloned into S. oneidensis strain MR-1. The heterologous strain grew normally but produced a much wider range of nonpolar extractable products (Fig. 4). The endogenous compound I was present and readily identified by GC retention time and mass spectrum and is shown in Fig. 4 with an asterisk and the chemical formula C31H46. The recombinant Shewanella strain produced at least 17 additional long-chain compounds, of which 13 were monoketones (Fig. 4). The chemical formulas are shown, indicating the degree of unsaturation of the hydrocarbon chains. All of the compounds are significantly more saturated than the endogenous C31H46 hydrocarbon, suggesting that the Stenotrophomonas OleA protein, unlike the Shewanella OleA protein, condenses fatty acids not derived from the polyunsaturated fatty acid pathway. The ketones were identified from their characteristic mass spectra; both the parent ions and ion fragments were consistent with these assignments. Moreover, the observation of a single major carbonyl ion, or two such ions of similar molecular weight, is consistent with the carbonyl functional group being present at the median carbon for odd-numbered chain lengths. This observation is consistent with these products arising from a head-to-head fatty acid condensation mechanism.
FIG. 4.
GC results for a solvent extract from recombinant S. oneidensis expressing the heterologous S. maltophilia OleA protein. Compounds were identified as hydrocarbons or ketones by mass spectrometry as described in the text and are designated by the molecular formula next to each major GC peak. The asterisk indicates compound I, which is endogenously produced by wild-type S. oneidensis MR-1.
The data shown in Fig. 4 were striking because the native Shewanella only made a single endogenous C31H46 hydrocarbon, compound I. In contrast, S. maltophilia is known to produce a large number of different hydrocarbons with chain lengths of C26 to C30 (28), and the S. maltophilia oleA gene alone directed the formation of a much wider range of products in Shewanella. The observation here of diverse hydrocarbons and ketones has implications for the production of molecules for fuel or specialty chemical applications via the heterologous expression of different oleA genes in Shewanella.
Ketone formation could potentially result from the OleA protein alone, and this would be consistent with the data presented here. OleA is in the thiolase superfamily, which catalyzes both decarboxylative and nondecarboxylative acyl group condensation reactions (14, 15). A nondecarboxylative thiolytic condensation would produce an intermediate that could give rise to ketones (Fig. 5). Figure 5 shows the structure of the natively produced polyolefin, compound I. Hydrocarbons and ketones could both be derived from an intermediate generated by OleA, and that is consistent with reactions catalyzed by thiolase superfamily members, of which OleA is a member. Thioester cleavage could occur by the action of (i) OleA, (ii) a thioesterase, or (iii) spontaneous hydrolysis (12) to generate a β-keto acid (Fig. 5C, compound II). β-Keto acids are known to be unstable and decarboxylate spontaneously (23). Spontaneous decarboxylation of β-keto acids in biological systems is well known and underlies the production of ketone bodies in mammalian liver (17). In the case of the S. oneidensis oleC mutant, intermediate compound II would be generated and decarboxylate to generate compound III, the observed ketone. When the OleA from Stenotrophomonas was expressed in Shewanella, a narrower specificity for the Shewanella enzymes could lead to the buildup of different intermediates that undergo hydrolysis and decarboxylation to yield the ketones. An alternative mechanism for the OleA-catalyzed condensation reaction has been proposed in the literature (5). Further studies will be required to discern between that and the role for OleA proposed here.
FIG. 5.
Product structures and proposed pathways in S. oneidensis MR-1 wild-type and mutant strains for head-to-head hydrocarbon and ketone formation, respectively. (A) Structure of compound I, identified as described in the text. (B) Proposed role of OleA in the head-to-head biosynthetic pathway. (C) A proposed pathway to ketones in the presence of the OleA protein alone.
Potential role of an ole gene product(s) in cold adaption.
A hydrocarbon that appears to be identical to compound I was previously identified in a significant number of Antarctic bacterial isolates (22). The hypothesis that long-chain olefins might contribute to cold adaption was tested directly with S. oneidensis strain MR-1, which grows within the temperature range of 4 to 37°C (optimal growth at 30°C). The first observation in this study supporting the cold adaption hypothesis was that decreasing the growth temperature led to significant increases in the amount of compound I and compound III present in cells (Fig. 6a).
FIG. 6.
Long-chain polyunsaturated compounds as a function of growth temperature in S. oneidensis MR-1 wild type and an oleABCD deletion mutant. (a) Hydrocarbon (blue) and ketone (red) contents at different temperatures relative to the maximum observed (at 4°C). (b) Wild-type MR-1 (black) and the corresponding oleABCD-deficient mutant (green) were downshifted from 30°C to 4°C, and the cold temperature growth curves are shown. Experimental points are average triplicate samplings from six treatments. Variation is shown as the standard deviation.
In other experiments, wild-type and olefin-deficient strains were grown at 30°C and then inoculated into medium at 4°C (Fig. 6b). Although there was not much difference in the growth rate during exponential phase, the olefin-deficient oleABCD mutant strain showed a significantly longer lag phase prior to exponential growth (Fig. 6b). When the oleABCD mutant was pregrown at 4°C, this lag in growth following transfer was not observed. These data suggested at least one role for long-chain olefins in facilitating growth following a shift to colder temperatures. We expect that the polyolefin would increase membrane fluidity and contribute to a maintenance of proper membrane function following a sudden decrease in temperature.
Structurally analogous long-chain alkadienes and alkatrienes are prominent in the lipids of marine photosynthetic eukaryotes, such as Isochrysis galbana, that grow at cold oceanic temperatures (25). They are also present, along with long-chain alkenones, in the lipid fractions of Emiliania huxleyi (25), a photosynthetic eukaryote which is so common that oceanic algal blooms of this organism are observable by satellite photographs (7). The mechanism of hydrocarbon formation in these eukaryotes remains open, but our findings here, coupled with ongoing genome sequencing of these organisms, may help provide insight. It is interesting that the amount and degree of unsaturation of the long-chain hydrocarbons and alkenones increase with decreasing temperature (24). This suggests that long-chain hydrocarbons and ketones could be involved in cold adaption in both bacteria and eukaryotes.
ADDENDUM IN PROOF
While our paper was under review, we became aware of a new paper describing a C31:9 hydrocarbon in a marine bacterium tentatively identified as a Shewanella sp. (S. Sugihara, R. Hori, H. Nakanowatari, Y. Takada, I. Yumoto, N. Morita, Y. Yano, K. Watanabe, and H. Okuyama, Lipids 45:167-177, 2010). This hydrocarbon appears to be identical to the one in S. oneidensis strain MR-1 we describe here.
Supplementary Material
Acknowledgments
This work was supported by grant LG-B13 from the Initiative for Renewable Energy and the Environment (to L.P.W.) and a Watson Fellowship (to D.J.S.).
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry 8:394-405. [DOI] [PubMed] [Google Scholar]
- 2.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain, nonisoprenoid hydrocarbons. II. The incorporation of acetate and the aliphatic chain is isoleucine and valine into fatty acids and hydrocarbons by Sarcina lutea in vivo. Biochemistry 8:953-959. [DOI] [PubMed] [Google Scholar]
- 3.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. III. The metabolic relationship of long-chain fatty acids and hydrocarbons and other aspects of hydrocarbon metabolism in Sarcina lutea. Biochemistry 8:1913-1918. [DOI] [PubMed] [Google Scholar]
- 4.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry 8:3317-3324. [DOI] [PubMed] [Google Scholar]
- 5.Beller, H. R., E. B. Goh, and J. D. Keasling. 2010. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, J. P., S. A. McCammon, D. S. Nichols, J. H. Skerratt, S. M. Rea, P. D. Nichols, and T. A. McMeekin. 1997. Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5 omega 3) and grow anaerobically by dissimilatory Fe(III) reduction. Int. J. Syst. Bacteriol. 47:1040-1047. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. W., and J. A. Yoder. 1994. Coccolithophorid blooms in the global ocean. J. Geophys. Res. 99(C4):7467-7482. [Google Scholar]
- 8.Caudales, R., C. Forni, and J. M. Wells. 1998. Cellular fatty acid composition of rod and coccus forms of Arthrobacter globiformis, A. crystallopoietes and A. nicotianae isolated from the water fern Azolla. J. Appl. Microbiol. 84:784-790. [Google Scholar]
- 9.Channon, H. J., and A. C. Chibnall. 1929. The ether-soluble substance of cabbage leaf cytoplasm. V. The isolation of n-nonacosane and di-n-tetradecylketone. Biochem. J. 23:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis, M., and P. E. Kolattukudy. 1992. A cobalt-porphyrin enzyme converts a fatty aldehyde to a hydrocarbon and CO. Proc. Natl. Acad. Sci. U. S. A. 89:5306-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrickson, J. K., M. F. Romine, A. S. Beliaev, J. M. Auchtung, M. E. Driscoll, T. S. Gardner, K. H. Nealson, A. L. Osterman, G. Pinchuk, J. L. Reed, D. A. Rodionov, J. L. Rodrigues, D. A. Saffarini, M. H. Serres, A. M. Spormann, I. B. Zhulin, and J. M. Tiedje. 2008. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6:592-603. [DOI] [PubMed] [Google Scholar]
- 12.Fredslund, F., L. Jenner, L. B. Husted, J. Nyborg, G. R. Andersen, and L. Sottrup-Jensen. 2006. The structure of bovine complement component 3 reveals the basis for thioester function. J. Mol. Biol. 361:115-127. [DOI] [PubMed] [Google Scholar]
- 13.Frias, J. A., J. E. Richman, and L. P. Wackett. 2009. C29 olefinic hydrocarbons biosynthesized by Arthrobacter species. Appl. Environ. Microbiol. 75:1774-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haapalainen, A. M., G. Meriläinen, and R. K. Wierenga. 2005. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem. Sci. 31:64-71. [DOI] [PubMed] [Google Scholar]
- 15.Heath, R. J., and C. O. Rock. 2002. The Claisen condensation in biology. Nat. Prod. Rep. 19:581-596. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 17.Hird, F. J. R., and R. H. Symons. 1962. The mechanism of ketone-body formation from butyrate in rat liver. Biochem. J. 84:212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong, Y. S., S. K. Song, S. J. Lee, and B. K. Hur. 2006. The growth and EPA synthesis of Shewanella oneidensis MR-1 and expectation of EPA biosynthetic pathway. Biotechnol. Bioprocess Eng. 11:127-133. [Google Scholar]
- 19.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. K., H. Chou, T. S. Ham, T. S. Lee, and J. D. Keasling. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556-563. [DOI] [PubMed] [Google Scholar]
- 21.Metz, J. G., P. Roessler, D. Facciotti, C. Levering, F. Dittrich, M. Lassner, R. Valentine, K. Lardizabal, F. Domergue, A. Yamada, K. Yazawa, V. Knauf, and J. Browse. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290-293. [DOI] [PubMed] [Google Scholar]
- 22.Nichols, D. S., P. D. Nichols, and T. A. McMeekin. 1995. A new n-C31:9 hydrocarbon from Antarctic bacteria. FEMS Microbiol. Lett. 125:281-286. [Google Scholar]
- 23.Pedersen, K. J. 1929. The ketonic decomposition of beta-keto carboxylic acids. J. Am. Chem. Soc. 51:2098-2107. [Google Scholar]
- 24.Prahl, F. G., and S. G. Wakeham. 1987. Calibration of unsaturation patterns in long-chain ketone compositions for palaeotemperature assessment. Nature 330:367-369. [Google Scholar]
- 25.Rieley, G., M. A. Teece, T. M. Peakman, A. M. Raven, K. J. Greene, T. P. Clarke, M. Murray, J. W. Leftley, C. N. Campbell, R. P. Harris, R. J. Parkes, and J. R. Maxwell. 1998. Long-chain alkenes of the haptophytes Isochrysis galbana and Emiliania huxleyi. Lipids 33:617-625. [DOI] [PubMed] [Google Scholar]
- 26.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 19:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steen, E. J., Y. Kang, G. Bokinsky, Z. Hu, A. Schirmer, A. McClure, S. B. Del Cardayre, and J. D. Keasling. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559-562. [DOI] [PubMed] [Google Scholar]
- 28.Suen, Y., G. U. Holzer, J. S. Hubbard, and T. G. Tornabene. 1988. Biosynthesis of acyclic methyl branched poly-unsaturated hydrocarbons in Pseudomonas maltophilia. J. Ind. Microbiol. 2:337-348. [Google Scholar]
- 29.Tornabene, T. G., E. O. Bennett, and J. Oro. 1967. Fatty acid and aliphatic hydrocarbon composition of Sarcina lutea grown in three different media. J. Bacteriol. 94:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornabene, T. G., E. Gelphi, and J. Oro. 1967. Identification of fatty acids and aliphatic hydrocarbons in Sarcina lutea by gas chromatography and combined gas chromatography-mass spectrometry. J. Bacteriol. 94:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tornabene, T. G., and S. P. Markey. 1971. Characterization of branched monounsaturated hydrocarbons of Sarcina lutea and Sarcina flava. Lipids 3:190-195. [DOI] [PubMed] [Google Scholar]
- 32.Tornabene, T. G., and S. L. Peterson. 1978. Pseudomonas maltophilia: identification of the hydrocarbons, glycerides, and glycolipoproteins of cellular lipids. Can. J. Microbiol. 254:525-532. [PubMed] [Google Scholar]
- 33.Wackett, L. P., J. A. Frias, J. L. Seffernick, D. J. Sukovich, and S. M. Cameron. 2007. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl. Environ. Microbiol. 73:7192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wackett, L. P. 2008. Microbial-based motor fuels: science and technology. Microb. Biotechnol. 1:211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.