Abstract
Understanding ruminal methanogens is essential for greenhouse gas mitigation, as well as for improving animal performance in the livestock industry. It has been speculated that ruminal methanogenic diversity affects host feed efficiency and results in differences in methane production. This study examined methanogenic profiles in the rumen using culture-independent PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analysis for 56 beef cattle which differed in feed efficiency, as well as diet (the cattle were fed a low-energy diet or a high-energy diet). The methanogenic PCR-DGGE profiles detected were greatly affected by diet, and the major pattern changed from a community containing predominantly Methanobrevibacter ruminantium NT7 with the low-energy diet to a community containing predominantly Methanobrevibacter smithii, Methanobrevibacter sp. AbM4, and/or M. ruminantium NT7 with the high-energy diet. For each diet, the methanogenic PCR-DGGE pattern was strongly associated with the feed efficiency of the host. Diet-associated bands for Methanobrevibacter sp. AbM4 and M. smithii SM9 and a feed efficiency-related band for M. smithii PS were identified. The abundance of total methanogens was estimated by determining the numbers of copies of the 16S rRNA genes of methanogens. However, the size of the methanogen population did not correlate with differences in feed efficiency, diet, or metabolic measurements. Thus, the structure of the methanogenic community at the species or strain level may be more important for determining host feed efficiency under different dietary conditions.
Ruminal methanogens use methanogenesis pathways to maintain low hydrogen partial pressure and to facilitate fiber digestion in the rumen by converting hydrogen into methane gas (24, 37). However, although it is necessary, this process also has adverse effects because the released methane represents a significant loss of dietary energy for the host animal (14) and it constitutes a large proportion of the agricultural greenhouse gas emitted (4, 10). Many studies to obtain a better understanding of rumen methanogens have been conducted in order to improve the efficiency of ruminal function and to mitigate methane release. Assessments by both cultivation-dependent and cultivation-independent methods have found that members of the genus Methanobrevibacter account for the majority of the methanogens in the rumens of sheep and cattle (11, 18, 21-23, 28, 31, 33, 34). In addition, Methanosphaera stadtmanae, Methanobacterium species, and Methanosarcina barkeri have also been found in some studies (13, 32). Although the phylogenetic positions of the methanogens in the rumen are diverse, these organisms utilize only three major pathways for methanogenesis: the CO2 reduction pathway, the C1 compound (e.g., methanol and methylamine) conversion pathway, and the acetate fermentation pathway. Each methanogen species has a substrate preference, and most methanogens can use only one or two substrates (37).
Previous studies of rumen methanogens focused primarily on determining the methanogen species composition in different samples and developing strategies to reduce the methane yield from ruminants. Recently, there has been a strong desire to understand the impact of methanogens on host biology. Two primary studies found that feedlot beef cattle with higher feed efficiency (designated “efficient” animals) produced about 20% less methane gas than animals with lower feed efficiency (designated “inefficient” animals) (8, 19). The methanogenic communities of efficient and inefficient animals fed a low-energy diet have been compared, and divergence between the two communities has been reported (36). However, it is not clear how the methanogens in the rumen of cattle change when the animals are fed a different diet.
The aims of this study were to describe the methanogenic communities in 56 steers with different feed efficiencies that were fed two distinct diets (a low-energy diet and a high-energy diet) and to understand how methanogenic communities change in response to diet modification using PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and sequence analysis. Multivariate analysis was used to analyze the association of PCR-DGGE bands with the daily dry matter intake (DMI), average daily gain (ADG), feed conversion ratio (FCR), and residual feed intake (RFI). Methanogens that were associated with diet and with host feed efficiency were identified. In addition, the methanogen population of each rumen sample was examined by quantitative real-time PCR (qRT-PCR), and the results for different RFI groups and both diets were compared.
MATERIALS AND METHODS
Animal experiment and sample collection.
All experimental procedures were approved by the Animal Care and Use Committee for Livestock at University of Alberta. The steers involved in this study (n = 56; 10 months old; Hereford × Aberdeen Angus) were raised using the guidelines of the Canadian Council on Animal Care (Ottawa, Ontario, Canada) at the Kinsella Research Station, University of Alberta. Initially, the animals were fed a low-energy-density feedlot diet (74% oats, 20% hay, and 6% feedlot supplement) for 90 days, and the RFI was measured using the GrowSafe system (GrowSafe Systems Ltd., Airdrie, Alberta, Canada). Ruminal fluid samples were collected from the 56 steers on the same day before feeding and within 1 week after evaluation of the RFI by inserting a flexible plastic tube into the rumen and transferring the fluid into sterile 200-ml containers. Approximately 100 ml of rumen fluid was obtained from each animal, immediately frozen in dry ice, and stored at −80°C until the next processing step was performed. The animals were then fed a high-energy-density feedlot diet (28.3% oats, 56.7% barley, 10% alfalfa pellets, and 5% feedlot supplement) for 90 days. The same RFI measurement and sample collection procedures were performed using the methods described above. In this study, animals were also classified for each variable using the following criteria: animals with a value greater than the mean plus 0.5 standard deviation (SD) were placed in the H group, while animals with a value between the mean minus 0.5 SD and the mean plus 0.5 SD were placed in the M group and animals with a value less than the mean minus 0.5 SD were placed in the L group. Thus, all of the animals were first classified using RFI values and were placed in the H-RFI (n = 20; 0.76 ± 0.05 kg/day), M-RFI (n = 14; 0.14 ± 0.11 kg/day), and L-RFI (n = 22; −0.75 ± 0.05 kg/day) (P < 0.0001) groups and fed the low-energy diet. After the diet was changed, all of the animals were reclassified based on the new RFI values, as follows: H-RFI group (n = 14), 0.94 ± 0.11 kg/day; M-RFI group (n = 23), 0.02 ± 0.05 kg/day; and L-RFI group (n = 19), −1.25 ± 0.11 kg/day) (P < 0.0001). Additionally, data for the daily dry matter intake (DMI), average daily gain (ADG), and feed conversion ratio (FCR) were also evaluated and used for both trials described above.
DNA extraction.
Total DNA was extracted from 56 rumen fluid samples using the methods described by Guan et al. (7). Briefly, 0.5 ml of frozen rumen fluid was thawed on ice and washed with 4.5 ml of TN150 buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl). The liquid was vortexed for 30 s and then centrifuged at 200 × g at 4°C for 5 min. After this, 1 ml of the supernatant was transferred to a new microcentrifuge tube containing 0.3 g of autoclaved zirconium-silica beads (diameter, 0.1 mm). The cells were lysed by physical disruption with a BioSpec Mini Bead-Beater-8 at 4,800 rpm for 3 min. The supernatant obtained from each sample was then transferred to a new sterile tube for phenol-chloroform-isoamyl alcohol (25:24:1) extraction. The extracted DNA was precipitated with cold ethanol and resuspended in 20 μl of nuclease-free water. The concentration and quality of DNA were determined at A260 nm and A280 nm using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
PCR amplification of methanogenic 16S rRNA genes.
Total DNA was extracted from each rumen fluid sample, diluted to obtain a concentration of 50 ng/μl, and used as a template in PCRs. The universal primer pair Met 86f/Met 915r (Table 1) was used for the initial PCR to amplify a partial 16S rRNA gene fragment (∼800 bp) using the following program: initial denaturation for 5 min at 94°C; 30 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 60 s; and final elongation for 7 min at 68°C. The PCR solution (50 μl) contained 20 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 2.5 U Taq polymerase (Invitrogen, Carlsbad, CA), 1× PCR buffer, 100 mM MgCl2, and 50 ng of DNA template. The PCR products were then used as templates for nested PCR amplification using the universal primer pair GC-ARC344f/519r (with a GC clamp added to the 5′ end of ARC344f), which targeted the V3 region of the 16S rRNA gene (2) (Table 1). Since different PCR primer sets generate different amplicons and thus influence the observed diversity of a community, proper primers have to be used in ecological studies. As reported by Yu et al. (35a), the V3 region of the 16S rRNA gene is the preferred target in PCR-DGGE analysis when ruminal archaeal communities are profiled. Therefore, primers targeting the V3 region of the 16S rRNA gene were used for ruminal methanogenic community profiling in this study. The amplification conditions were as follows: denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 s, 56.5°C for 30 s, and 72°C for 30 s; and final elongation for 7 min at 72°C.
TABLE 1.
Primers used in this study to target methanogen 16S rRNA genes
| Primera | Sequence (5′ to 3′) | Annealing positionsb | Reference |
|---|---|---|---|
| Met 86f | GCTCAGTAACACGTGG | 86-101 | 35 |
| Met 915r | GTGCTCCCCCGCCAATTCCT | 915-935 | 30 |
| GC-ARC344fc | ACGGGGYGCAGCAGGCGC GA | 344-363 | 2 |
| 519r | GWATTACCGCGGCKGCTG | 519-534 | 2 |
| uniMet1-F | CCGGAGATGGAACCTGAGAC | 36 | |
| uniMet1-R | CGGTCTTGCCCAGCTCTTATTC | 36 |
f or F, forward primer; r or R, reverse primer.
Escherichia coli rrs gene numbering.
Primer with a 40-bp GC clamp (CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG).
PCR-DGGE analysis of methanogens.
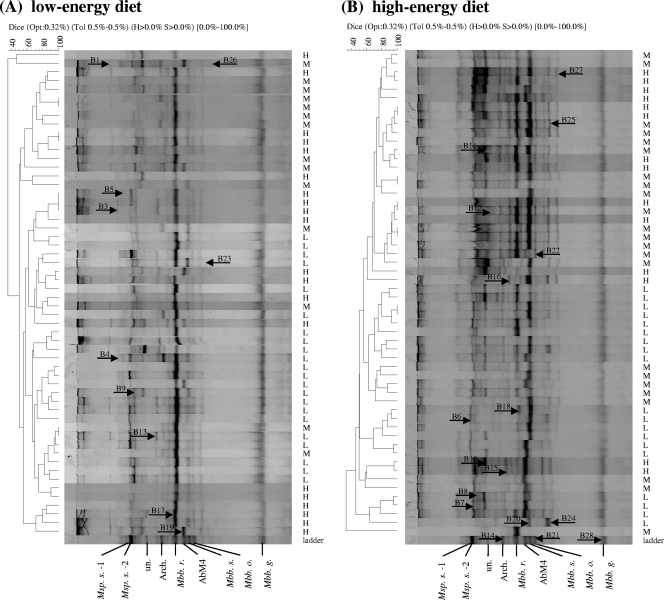
The nested PCR products were subjected to DGGE using the DCode universal mutation detection system (Bio-Rad Laboratories, Inc., Hercules, CA). PCR amplicons were separated using a 6% polyacrylamide gel in 1× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA) with a 35 to 45% linear denaturing gradient. The gel was run at 60°C and 150 V for 4 h and stained with 300 ml of 0.1% (vol/vol) ethidium bromide for 15 min. Then the gel was washed with 400 ml water for 30 min and photographed by using UV transillumination. To effectively assign the band positions for each gel, a common ladder was included in each gel as an internal control. The ladder was generated by mixing the amplicons of plasmid DNA obtained in our previous 16S rRNA sequence analysis (36). As shown in all PCR-DGGE gels (see Fig. 1 to 3), the ladder (from top to bottom) included two bands that resembled Methanosphaera stadtmanae bands (bands Msp. s.-1, Msp. s.-2), a band that resembled a Methanobacteriales archaeon CSIRO1.33 clone band (band Arch.), a Methanobrevibacter ruminantium NT7 band (band Mbb. r.), a Methanobrevibacter sp. AbM4 band (band Mbb. AbM4), a Methanobrevibacter smithii band (band Mbb. s.), and a Methanobrevibacter olleyae band (band Mbb. o.). In addition, two other PCR by-product bands were identified; the sequence of one of these bands was unknown (band un.), and another band resembled a Methanobrevibacter gottschalkii band (band Mbb. g.). The DGGE band patterns obtained were analyzed using the BioNumerics software (version 5.1; Applied Maths, Inc., Austin, TX). Since the clustering analysis of the PCR-DGGE patterns could be affected by various factors, such as position bias in gels, band assignment, and different settings in the BioNumerics software, the optimal position tolerance and optimization setting were calculated using the tolerance and optimization analysis program supplied with the BioNumerics software package to ensure that there were better matches for the band patterns. The similarity of the DGGE profiles was calculated using the average Dice similarity coefficient (Dsc) index and the 0.32% optimization and 0.48% position tolerance settings based on the program analysis described above.
FIG. 1.
Methanogenic PCR-DGGE profiles generated using ruminal fluid from 56 animals and primers GC-ARC344f and 519r (35 to 45% DGGE). The indices for the clustering analysis are indicated at the top for each comparison. Opt, optimization (original setting, 0.32%); Tol, position tolerance, expressed as a rounded-up value (0.5%); H and S, minimum height and minimum surface, respectively (0% used for the comparison); 0.0%-100%, indicating the entire length of each lane. RFI was a variable used to identify the cattle's feed efficiency (1). The RFI groups are indicated on the right (H, H-RFI; M, M-RFI; L, L-RFI). The comparison of the PCR-DGGE profiles was generated by using BioNumerics software (as described in the text). The band pattern for the major bands assigned to species or strains in the ladder is shown at the bottom. Msp. s.-1 and Msp. s.-2, Methanosphaera stadtmanae; Arch., Methanobacteriales archaeon clone CSIRO1.33; Mbb. r., Methanobrevibacter ruminantium NT7; AbM4, Methanobrevibacter sp. AbM4; Mbb. s., Methanobrevibacter smithii; Mbb. o., Methanobrevibacter olleyae; Mbb. g., Methanobrevibacter gottschalkii HO. The 28 distinct bands are indicated by arrows (B, band). (A) PCR-DGGE profiles for animals fed the low-energy diet. (B) PCR-DGGE profiles for animals switched to the high-energy diet.
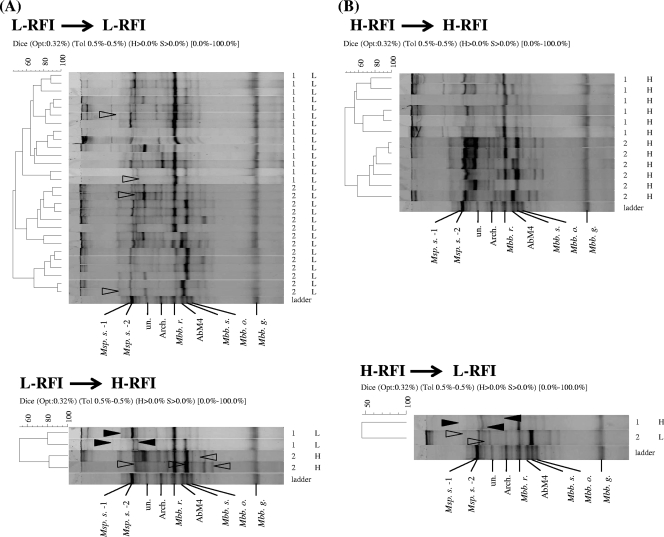
FIG. 3.
Comparison of methanogenic PCR-DGGE profiles for samples grouped based on RFI conditions. The indices for the clustering analysis are indicated at the top for each comparison. Opt, optimization (original setting, 0.32%); Tol, position tolerance, expressed as a rounded-up value (0.5%); H and S, minimum height and minimum surface, respectively (0% used for the comparison); 0.0%-100%, indicating the entire length of each lane. The numbers on the right indicate the diets used (1, low-energy diet; 2, high-energy diet). The letters on the right indicate the RFI groups (H, H-RFI; M, M-RFI; L, L-RFI). The triangles indicate the bands that shifted for the two sets of samples (described in the text). (A) PCR-DGGE profiles for animals in the L-RFI group when the first diet was used. (B) PCR-DGGE profiles for animals in the H-RFI group when the first diet was used. For an explanation of other abbreviations, see the legend to Fig. 1.
Cloning and sequence analysis of PCR-DGGE fragments.
A total of 28 distinct bands were excised aseptically from the gels and transferred to diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], 0.1% SDS). DNA fragments were extracted using a QIAEX II gel extraction kit (Qiagen Sciences, MD) and the polyacrylamide gel extraction protocol. The extraction products were reamplified using the ARC344f/519r primers without a GC clamp as described above. The fresh PCR products were then cloned into the TOP10 vector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA) using the manufacturer's chemical transformation procedures and were screened using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma, St. Louis, MO) medium containing ampicillin. Colonies with insertions (white colonies) were randomly selected and used for extraction of plasmid DNA with a Millipore plasmid extraction kit (Millipore, Billerica, MA). A sequencing reaction was performed with a 10-μl solution containing 0.5 μl of BigDye solution, 3.2 pmol of M13 Forward primer (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′), 1 × sequencing buffer, and 20 ng of plasmid DNA as the template using the ABI 3730 sequencing system and an ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequence compositions for the animals were compared using the UniFrac online comparison tool (16).
qRT-PCR analysis.
The total methanogen population in each ruminal sample was determined by determining 16S rRNA gene copy numbers. A universal primer pair targeting methanogens was used as described in a previous study (35). qRT-PCR was performed with the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) using SYBR green master mixture (Fast SYBR green master mixture; Applied Biosystems, Foster City, CA) with a fast cycle and melting curve and the following program: 95°C for 10 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. At the melting curve detection stage, the temperature was increased from 60°C to 95°C at a rate of 0.3°C every 20 s. Standard curves were constructed using serial dilutions of plasmid DNA from a clone identified as Methanobrevibacter sp. AbM4. The copy numbers for each standard curve were calculated based on the following equation: copy number = (NL × A × 10−9)/(660 × n), where NL is the Avogadro constant (6.02 × 1023 molecules per mol), A is the molecular weight of the molecule in the standard, and n is the length of the amplicon (in bp). The copy number of a targeted methanogen 16S rRNA gene per ml of rumen fluid was calculated using the following equation: copy number = (QM × C × DV)/(S × V), where QM is the quantitative mean copy number, C is the DNA concentration of the sample, DV is the dilution volume of extracted DNA, S is the amount of DNA (in ng) subjected to analysis, and V is the rumen fluid volume used for DNA extraction. The PCR efficiency (E) was calculated using the following equation: E = [10(−1/slope) − 1] × 100%. The data generated for reactions with efficiencies between 90 and 110% were used for further analysis.
Statistical analysis.
Acetate concentration and feed efficiency data used in this study were obtained previously (E. Hernandez-Sanabria et al., unpublished data). All statistical analyses were performed using SAS (SAS System, version 9.2; SAS Institute, Cary, NC). The band pattern for each rumen was first analyzed using SAS and a categorical model to identify the effects of different factors on the band distribution. When the animals were classified using category variables (e.g., diet), all samples were taken into account. In contrast, when the impact of numerical variables (e.g., DMI, FCR, acetate concentration, etc.) on the band patterns was tested, animals were placed in different classes based on the following criteria: animals with a value greater than the mean value plus 0.5 standard deviation were placed in the high group (H group), and animals with a value less than the mean value minus 0.5 standard deviation were placed in the low group (L group). Only the H and L groups were used for the analysis. When the impact of RFI on methanogenic patterns was examined, only the L-RFI and H-RFI groups were used.
In the correlation analysis, PCR-DGGE patterns were converted to categorical data, and the metabolite measurements and qRT-PCR measurements were used as numerical variables. All statistical analyses were performed by using SAS (SAS System, version 9.1; SAS Institute, Cary, NC). A mixed model was used to test the differences among the RFI values and the possible interactions between indexes. A covariation model was used to identify covariation for all of the numeric measurements. Principal component analyses (PCA) and categorical models were used to measure the linkage between DGGE profiles and metabolic data. A P value of <0.05 was considered statistically significant.
RESULTS
Comparison of methanogenic PCR-DGGE profiles.
PCR-DGGE profiles were obtained for samples and were first compared using each diet. For the low-energy diet, 24 dominant and faint methanogen DGGE bands were identified (Fig. 1A); the predominant band for most of the samples was at the location of the Methanobrevibacter ruminantium NT7 band in the ladder, while the other bands were much less intense. The band patterns for the L-RFI group tended to group together and separate from those for the H-RFI group, while the band patterns for the M-RFI group were more likely to group with the band patterns for either the L-RFI or H-RFI group instead of generating a distinct cluster. The overall average Dsc for the DGGE patterns for the low-energy diet was 56.4%, and the average Dsc values for the H-RFI group, the M-RFI group, and the L-RFI group were 59.3%, 59.5%, and 65.4%, respectively.
For the high-energy diet, 22 distinct bands were identified (Fig. 1B). Unlike the PCR-DGGE profiles obtained for the low-energy diet, the predominant bands for the animals were different. In general, there was only one predominant band for most animals in the L-RFI group, which corresponded to the band representing Methanobrevibacter smithii or Methanobrevibacter sp. AbM4 in the ladder. However, for most animals belonging to the M-RFI and H-RFI groups there were multiple prominent bands corresponding to the Methanobrevibacter smithii and Methanobrevibacter ruminantium NT7 bands. The clustering of the band patterns for this diet was similar to that for the low-energy diet. The overall level of similarity of all profiles was 56.9%, and the levels of similarity of the band patterns for the three groups were 70.3% for the H-RFI group, 62.0% for the M-RFI group, and 56.7% for the L-RFI group.
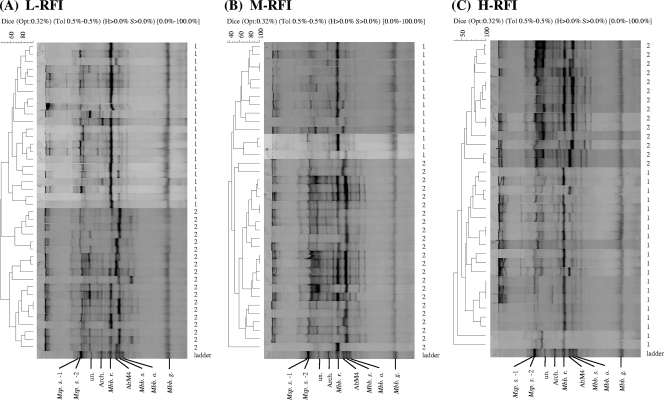
All of the PCR-DGGE profiles were also compared for each RFI group. As shown in Fig. 2, all three comparisons generated the same clustering pattern with two large clusters depending on the diet. The levels of similarity of the band patterns were 51.4% for the L-RFI group, 52.6% for the M-RFI group, and 55.4% for the H-RFI group.
FIG. 2.
Comparison of methanogenic PCR-DGGE profiles for different RFI groups. The indices for the clustering analysis are indicated at the top for each comparison. Opt, optimization (original setting, 0.32%); Tol, position tolerance, expressed as a rounded-up value (0.5%); H and S, minimum height and minimum surface, respectively (0% used for the comparison); 0.0%-100%, indicating the entire length of each lane. The numbers on the right indicate the diets used (1, low-energy diet; 2, high-energy diet). (A) Animals from the L-RFI group. (B) Animals from the M-RFI group. (C) Animals from the H-RFI group. The ladder shows the positions of the major bands (see the legend to Fig. 1).
Sequence analysis of methanogenic PCR-DGGE bands.
A total of 28 distinct PCR-DGGE bands were identified for the entire set of samples. To characterize the taxonomic relationships of the bands, all of the bands were cloned and sequenced, and 20 bands were successfully identified (Table 2). Of the 20 bands identified, 17 generated a single reading sequence, while 3 bands generated multiple reading sequences. Sequencing bias was eliminated by performing DGGE again and excluding the redundant sequences from the analysis (see Fig. S1 in the supplemental material).
TABLE 2.
Identification of PCR-DGGE bands
| PCR-DGGE band | Most closely related taxon (GenBank accession no.) | % Similarity |
|---|---|---|
| 1 | —a | |
| 2 | — | |
| 3 | Methanobrevibacter thaueri strain CW (U55236) | 100 |
| 4 | — | |
| 5 | Methanobrevibacter gottschalkii strain HO (U55238) | 94 |
| 6 | Methanosphaera stadtmanae (AY196684) | 96 |
| 7 | Methanosphaera stadtmanae (AY196684) | 96 |
| 8 | Methanobrevibacter gottschalkii strain HO (U55238) | 94 |
| 9 | Methanogenic archaeon SRmetG36 (EU413657) | 99 |
| 10 | Methanobrevibacter ruminantium strain NT7 (AJ009959) | 94 |
| 11 | Methanobrevibacter sp. AbM4 (AJ550156) | 94 |
| 12 | — | |
| 13 | Methanobrevibacter gottschalkii strain HO (U55238) | 94 |
| 14 | Methanobacteriales archaeon clone CSIRO1.33 (AY351466) | 96 |
| 15 | — | |
| 16 | Methanobrevibacter gottschalkii strain HO (U55238) | 93 |
| 17 | Methanobrevibacter ruminantium strain NT7 (AJ009959) | 100 |
| 18 | Methanobrevibacter smithii PS (U55233) | 100 |
| 19 | Methanobrevibacter sp. AbM4 (AJ550156) | 100 |
| 20 | Methanobrevibacter smithii (AY196669) | 99 |
| 21 | Methanobrevibacter olleyae (AY615201) | 99 |
| 22 | Methanobrevibacter smithii ATCC 35061 (CP000678) | 100 |
| 23 | — | |
| 24 | Methanobrevibacter sp. AbM4 (AJ550156) | 99 |
| 25 | — | |
| 26 | — | |
| 27 | Methanobrevibacter smithii SM9 (AJ009958) | 99 |
| 28 | Methanobrevibacter gottschalkii strain HO (U55238) | 92 |
—, band could not be successfully cloned and sequenced.
The sequences obtained from the 20 PCR-DGGE bands represented seven different known species and two methanogen clones, including Methanobrevibacter thaueri strain CW (band 3), Methanobrevibacter smithii strain PS (band 18), Methanobrevibacter smithii (band 20), Methanobrevibacter olleyae strain KM1H5-1P (band 21), Methanobrevibacter smithii ATCC 35061 (band 22), and Methanobrevibacter smithii SM9 (band 27). Methanogenic archaeon SRmetG36 (band 9) and Methanobacteriales archaeon clone CSIRO1.33 (band 14) were represented by a single phylotype; Methanosphaera stadtmanae (bands 6 and 7) and Methanobrevibacter ruminantium NT7 (bands 10 and 17) were represented by two phylotypes; Methanobrevibacter sp. AbM4 was represented by three phylotypes (bands 11, 19, and 24); and Methanobrevibacter gottschalkii strain HO was represented by five phylotypes (bands 5, 8, 13, 16, and 28).
Changes in PCR-DGGE band patterns in response to changes in host feed efficiency.
To investigate potential associations between the methanogenic PCR-DGGE profiles and host feed efficiency and to examine the influence of the two diets on the microbial community, the band patterns generated for the H- and L-RFI groups were compared further. As shown in Fig. 3A, for the animals that were in the L-RFI group when both diets were used, 15 common bands were found. However, for the animals that switched from the L-RFI group to the H-RFI group, three new bands were identified: band 18 (Methanobrevibacter smithii PS), band 24 (Methanobrevibacter sp. AbM4), and band 27 (Methanobrevibacter smithii SM9). Also, the intensities of two bands, bands 1 and 9 corresponding to methanogenic archaeon clone SRmetG36 bands, decreased. Figure 3B shows the 15 bands shared by the animals that were in the H-RFI group when both diets were used; for the animals that changed from the H-RFI group to the L-RFI group, band 3 (Methanobrevibacter thaueri strain CW) appeared, whereas bands 1 and 9 (methanogenic archaeon clone SRmetG36) vanished. The changes in the band pattern for the M-RFI group were also compared, and no specific band was identified for this group of animals when they switched from the L-RFI or H-RFI group and vice versa (see Fig. S2 in the supplemental material).
In addition, all of the PCR-DGGE band patterns were analyzed further to determine the impact of diet. When the 28 bands were examined, bands 1, 5 (Methanobrevibacter gottschalkii strain HO), and 9 (methanogenic archaeon clone SRmetG36) were found in the rumen samples only when the low-energy diet was used, while band 24 (Methanobrevibacter sp. AbM4) and band 27 (Methanobrevibacter smithii SM9) were observed in the rumen samples only when the high-energy diet was used.
Associations between PCR-DGGE patterns, host feed efficiency, and changes in diet.
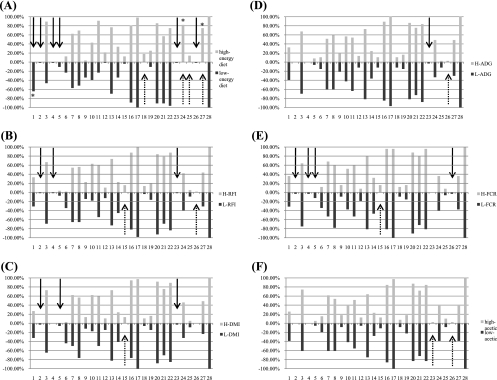
To determine the frequency of the presence of the bands and to evaluate whether diet or host RFI group influenced the band distribution for the population examined, a multivariate analysis was performed. Diet-specific bands were identified for both diets (Fig. 4A). Bands 1, 2, 4, 5 (Methanobrevibacter gottschalkii strain HO), 23, and 26 were distinctively linked to the low-energy diet (band 1 was found in about 80% of the samples, while the other bands were found in less than 10% of the animals), whereas bands 18 (Methanobrevibacter smithii PS), 24 (Methanobrevibacter sp. AbM4), 25, and 27 (Methanobrevibacter smithii SM9) were found to be associated with the high-energy diet (bands 24 and 27 were both detected in about 80% of the samples).
FIG. 4.
Band plot of the PCR-DGGE band frequencies. The percentages indicate the frequencies of appearance of the bands. The numbers indicate the bands from the top to the bottom of the gel. The solid arrows indicate L-group-specific bands; the dotted arrows indicate the H-group-specific bands. Group-specific bands for which the level of appearance was more than 20% are indicated by an asterisk. (A) Band plot for different diets. (B) Band plot for different RFI groups. (C) Band plot for different DMI groups. (D) Band plot for different ADG groups. (E) Band plot for different FCR groups. (F) Band plot for different acetate concentration groups.
For the RFI groups, certain trends for band distribution were identified (Fig. 4B). More than 10 bands were observed for most of the animals in both the H- and L-RFI groups (bands 3, 7, 8, 11, 13, 16, 17, 20, 21, 22, and 28), while the frequency of another set of bands was low in both RFI groups (bands 5, 12, 18, 19, and 25). Also, band 6 (Methanosphaera stadtmanae) was detected more frequently for L-RFI group animals, and band 10 (Methanobrevibacter ruminantium strain NT7) was more likely to appear for H-RFI group animals. Additionally, five bands were found to be RFI group specific; bands 2, 4, and 23 were observed only for L-RFI group animals, and bands 15 and 18 (Methanobrevibacter smithii PS) were observed only for H-RFI group animals. However, the abundance of each of these four bands was relatively low; band 15 was the only band that was identified for more than 20% of the entire population. Furthermore, samples from neither the H-RFI group animals nor the L-RFI group animals produced band 23 or 26; these two bands were detected only for animals in the M-RFI group.
In addition to the comparisons based on diet and RFI group, all of the PCR-DGGE band patterns were also compared for other indexes related to ruminal fermentation, such as DMI, ADG, FCR, and acetate concentration. In general, more than one-half of the bands were observed for either the H group or the L group for each measurement (Fig. 4C to 4F). One of these four parameters, DMI, tended to be related to four bands; band 15 was observed only for an H-DMI animal, band 6 (Methanosphaera stadtmanae) was found to be more prevalent for L-DMI animals, and band 10 (Methanobrevibacter ruminantium NT7) and band 27 (Methanobrevibacter smithii SM9) were more prevalent for H-DMI animals. The band distribution for the FCR revealed that bands 2, 4, 5 (Methanobrevibacter gotschalkii HO), and 26 were found only for L-FCR animals (at low frequencies), while band 15 was found only for H-FCR animals. The band distributions for ADG and acetate concentration were similar to each other, and in particular, bands 23 and 26 were found for only one group. The significance of each grouping was tested, and the band distribution was found to be significantly different only for the RFI groups (P < 0.0001) and the DMI groups (P < 0.0001).
Comparison of total methanogen populations.
The results for the total methanogen population were compared for the two diets and different RFI groups (Table 3). When animals were fed the low-energy diet, the sizes of the total methanogen populations in the L-RFI and H-RFI group animals were 2.12 × 107 cells/ml and 2.52 × 107 cells/ml, respectively (34). When animals were fed the high-energy diet, the sizes of the total methanogen populations in the L-RFI and H-RFI group animals were 2.15 × 107 cells/ml and 2.18 × 107 cells/ml, respectively. The total methanogen population did not change in response to different diets (P > 0.05), and no difference between the L-RFI group animals and the H-RFI group animals was detected (P > 0.05).
TABLE 3.
Comparison of copy numbers of targeted methanogen 16S rRNA genes in L-RFI and H-RFI group animals fed the low-energy and high-energy diets
| Diet | No. of copies/mla |
|
|---|---|---|
| L-RFI group animalsb | H-RFI group animalsc | |
| Low energyd | (2.12 ± 0.29) × 107 | (2.52 ± 0.29) × 107 |
| High energy | (2.15 ± 0.50) × 107 | (2.18 ± 1.10) × 107 |
The values are means ± standard errors. The amplification efficiency was 93.5 to 108.5%.
The P value for L-RFI group animals was 0.96.
The P value for H-RFI group animals was 0.76.
Values for the low-energy diet were published previously by Zhou et al. (36).
DISCUSSION
Methane is an undesirable end product of ruminal fermentation because it is a greenhouse gas that has adverse effects and because it causes a notable loss of energy in cattle. Understanding the ecology of methanogens in animals with different feed efficiencies and/or different diets can help elucidate the role of methanogens in ruminal methanogenesis and the mechanisms of this process. Methanogens have fastidious nutritional requirements, and thus the availability of culturable ruminal methanogens is limited. PCR-DGGE analysis is a useful culture-independent tool for identifying the microbial components in diverse environmental samples and for observing adaptation of microbial communities to various conditions. In this study, 56 animals fed two different diets and with different feed efficiencies were used for PCR-DGGE analysis. A comparison of the results allowed detection of a complex methanogenic microbiota under various conditions that could be used to elucidate methanogenic ecological changes that may be associated with different diets and different levels of host performance.
Consistent with the results of previous 16S rRNA gene sequencing analyses (13, 18, 23, 27, 28, 33, 36), the predominant members of the ruminal methanogenic community found in our PCR-DGGE analysis were Methanobrevibacter species. In particular, the predominant band for the samples from animals fed the low-energy diet was a Methanobrevibacter ruminantium band, in accordance with a previous report (36). The PCR-DGGE profiles of most animals contained multiple bands resembling Methanobrevibacter gottschalkii strain HO bands, which were not identified in the previous 16S rRNA library analysis (Fig. 4), suggesting that this species may be common in the bovine rumen under various diet conditions. The difference between the 16S rRNA library and the PCR-DGGE analysis data for this species may have been due to its low abundance in the rumen of the animals examined. Since PCR-DGGE can detect microbial components that comprise as little as 1% of the total population, rare species may be more likely to be discovered by analysis of the PCR-DGGE bands for each animal. The bands detected with a low frequency indicate the complexity in individuals and demonstrate the challenges when ruminal microbial communities are identified and compared.
Similar to the findings of our previous study (36), various strains and/or genotypes of the same species were identified. It was not surprising to find multiple DGGE bands representing the same species. For example, five bands were found for Methanobrevibacter gottschalkii strain HO and two bands were found for Methanosphaera stadtmanae (Table 2). This may have been due to microbial adaptation to different host animals. An alternative explanation is that the multiple bands may have been a result of amplification of a heteroduplex of the 16S rRNA gene. PCR-DGGE band patterns were compared using several tools. The sequence compositions of samples were compared using UniFrac (16) based on a 97% similarity cutoff. No difference at the species level was found between the groups of animals for any classification (data not shown), indicating that the divergence in the methanogenic community tended to be at the strain or genotype level rather than at the species level.
As shown in Fig. 4, the very prevalent bands may represent the core species, which are commonly found in the majority of the animals despite changes in diet, while the less prevalent bands may represent species that adapt to host conditions or particular diets. The observed change in the predominant methanogen population when the two different diets were used may have been due to a substrate utilization preference for methanogenesis by the phylotypes of the methanogens present. For instance, Methanobrevibacter ruminantium, the predominant species detected in numerous rumen samples, produces methane by utilizing CO2 as the substrate (18), and Methanobrevibacter smithii PS utilizes CO2-H2 and/or formate for methanogenesis (20) but also contains enzymes involved in the methanol-ethanol pathways (3, 6). Methanobrevibacter sp. AbM4 was recently found in the bovine rumen (36), but its substrate preference for methanogenesis pathways is unknown. In this study, two bands (bands 11 and 19) representing Methanobrevibacter sp. AbM4 were found when both diets were used, while band 24 was found only when the high-energy diet was used. Sequence mutations were found for these three bands (Fig. 5), suggesting that the band 24 phylotype may preferentially inhabit the rumen of cattle fed the high-energy diet. Another example is the bands representing Methanobrevibacter smithii, bands 20 and 22, which represent two different strains of Methanobrevibacter smithii identified when both diets were used. However, in another case, bands 18 and 27 were present only when the high-energy diet was used, showing that there was a difference in the band distribution at the strain level. Accordingly, it can be speculated that diet has an impact on the methanogenic community structure in the rumen, resulting in selection of methanogens that have particular methanogenesis pathways. This supports our hypothesis that the differences at the strain or genotype level of methanogens may play an important role in differences in methane production and hence lead to variations in the energy lost in host animals.
FIG. 5.
Alignment of Methanobrevibacter sp. AbM4-associated bands (bands 11, 19, and 24). Mutations are indicated by boxes.
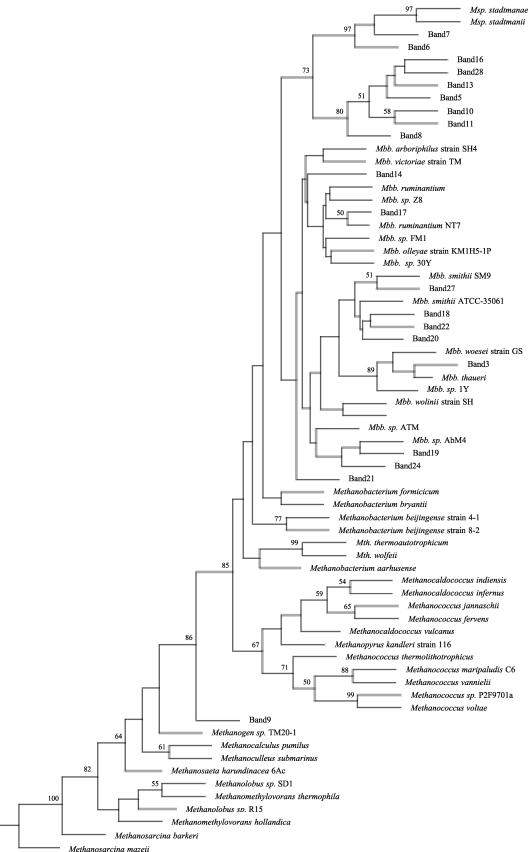
Potential correlations between the PCR-DGGE bands and some phenotypic data were analyzed by using PCA and multivariate analysis. PCA showed that band 10 was more likely to be associated with RFI and DMI when the high-energy diet was used and with RFI and FCR when the low-energy diet was used (data not shown). This band was classified as a Methanobrevibacter ruminantium strain NT7 band, but the level of identity to this species was low (94%). As shown in Fig. 6, this band did not cluster with band 17 corresponding to a Methanobrevibacter ruminantium strain NT7 band; instead, it was more closely related to Methanosphaera species bands. Since Methanobrevibacter ruminantium strain NT7 and Methanosphaera species have different substrate utilization profiles for methanogenesis, the phylotype represented by band 10 may utilize substrates more similar to the substrates utilized by Methanosphaera species than to the substrates utilized by Methanobrevibacter species. Moreover, the distribution of band 10 showed that a higher proportion of the animals in the H-RFI group than in the L-RFI group produced this band. This suggests that this phylotype may prefer H-RFI group animals over L-RFI group animals and that a possible substrate difference may be the reason for this preference. Therefore, strain and/or genotype diversity should not be neglected when the impact of methanogens on bovine rumen performance is considered. For instance, Methanobrevibacter gottschalkii strain HO was represented by five different bands (bands 5, 8, 13, 17, and 28), each with a different strain sequence type. Only one of these bands, band 5, was found to be associated with the L-DMI group (Fig. 4B). Methanobrevibacter gottschalkii has been reported to form a clade with Methanobrevibacter thaueri and Methanobrevibacter millerae and to occur in the rumen of lambs (12), and it has also been identified in the rumen of feedlot cattle (33). This species utilizes a CO2-H2 methanogenic pathway and requires acetate for growth (17). Further understanding of this rumen species and the methanogenic pathways utilized by it may help explain its distribution based on the DMI classification.
FIG. 6.
Phylogenic analysis of methanogen partial 16S rRNA sequences obtained from PCR-DGGE bands and identified species. Bootstrap values of >50% based on 100 replications are indicated at the nodes. Msp., Methanosphaera; Mbb., Methanobrevibacter; Mth., Methanothermobacter.
As indicated in our previous study, the total methanogen populations of animals with high and low feed efficiencies were not different when a low-energy diet was used (36). Similarly, no significant difference in the total methanogen populations was detected when the diet was switched from a low-energy diet to a high-energy diet. Comparable results were reported by Hook et al., who found that after a long period of monensin supplementation, the quantity of methanogens did not change significantly (9). In both studies, the periods after the diet was changed were long enough for the host animal to acclimate to the new conditions; thus, the total population of methanogens recovered so that the level was the same as the original level. Accordingly, the total ruminal methanogen population may not be the key factor that affects methane production. The observed differences in the methanogenic communities with different methanogenesis pathways may be a fundamental characteristic of the rumen ecosystem when different feeding strategies are used, as well as of individuals.
In addition, band 9, which resembled the methanogenic archaeon clone SRmetE18 (accession no. EU413577) band, was detected for animals in all RFI groups (H-RFI, L-RFI, and M-RFI) when the low-energy diet was used, as well as for animals in the M-RFI group (n = 2) when the high-energy diet was used. This clone was described in a study of Svalbard reindeer, in which it clustered with other ruminal archaeal clones and acidophilic archaea, forming a new phylogenetic clade (26). However, the known archaeal species most closely related to the SRmetE18 clone were Aciduliprofundum boonei and Thermogymnomonas acidicola, both of which are unlikely to be ruminal species; thus, the physiology of the species which clone SRmetE18 represented could not be predicted. The identification of a PCR-DGGE band having a sequence similar to a sequence of this clone suggests that there may be archaea other than methanogens in the bovine rumen that have not been reported previously, and the functions of these archaea should be determined.
This study was a preliminary study which showed that there was a change in the ruminal methanogenic community when the diet was changed from a low-energy diet to a high-energy diet. We are performing an experiment with multiple sampling points and a diet swap design to further confirm the impact of diet on ruminal methanogenic ecology and to investigate methanogen adaptation in response to diet modification. In the rumen, methanogens rely on bacteria, protozoa, and fungi to provide digestive products for methanogenesis. Therefore, the variation in the methanogen community may also be related to these other microbial components. For example, some rumen methanogens have been reported to be associated with protozoa (25) and to account for 37% of the total ruminal methane production (5). Thus, our results may underestimate the complexity of the methanogen community and the interaction of methanogens with other ruminal microbes. As a result, further studies identifying protozoan-associated methanogens are necessary to determine the effect of the associations on rumen methanogen ecology, host feed efficiency, and methane production. Additionally, the results of PCR-DGGE and sequence analysis obtained in this study could have been biased by the quality of the DNA, PCR amplification, and limitations of the sequence information in the database. Use of other technologies, such as multiplex qRT-PCR assays, should increase the spectrum and quantity of the target methanogens detected and should help identify low-abundance species that we were unable to identify in this study.
In conclusion, the methanogenic community varied in the rumens of steers with different feed efficiencies that were fed different diets. The ruminal methanogenic structure was found to correlate strongly with diet, and it may be associated with RFI in beef cattle. This is the first study to report a link between the ruminal methanogenic community profile, host metabolic variables, and host feed efficiency. Recent studies have reported that Methanobrevibacter smithii interacts with bacteria in the gut of mammalian species, such as mice (29) and humans (15). However, the role of methanogens in the gut remains unclear. Our study provides model for investigating the interactions between methanogens and hosts, as well as the interactions with other microorganisms, and for elucidating how these interactions could be impacted by nutrients in the gut. The demonstrated variation in the methanogenic community in individuals at the strain or genotype level indicates the importance of the microbial adaptation relationship with the host and its impact on animal performance. Advanced technologies and further studies are required to obtain a worldwide perspective for cattle and to generate a reference database for prediction of methane production, as well as animal performance.
Supplementary Material
Acknowledgments
This study was supported by ALIDF/AARI (2007041R) and by an NSERC discovery grant to L. L. Guan.
We thank S. Moore for supplying the animal information and B. Irving and all of the technical staff that assisted with animal management and sampling.
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Archer, J. A., E. C. Richardson, R. M. Herd, and P. F. Arthur. 1999. Potential for selection to improve efficiency of feed use in beef cattle: a review. Aust. J. Agric. Res. 50:147-161. [Google Scholar]
- 2.Bano, N., S. Ruffin, B. Ransom, and J. T. Hollibaugh. 2004. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk, H., and R. K. Thauer. 1997. Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch. Microbiol. 168:396-402. [DOI] [PubMed] [Google Scholar]
- 4.Environment Canada. 2004. Canada's greenhouse gas inventory, 1990-2001. Environment Canada, Ottawa, Ontario, Canada. http://www.ec.gc.ca/Publications/default.asp?lang-En&xml-FBDDD1E2-0183-4FD8-9B7E-247D92BD7348.
- 5.Finlay, B. J., G. Esteban, K. J. Clarke, A. G. Williams, T. M. Embley, and R. P. Hirt. 1994. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 117:157-161. [DOI] [PubMed] [Google Scholar]
- 6.Fricke, W. F., H. Seedorf, A. Henne, M. Kruer, H. Liesegang, R. Hedderich, G. Gottschalk, and R. K. Thauer. 2006. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan, L. L., J. D. Nkrumah, J. A. Basarab, and S. S. Moore. 2008. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. FEMS Microbiol. Lett. 288:85-91. [DOI] [PubMed] [Google Scholar]
- 8.Hegarty, R. S., J. P. Goopy, R. M. Herd, and B. McCorkell. 2007. Cattle selected for lower residual feed intake have reduced daily methane production. J. Anim. Sci. 85:1479-1486. [DOI] [PubMed] [Google Scholar]
- 9.Hook, S. E., K. S. Northwood, A. D. Wright, and B. W. McBride. 2009. Long-term monensin supplementation does not significantly affect the quantity or diversity of methanogens in the rumen of the lactating dairy cow. Appl. Environ. Microbiol. 75:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Intergovernmental Panel on Climate Change. 2001. Climate change 2001: a scientific basis. University Press, Cambridge, United Kingdom.
- 11.Irbis, C., and K. Ushida. 2004. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 12.Janssen, P. H., and M. Kirs. 2008. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74:3619-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis, G. N., C. Strompl, D. M. Burgess, L. C. Skillman, E. R. Moore, and K. N. Joblin. 2000. Isolation and identification of ruminal methanogens from grazing cattle. Curr. Microbiol. 40:327-332. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, K. A., and D. E. Johnson. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483-2492. [DOI] [PubMed] [Google Scholar]
- 15.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, T. L., and C. Lin. 2002. Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int. J. Syst. Evol. Microbiol. 52:819-822. [DOI] [PubMed] [Google Scholar]
- 18.Miller, T. L., M. J. Wolin, H. X. Zhao, and M. P. Bryant. 1986. Characteristics of methanogens isolated from bovine rumen. Appl. Environ. Microbiol. 51:201-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nkrumah, J. D., E. K. Okine, G. W. Mathison, K. Schmid, C. Li, J. A. Basarab, M. A. Price, Z. Wang, and S. S. Moore. 2006. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 84:145-153. [DOI] [PubMed] [Google Scholar]
- 20.Samuel, B. S., E. E. Hansen, J. K. Manchester, P. M. Coutinho, B. Henrissat, R. Fulton, P. Latreille, K. Kim, R. K. Wilson, and J. I. Gordon. 2007. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. U. S. A. 104:10643-10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp, R., C. J. Ziemer, M. D. Stern, and D. A. Stahl. 1998. Taxon-specific association between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol. Ecol. 26:71-78. [Google Scholar]
- 22.Skillman, L. C., P. N. Evans, G. E. Naylor, B. Morvan, G. N. Jarvis, and K. N. Joblin. 2004. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10:277-285. [DOI] [PubMed] [Google Scholar]
- 23.Skillman, L. C., P. N. Evans, C. Strompl, and K. N. Joblin. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222-228. [DOI] [PubMed] [Google Scholar]
- 24.Stewart, C. S., H. J. Flint, and M. P. Bryant. 1997. The rumen bacteria, p. 10-72. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial system, 2nd ed. Blackie Academic and Professional, New York, NY.
- 25.Stumm, C. K., H. J. Gijzen, and G. D. Vogels. 1982. Association of methanogenic bacteria with ovine rumen ciliates. Br. J. Nutr. 47:95-99. [DOI] [PubMed] [Google Scholar]
- 26.Sundset, M. A., J. E. Edwards, Y. F. Cheng, R. S. Senosiain, M. N. Fraile, K. S. Northwood, K. E. Praesteng, T. Glad, S. D. Mathiesen, and A. D. Wright. 2009. Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture. Microb. Ecol. 57:335-348. [DOI] [PubMed] [Google Scholar]
- 27.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 28.Tokura, M., I. Chagan, K. Ushida, and Y. Kojima. 1999. Phylogenetic study of methanogens associated with rumen ciliates. Curr. Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 29.Turnbaugh, P. J., F. Backhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, T., S. Asakawa, A. Nakamura, K. Nagaoka, and M. Kimura. 2004. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol. Lett. 232:153-163. [DOI] [PubMed] [Google Scholar]
- 31.Whitford, M. F., R. M. Teather, and R. J. Forster. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolin, M. J., T. L. Miller, and C. S. Stewart. 1997. Microbe-microbe interactions, p. 467-491. In P. N. Hobson and C. S. Steward (ed.), The rumen microbial ecosystem. Blackie Academic and Professional, New York, NY.
- 33.Wright, A. D., C. H. Auckland, and D. H. Lynn. 2007. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73:4206-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright, A. D., X. Ma, and N. E. Obispo. 2008. Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb. Ecol. 56:390-394. [DOI] [PubMed] [Google Scholar]
- 35.Wright, A. D., and C. Pimm. 2003. Improved strategy for presumptive identification of methanogens using 16S riboprinting. J. Microbiol. Methods 55:337-349. [DOI] [PubMed] [Google Scholar]
- 35a.Yu, Z., R. Garcia-Gonzalez, F. L. Schanbacher, and M. Morrison. 2008. Evaluation of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by Archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 74:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, M., E. Hernandez-Sanabria, and L. L. Guan. 2009. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 75:6524-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman & Hall, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.