Abstract
Previous studies identified the oleABCD genes involved in head-to-head olefinic hydrocarbon biosynthesis. The present study more fully defined the OleABCD protein families within the thiolase, α/β-hydrolase, AMP-dependent ligase/synthase, and short-chain dehydrogenase superfamilies, respectively. Only 0.1 to 1% of each superfamily represents likely Ole proteins. Sequence analysis based on structural alignments and gene context was used to identify highly likely ole genes. Selected microorganisms from the phyla Verucomicrobia, Planctomyces, Chloroflexi, Proteobacteria, and Actinobacteria were tested experimentally and shown to produce long-chain olefinic hydrocarbons. However, different species from the same genera sometimes lack the ole genes and fail to produce olefinic hydrocarbons. Overall, only 1.9% of 3,558 genomes analyzed showed clear evidence for containing ole genes. The type of olefins produced by different bacteria differed greatly with respect to the number of carbon-carbon double bonds. The greatest number of organisms surveyed biosynthesized a single long-chain olefin, 3,6,9,12,15,19,22,25,28-hentriacontanonaene, that contains nine double bonds. Xanthomonas campestris produced the greatest number of distinct olefin products, 15 compounds ranging in length from C28 to C31 and containing one to three double bonds. The type of long-chain product formed was shown to be dependent on the oleA gene in experiments with Shewanella oneidensis MR-1 ole gene deletion mutants containing native or heterologous oleA genes expressed in trans. A strain deleted in oleABCD and containing oleA in trans produced only ketones. Based on these observations, it was proposed that OleA catalyzes a nondecarboxylative thiolytic condensation of fatty acyl chains to generate a β-ketoacyl intermediate that can decarboxylate spontaneously to generate ketones.
There is currently great interest in elucidating the means by which microbes produce nongaseous hydrocarbons for use as specialty chemicals and fuels (8, 40). While many details remain to be revealed, there appear to be several different pathways by which microbes biosynthesize long-chain hydrocarbons. The most studied of the pathways (16) involves the condensation of isoprene units to generate hydrocarbons with a multiple of five carbon atoms (C10, C15, C20, etc.). A more obscure biosynthetic route is a reported decarbonylation of fatty aldehydes to generate a Cn−1 hydrocarbon chain (19). A third mechanism that has received some attention is what has been denoted head-to-head condensation of fatty acids (2, 3, 4, 8, 64). In this pathway, the hydrocarbons are described to arise from the formation of a carbon-to-carbon bond between the carboxyl carbon of one fatty acid and the α-carbon of another fatty acid (3). This condensation results in a particular type of hydrocarbon with chain lengths of C23 to C33 and containing one or more double bonds. One double bond involves the median carbon in the chain at the point of fatty acid condensation. An example of this overall biosynthetic pathway leading to the formation of specific C29 olefinic hydrocarbon isomers from fatty acid precursors has been demonstrated in vivo (61, 63, 64) and in vitro (4, 8).
The condensation, elimination of carbon dioxide, and loss of the other carboxyl group oxygen atoms likely require multiple enzyme-catalyzed reactions. Recent patent applications by L. Friedman et al. describe a role for three or four proteins in this biosynthetic pathway (18 September 2008, WO2008/113041; 4 December 2008, WO2008/147781). Most recently, Beller et al. demonstrated the requirement for three genes from Micrococcus luteus in the biosynthesis of long-chain olefins (8). That study also demonstrated in vitro production of olefins by recombinant proteins in the presence of crude cell extracts from Escherichia coli. In another study, Shewanella oneidensis strain MR-1 was shown to produce a head-to-head hydrocarbon (59). A cluster of four genes, oleABCD, was shown to be involved in olefin biosynthesis by that organism.
While genetic and biochemical data have provided evidence for Ole proteins producing long-chain olefins in M. luteus and S. oneidensis, there are many outstanding details of the biosynthesis that remain to be elucidated. Moreover, the extent to which microbial and other species produce head-to-head olefins is unclear. A recent patent application by Friedman and Rude (WO2008/113041) presented tables listing genes homologous to the ole genes described by Beller et al. (8). However, the homologs identified included genes from mouse and tree frog, organisms not known to produce head-to-head hydrocarbons. Additionally, hydrocarbon biosynthetic genes from Arthrobacter sp. FB24 were claimed, and that strain was later shown not to produce hydrocarbons under identical conditions for which other Arthrobacter strains did (22). In that context, the present study closely examined the protein sequence families of Ole proteins and the configurations of putative ole genes within genomes to identify those most likely to be involved in head-to-head hydrocarbon biosynthesis. This was followed by experimental testing for the presence of long-chain head-to-head hydrocarbons in representative bacteria from diverse phyla. This study also found that, of closely related bacteria, some produce head-to-head hydrocarbons and others do not.
A previous publication investigated in vitro olefin biosynthesis from myristyl-coenzyme A (CoA) (8). That study showed ketone and olefin biosynthesis in vitro and proposed a mechanism requiring the participation of ancillary proteins not encoded in the oleABCD gene cluster. The mechanism proposed fatty acyl oxidation to generate a β-keto acid that is the substrate for the OleA protein.
In fact, different mechanisms have been suggested previously for the biosynthesis of head-to-head olefins (3; Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781), and different roles for the OleA protein have been proposed (8; Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781). It is not possible to deduce the olefinic biosynthetic pathway or individual reaction types based on protein sequence alignments alone, because this pathway is unique, differing markedly from isoprenoid or decarbonylation hydrocarbon biosynthesis pathways. Moreover, the individual Ole proteins are each homologous to proteins that collectively catalyze diverse reactions. In that context, we (i) initiated a more detailed study of the Ole protein superfamilies, (ii) identified likely olefin (ole) biosynthetic genes out of thousands of homologs, (iii) experimentally tested bacteria from different phyla for long-chain olefins, (iv) developed insights into the role of OleA in head-to-head olefin biosynthesis, and (v) propose an alternative mechanism for head-to-head condensation of fatty acyl groups.
MATERIALS AND METHODS
Strains and culture conditions.
Wild-type and recombinant bacteria used in this study are listed in Table 1. All organisms, including recombinant strains, were grown aerobically in 50-ml culture flasks on a rotary shaker at 225 rpm, except for Geobacter strains, which were grown in a 100-ml anaerobic culture flask flushed for 30 min with a nitrogen/carbon dioxide gas mix prior to culture inoculation (50). All organisms were grown at 30°C (7, 11, 35, 37, 50) except for Shewanella amazonensis (35°C) (69), Shewanella frigidimarina (22°C) (70), Opitutaceae bacterium TAV2 (22°C) (57), Brevibacterium fuscum (22°C) (35), Colwellia psychrerythraea (4°C) (75), Chloroflexus aurantiacus (55°C) (48), and all Escherichia coli strains (37°C) (52). The organisms were allowed to achieve stationary phase prior to hydrocarbon extraction and analysis. All organisms were grown in Luria broth (Difco) (35, 37, 69, 70) except for S. frigidimarina (9), C. psychrerythraea (75), and Planctomyces maris (Marine broth; Difco) (73), Geobacter species (Geobacter medium; DSMZ) (50), C. aurantiacus (Chloroflexus medium; DSMZ) (48), Opitutaceae bacterium TAV2 (R2A medium; Difco) (T. D. Schmidt, personal communication), and X. campestris (nutrient broth; Difco) (11).
TABLE 1.
Organisms, plasmids, and primers used in this study
| Organism,a plasmid, or primer | Genotype, relevant characteristic(s), or sequence | Source or reference |
|---|---|---|
| Genetically modified organisms | ||
| S. oneidensis ΔoleA | S. oneidensis MR-1, ΔoleA; hydrocarbon minus | This study |
| S. oneidensis Δole | S. oneidensis MR-1, ΔoleABCD; hydrocarbon minus | 59 |
| E. coli UQ950 | E. coli DH5α λ(pir) host for cloning, F−Δ(argF-lac)169 φ80dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96 (NalR) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | 52 |
| E. coli WM3064 | Donor strain for conjugation, thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | 52 |
| Plasmids | ||
| pSMV3 | 9.5-kb vector; Kmr version of pSMV8; lacZ sacB | 6 |
| pSMV3-ΔoleA | 0.9-kb fusion PCR fragment containing ΔoleA cloned into SpeI/BamHI site of pSMV3; used to make S. oneidensis ΔoleA strain | This study |
| pBBR1MCS-2 | 5.1-kb broad-host-range plasmid; lacZ; Kmr | 36 |
| pOleA-S.m. | 1.1-kb PCR fragment containing S. maltophilia olnA, cloned into SpeI/SacI site of pBBR1MCS-2 | 59 |
| pOleA | 1.9-kb PCR fragment containing S. oneidensis oleA, cloned into SpeI/SacI site of pBBR1MCS-2 | This study |
| Primers | ||
| oleASoF1 | ACTAGTTACATGTGCGTTTATTGCAACTGGCC | |
| oleASoR1 | CCAGAGATATAGAGGCGCGAGGCGAGATTC | |
| oleASoF2 | GGTCTCATGGCACACGATCAAGGCTTTTTAC | |
| oleASoR2 | GGATCCCCAACAAATCAGTGTCGGCACC | |
| SooleACompF | ACTAGTTACATGTGCGTTTATTGCAACTGGCC | |
| SooleACompR | GAGCTCGTTAAAGCATCGGCTAAGGCAGATAACAA |
The following wild-type organisms were also used in this study (reference number[s]) shown in parentheses: Shewanella oneidensis MR-1 (20, 70), Shewanella putrefaciens CN-32 (33), Shewanella baltica OS185 (76), Shewanella frigidimarina NCIMB 400 (9), Shewanella amazonensis SB2B (69), Shewanella denitrificans OS217 (10), Colwellia psychrerythraea 34H (18), Geobacter bemidjiensis Bem (45), Geobacter sulfurreducens PCA (12), Opitutaceae bacterium TAV2 (57), Planctomyces maris DSM 8797 (7), Chloroflexus aurantiacus J-10-fl (48), Kocuria rhizophila DC2201 (37), Brevibacterium fuscum ATCC 15993 (51), Xanthomonas campestris (68), Vibrio furnissii M1 (46), and E. coli K-12 (lab supply).
Hydrocarbon and ketone extraction, chromatography, and characterization.
Early-stationary-phase cultures were extracted as previously described (72). Briefly, both cells and medium from a 50-ml bacterial culture that had reached stationary phase were extracted using a mixture of spectrophotometric-grade methanol (Sigma-Aldrich), high-performance liquid chromatography (HPLC)-grade chloroform (Sigma-Aldrich), and distilled water in a 5:5:4 ratio. The resulting nonpolar phase was collected and dried under vacuum. Evaporated residue was recovered in 1 ml of methyl-tert-butyl ether (MTBE), applied to a 4.0-g silica gel column (Sigma-Aldrich), and eluted with 35 ml of HPLC-grade hexanes (Fischer Scientific), followed by 35 ml of MTBE and 25 ml of HPLC-grade ethyl acetate (Sigma). Each solvent fraction was concentrated and subjected to GC-mass spectrometry (MS) analysis using an HP6890 gas chromatograph connected to an HP5973 mass spectrometer (Hewlett Packard, Palo Alto, CA). GC conditions consisted of the following: helium gas, 1 ml/min; HP-1ms column (100% dimethylpolysiloxane capillary, 30 m by 0.25 mm by 0.25 μm); temperature ramp, 100 to 320°C, 10°C/min, with a 5-min hold at 320°C. The mass spectrometer was run in electron impact mode at 70 eV and 35 μA. Alkene and ketone products were identified from the parent ions and corresponding fragmentation patterns. Major compounds were further analyzed by hydrogenation over palladium on carbon (Sigma-Aldrich) and observation of the corresponding increase in mass to confirm the number of double bonds present.
Gene deletion and oleA gene complementation.
All deletion strains, plasmids, and primers used are listed in Table 1. Gene deletions were made using homologous recombination between flanking regions of oleA cloned into a suicide vector, pSMV3 (52). Briefly, by using oleASoF1, oleASoR1, oleASoF2, and oleASoR1, the upstream and downstream regions surrounding the gene were cloned using the restriction sites SpeI and BamHI into the suicide vector in a compatible E. coli cloning strain (UQ950) (52). This plasmid was transformed into an E. coli mating strain (WM3064) (52) and then conjugated into MR-1. While E. coli was commonly grown at 37°C, when S. oneidensis was present cells were incubated at 30°C. The initial recombination event was selected for by resistance to kanamycin. Cells containing the integrated suicide vector grew in the absence of selection overnight at 30°C and then were plated onto LB plates containing 5% sucrose (52). Cells retaining the suicide vector were unable to grow due to the activity of SacB, encoded on the vector, while cells that underwent a second recombination event formed colonies. Colonies were then screened by PCR to determine strains containing the deletion. The oleABCD gene cluster deletion of S. oneidensis MR-1 was created as described previously (59).
Complementation of the S. oneidensis oleA mutant was performed using the pBBR1MCS-2 expression vector (36) and the endogenous lac promoter (which is constitutive in MR-1 due to the absence of lacI). Primers oleASoFcomp and oleASoRcomp containing SacI and SpeI restriction sites were designed for the regions flanking the ends of oleA. Resulting PCR products were ligated into the Strataclone cloning system (Agilent Technologies), followed by digestion and ligation of the product into the pBBR1MCS-2 expression vector. The Stenotrophomonas maltophilia oleA gene was introduced into pBBR1MCS-2 as described previously (59). Constructs were introduced into E. coli WM3064 prior to conjugation with the oleA deletion, the ole cluster deletion, or wild-type MR-1 strains. All constructs were verified through PCR and sequencing analysis. Following conjugation, all constructs were maintained using 50 μg/ml kanamycin.
Identification of oleABCD-containing organisms.
The oleABCD genes in S. oneidensis MR-1 were used to find homologous gene clusters in the GenBank nonredundant database using the BLAST algorithm (5). Subsequently, the OleA homologs in Stenotrophomonas maltophilia strain R551-3 (gi 194346749), Arthrobacter aurescens TC1 (gi 119962129), and Micrococcus luteus NCTC 2665 (gi 239917824) were used as additional queries to the database. Other homologous thiolases were identified. The genome context of each of these thiolases was investigated and allowed for the assembly of a set of organisms with either a four- or three-gene cluster, encoding OleA, -B, -C, and -D protein domains. A lack of clustering did not preclude the existence of the pathway in an organism. Therefore, those organisms that lacked clustered genes were searched for oleBCD genes in other locations of their genome. Organisms with clustering of at least two identifiable ole homologs and which had all four genes in their genome were included as potential hydrocarbon producers and investigated experimentally.
Superfamily sequence identification and alignments.
The PSI-BLAST algorithm with default conditions (53) was used with S. oneidensis MR-1 or A. aurescens TC1 Ole protein sequences as queries. Thousands of homologous sequences were found. The sequence and catalytic diversity within each superfamily were sufficiently broad that standard sequence alignment tools did not align amino acid residues that are known to comprise the active sites in proteins for which X-ray structures are available (15, 25, 27, 28, 32, 34, 44, 49, 56, 61, 71, 74). Thus, to properly align Ole protein sequences with other proteins in their respective superfamilies, it was necessary to generate structure-based alignments. For each OleABCD alignment, 6 to 10 homologous proteins from previously described high-resolution X-ray structures were structurally superimposed, using the Match command in Chimera (42). Conserved residues within each superfamily of homologs were derived from the literature (15, 25, 27, 28, 32, 34, 44, 49, 56, 61, 71, 74), and their locations were plotted onto the protein backbone to confirm alignments. Sequence alignments based on the structure alignments were exported. Multiple sequence alignments of each of the OleABCD families were made with 41 to 55 sequences, using ClustalW (13). In the case of the OleA alignments, 14 OleA homologs with genes that did not cluster with oleBCD genes were also included for sequence comparison purposes. A profile-profile alignment between the structural superfamily alignments and the family sequence alignments was produced, using ClustalW (13). These superfamily Ole sequence alignments were viewed in Chimera with the overlaid superfamily crystal structures linked to the alignments so that the positions of residues in the alignment could be viewed (42). For OleBC fusion proteins, the individual domains were used for alignments with the appropriate superfamilies.
Analysis of protein superfamilies.
The Superfamily database (24) was searched with each of the S. oneidensis MR-1 Ole protein sequences. The superfamilies identified by these searches confirmed assignments made independently as described above. The number of distinct proteins in each superfamily was kindly provided from the Superfamily database (Derek Wilson, personal communication). The relevant superfamily categories in the Superfamily database are thiolase-like, α/β-hydrolases, acetyl-CoA synthetase-like, and NAD(P) Rossman fold domains. It should be noted that the NAD(P) Rossman fold domains superfamily, as listed in the Superfamily database, consists of a number of families in which the proteins share the ability to bind NAD(P), and it contained a total of 136,722 proteins as of 1 February 2010. These proteins have a second domain involved in substrate binding and which confers the catalytic residues. These differentiations are made in the Superfamily database at what is denoted as the family level. The OleD proteins belong to the tyrosine-dependent oxidoreductase domain family. This set was used for our analysis and was equivalent to the set given superfamily status by Jornvall et al. and described as the short-chain dehydrogenase/reductase superfamily (34).
Network clustering of OleABCD proteins.
Network clustering of each of the OleABCD proteins was analyzed using previously described procedures (43, 55). This method was used to make an all-by-all BLASTp library for each of the OleABCD proteins using sequences from 15 organisms. The sequences used were (i) S. oneidensis MR-1, OleA gi24373309, OleB gi24373310, OleC gi24373311, and OleD gi24373312; (ii) Shewanella amazonensis SB2B, OleA gi119774319, OleB gi119774320, OleC gi119774321, and OleD gi119774322; (iii) Shewanella baltica OS185, OleA gi153000075, OleB gi153000076, OleC gi153000077, and OleD gi153000078; (iv) Shewanella denitrificans OS217, OleA gi91792727, OleB gi91792728, OleC gi91792728, and OleD gi91792730; (v) Shewanella frigidimarina NCIMB 400, OleA gi114562543, OleB gi114562544, OleC gi114562545, and OleD gi114562546; (vi) Shewanella putrefaciens CN-32, OleA gi146292545, OleB gi 146292546, OleC gi146292547, and OleD gi146292548; (vii) Colwellia psychrerythraea 34H, OleA gi71279747, OleB gi71279056, OleC gi71281286, and OleD gi71280771; (viii) Geobacter bemidjiensis Bem, OleA gi197118484, OleB gi197118483, OleC gi197118482, and OleD gi197118481; (ix) Planctomyces maris DSM 8797, OleA gi149174448, OleB gi149178001, OleC gi149178707, and OleD gi149178706; (x) Opitutaceae bacterium TAV2, OleA gi225164858, OleB gi225164858, OleC gi225155590, and OleD (no cluster); (xi) Stenotrophomonas maltophilia R551-3, OleA gi194363945, OleB gi194363946, OleC gi194363948, and OleD gi194363949; (xii) Xanthomonas campestris pv. campestris strain B100, OleA gi188989629, OleB gi188989631, OleC gi188989633, and OleD gi188989637; (xiii) Chloroflexus aurantiacus J-10-fl, OleA gi163849058, OleB gi163849062, OleC gi163849060, and OleD gi163849059; (xiv) Arthrobacter aurescens TC1, OleA gi119962129, OleB gi119960515 (residues 1 to 310), OleC gi119960515 (residues 389 to 921), and OleD gi119962242; (xv) Arthrobacter chlorophenolicus A6, OleA gi220911225, OleB domain gi220911226 (residues 1 to 296), OleC gi220911226 (residues 370 to 927), and OleD gi220911227; (xvi) Kocuria rhizophila DC2201, OleA gi184200698, OleB gi184200697 (residues 1 to 312), OleC gi184200697 (residues 392 to 909), and OleD gi184200696; (xvii) Micrococcus luteus NCTC 2665, OleA gi239917824, OleB gi239917825 (residues 1 to 330), OleC gi239917825 (residues 439 to 978), and OleD gi239917826. From these sequences, a network diagram was created. The nodes represent protein sequences and the edges represent a BLAST linkage that connects the two proteins. A shorter edge represents a lower e-score (greater relatedness). Expectation values from e−2 to e−200 were analyzed for connectivity and divergence, respectively, of OleA, -B, -C, and -D protein sequence clusters.
RESULTS AND DISCUSSION
Ole protein superfamily analysis.
Thousands of sequences were identified as being homologous to each of the OleA, OleB, OleC, and OleD sequences from S. oneidensis MR-1 (Table 2). OleA is homologous to members of the thiolase superfamily, also known as the condensing enzyme superfamily. The sequence relatedness between different OleA proteins and FabH, a thiolase superfamily member, has been noted previously even though sequence identities of OleA to FabH and other superfamily members are generally low, in the range of 20 to 30% (8; Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781). OleB is a member of the α/β-hydrolase superfamily. OleC is a member of the AMP-dependent ligase/synthase superfamily, also known as the acetyl-CoA synthetase-like superfamily. OleD is a member of the short-chain dehydrogenase/reductase superfamily.
TABLE 2.
Ole protein superfamily characteristics for this study
| Protein in this study | Superfamily name (alternative name[s]) | Enzymatic activities and biological functions in the superfamily | No. of homologs in superfamily | No. of Ole proteins identified |
|---|---|---|---|---|
| OleA | Thiolase (condensing enzymes) | Acyl-ACP synthase, thiolase (degradative), thiolase (biosynthetic), 3-hydroxyl-3-methylglutaryl-CoA synthase, fatty acid elongase, stage V sporulation protein, 6-methylsalicylate synthase, Rhizobium nodulation protein NodE, chalcone synthase, stilbene synthase, naringenin synthase, β-ketosynthase domains of polyketide synthase | 13,586 | 69 |
| OleB | α/β-Hydrolase | Esterase, haloalkane dehalogenase, protease, lipase, haloperoxidase, lyase, epoxide hydrolase, enoyl-CoA hydratase/isomease, MhpC C-C hydrolase (carbon-carbon bond cleavage) | 67,923 | 69 |
| OleC | AMP-dependent ligase/synthase (LuxE; acyl-adenylate/thioester-forming, acetyl-CoA synthetase-like) | Firefly luciferase, nonribosomal peptide synthase, acyl-CoA synthase (AMP forming), 4-chlorobenzoate:CoA ligase, acetyl-CoA synthetase, O-succinylbenzoic acid-CoA ligase, fatty acyl ligase, acetyl-CoA synthetase, 2-acyl-glycerophospho-ethanolamine acyl transferase, enterobactin synthase, amino acid adenylation domain, dicarboxylate-CoA ligase, crotonobetaine/carnitine-CoA ligase | 19,660 | 69 |
| OleD | Short-chain dehydrogenase/reductase | Nucleoside-diphosphate sugar epimerase/dehydratase/reductase, aromatic diol dehydrogenase, steroid dehydrogenase/isomerase, sugar dehydrogenase, acetoacetyl-CoA reductase, 3-oxoacyl-ACP reductase, alcohol dehydrogenase, carbonyl reductase, 4-α-carboxysterol-C3-dehydrogenase/C4-decarboxylase, flavonol reductase, cinnamoyl CoA reductase, NAD(P)-dependent cholesterol dehydrogenase | 25,454 | 69 |
Figure 1 shows conserved regions of a structure-based multiple sequence alignment for each of the OleA, -B, -C, and -D proteins with three of their respective superfamily members. Figure 1 focuses on regions containing catalytically important residues that are highly conserved among the homologous proteins. A more detailed set of alignments is available in the supplemental material. The superfamily members shown in Fig. 1 were selected to represent proteins serving quite different biological functions. So, while OleABCD are clearly seen to contain critical catalytic residues of each respective superfamily, a precise prediction of the biochemical reaction catalyzed is difficult due to the enormous functional diversity found within each Ole protein's superfamily.
FIG. 1.
Structure-based amino acid sequence alignments of OleA, OleB, OleC, and OleD from S. oneidensis MR-1 (denoted MR1 in the figure) with highly conserved regions of proteins catalyzing divergent reactions from each respective superfamily. Accession numbers or PDB identifiers of the proteins used are as follows. OleA thiolase superfamily: OleA from Shewanella oneidensis MR-1 (gi 24373309), β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli (1EBL), 3-hydroxy-3-methylglutaryl-CoA synthase (HMG_CoA) from Staphylococcus aureus (1XPK), and chalcone synthase (Chalcone) from Medicago sativa (1CGZ); blue residues indicate the glutamate that abstracts a proton to produce a carbanion for the nondecarboxylative condensation reaction, red indicates the absolutely conserved cysteine of the superfamily that forms a covalent bond with the substrate, and green residues are involved in formation of an oxyanion hole. OleB α/β-hydrolase superfamily: OleB from Shewanella oneidensis MR-1 (gi 24373310), haloalkane dehalogenase (HAD) from Xanthobacter autotrophicus GJ10 (1B6G), epoxide hydrolase (EH) from Agrobacterium radiobacter AD1 (1EHY), and prolyloligopeptidase (prolyl) from porcine brain (1H2W); red residues indicate the catalytic nucleotide (Ser, Asp, or Cys in the whole superfamily), green indicates the general acid, and blue indicates the conserved histidine that activates water. OleC AMP-dependent ligase/synthetase superfamily: OleC from Shewanella oneidensis MR-1 (gi 24373311), gramicidin synthetase (gramicidin) from Brevibacillus brevis (1AMU), acetyl-CoA synthetase (AcCoA) from Saccharomyces cervisiae (1RY2), and luciferase from the Japanese firefly (2D1Q); red indicates absolute conservation in the three consensus regions identified by Conti et al. ([STG]-[STG]-G-[ST]-[TSE]-[GS]-x-[PALIVM]-K, [YFW]-[GASW]-x-[TSA]-E, [STA]-[GRK]-D) (15); blue and green indicate Thr/Ser residues thought to be involved in binding the phosphoryl group in ATP and AMP. OleD short chain dehydrogenase/reductase superfamily: OleD from Shewanella oneidensis MR-1 (gi 24373312), UDP-galactose-4-epimerase (Udp-gal-4 epim) from humans (1EK6), 7-α-hydroxysteroid dehydrogenase (7a-HOstroid DH) from Escherichia coli (1AHH), and D-3-hydroxybutyrate dehydrogenase (D-3-HObut DH) from Pseudomonas fragi (3ZTL); blue identifies the tyrosine anion that abstracts the proton from the substrate, red is a lysine that stabilizes the tyrosine anion, green is a glycine-rich region involved in cofactor NAD(P)+ binding, and pink is the serine that orients the substrate or stabilizes intermediates.
The superfamilies to which Ole proteins belong each consist of between 104 and 105 curated protein members that have been identified for inclusion in the Superfamily database (Table 2). The present study suggested that only 0.1% to 1% of the proteins in each superfamily represents Ole proteins that participate in head-to-head hydrocarbon biosynthesis. The identification of these Ole proteins in the sequenced genomes of microorganism is discussed below.
Protein relatedness and gene organization used to identify ole genes among thousands of homologs.
Only a limited number of bacteria to date have been found to produce long-chain olefinic hydrocarbons. For example, among 10 Arthrobacter strains tested, 6 produced long-chain olefinic hydrocarbons and 4 did not (22). Of three closely related Arthrobacter strains for which genome sequences were available, two (A. aurescens TC1 and A. chlorophenolicus A6) were shown to produce hydrocarbons and one (Arthrobacter sp. FB24) was devoid of long-chain olefinic hydrocarbons. The FB24 strain that did not produce hydrocarbons contained ole gene homologs, but the percent identity was much lower and the genes were distributed within the genome differently. By examining such divergences, a strategy for identifying highly likely ole genes was developed as described in the Materials and Methods section. The example below is illustrative.
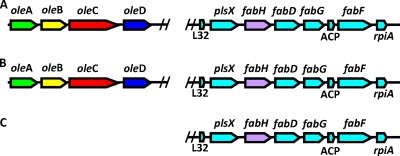
A putative oleA gene region was identified in Geobacter bemidjiensis Bem that, after translation, showed 58% amino acid sequence identity to the OleA protein in S. oneidensis MR-1. Directly downstream from the G. bemidjiensis Bem oleA gene, oleBCD gene homologs were present in a configuration that mirrored that of S. oneidensis MR-1 (Fig. 2). An OleA homolog was also identified in Geobacter sulfurreducens PCA. It showed significantly lower amino acid sequence identity, 28%, to the OleA from S. oneidensis MR-1. It lacked flanking ole gene neighbors. Closer examination of the two genomes revealed that the OleA homolog in G. sulfurreducens PCA was encoded by a gene region that matched a gene region with identical synteny in G. bemidjiensis Bem. This same gene region was also identified in S. oneidensis MR-1. From this analysis, it was concluded that the OleA homolog in G. sulfurreducens PCA was not involved in a head-to-head condensation reaction and it was suggested that this organism was genetically incapable of making head-to-head olefins. Cells of G. sulfurreducens PCA were tested experimentally for the presence of long-chain olefinic hydrocarbons. Hydrocarbons were absent under identical growth conditions in which they were present in G. bemidjiensis Bem (see the discussion on hydrocarbon identification below).
FIG. 2.
Analysis of the gene regions of S. oneidensis MR-1 (A), G. bemidijiensis Bem (B), and G. sulfurreducens PCA (C). Genes denoted oleA and fabH are homologs to the oleA from S. oneidensis MR-1. A predicted oleA gene region is shown for S. oneidensis (A) and G. bemidijiensis Bem (B), clustering with oleBCD genes. The fabH gene, which is an oleA homolog with highest percent identity with G. sulfurreducens PCA, fails to cluster with oleBCD homologs.
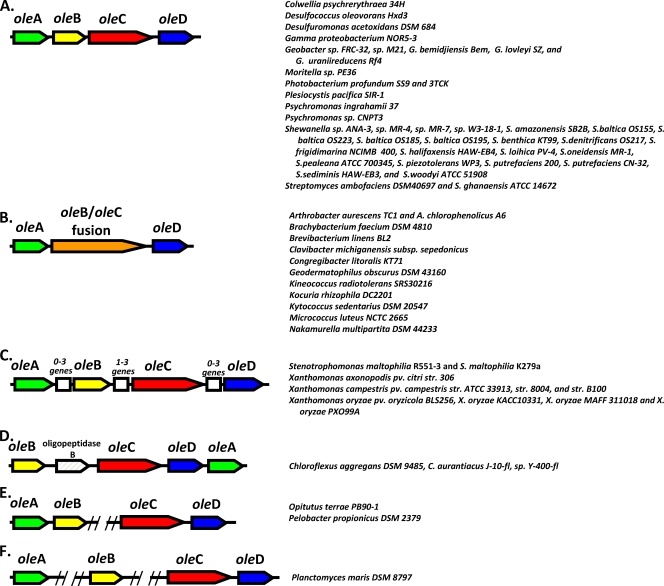
A collection of 3,558 genomes were examined using the described methods, leading to the identification of several different ole gene arrangements (Fig. 3). One major distinction in ole gene organization had been recognized previously by Friedman et al. (WO2008/113041 and WO2008/147781); a significant number of organisms contained either three or four separate ole genes. Of those characterized in this study, the largest set contained four contiguous oleABCD genes. However, some bacteria of the class Actinobacteria contained three ole genes, with the oleB and oleC gene regions fused into one gene (Fig. 3A and B). In 61 organisms either the four- or three-gene cluster was readily identifiable (Fig. 1A to D). Genomes that had a clear clustering of homologs of at least two of these genes were included as potential clusters. At least one sample organism from each of the gene clusterings in Fig. 3A to F was obtained, and the phenotype was confirmed experimentally by the presence of long-chain olefinic hydrocarbons in solvent extracts of growing cells (see below). As of 20 July 2009, highly likely ole genes were identified in 69 genomes. This was out of 3,558 total genomes. Thus, only 1.9% of the genomes examined contained evidence for ole genes based on the methods described here. Of the bacterial genomes, 69 out of 1,331, or 5.2%, showed bioinformatic evidence for ole genes. The genome analysis included 2,143 Eukaryota and 84 Archaea, none of which showed clear evidence of containing an ole gene cluster. This analysis does not rule out that the head-to-head hydrocarbon genes and pathway will be shown to be present in Archaea or Eukaryota, but only that our analysis could not identify them with confidence.
FIG. 3.
ole gene regions of different bacteria. The gene region configuration is shown on the left, and the bacteria containing each are listed at the right. The double slashes in panels E and F indicate that the genes on either side are not contiguous. Green, oleA; yellow, oleB; red, oleC; blue, oleD; orange, oleBC fusion; white, other genes not currently identified as being involved in hydrocarbon biosynthesis. The different parts represent the most common contiguous four-gene configuration (A), the three-gene cluster in which the oleB and oleC genes are in a single gene, oleBC fusion (B), and gene organization with various insertions between the identified ole genes (C). The white boxes indicate multiple genes that may be encoded in the same or opposite directions to the ole genes. In particular, various Xanthomonas spp. strains have different numbers of genes identified in the indicated locations. (D) Chloroflexi species that have an oligopeptidase inserted between oleB and oleC. Also, the oleA homolog is located after the other genes. (E) Configuration in which pairs of genes are in different parts of the genome. (F) A configuration in which oleA and oleB are located in different parts of the genome but oleC and oleD are clustered. Note: hydrocarbon production was confirmed in at least one organism in each class, A to F. Identifiers for each of the genes are listed in Table S1 of the supplemental material.
Hydrocarbon identification.
It could not be inferred from sequence analysis alone whether all of the gene configurations would give rise to hydrocarbon products. In this context, at least one organism from each class (A to F) of Fig. 3 was tested directly for long-chain olefin biosynthesis. In previous studies (2, 8, 22, 59, 62), olefins were produced under all growth conditions for all of the organisms tested; olefin production appears to be constitutive. In this context, each strain was grown under optimum conditions for the strain, as described in Table 1 and Materials and Methods. From each organism, nonpolar material was extracted with solvent and analyzed by chromatography and mass spectrometry. Controls were conducted with solvent blanks and organisms previously described not to produce head-to-head hydrocarbons (72) to exclude that olefins were derived from solvents or work-up procedures. This study showed that bacteria from the different types of gene clusterings shown in Fig. 3 produced hydrocarbons in direct experimental tests (Fig. 4 and Table 3). Different hydrocarbons were produced, but all were long chain (>C23) and contained at least one double bond, consistent with their formation by a head-to-head coupling of fatty acyl groups.
FIG. 4.
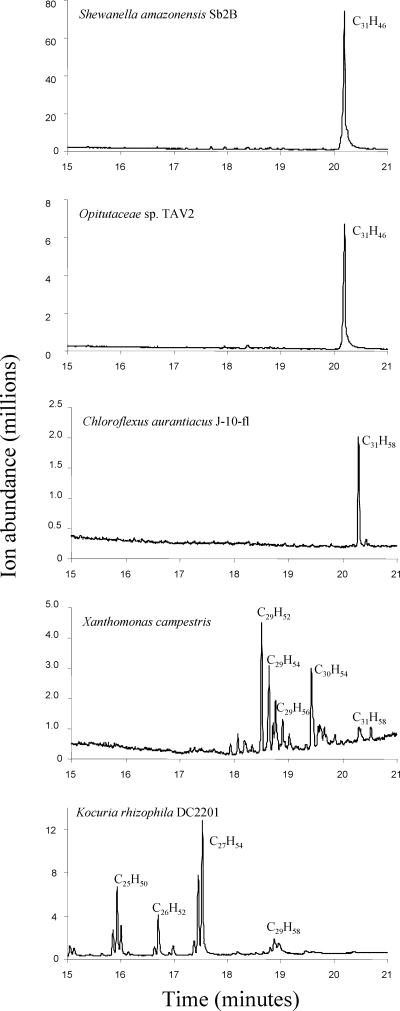
Gas chromatograms of extracts from different bacteria containing ole genes as identified by bioinformatics. Bacteria were extracted, and extracts were analyzed by GC-MS as described in Materials and Methods. The major products are labeled with their chemical formulas. No hydrocarbon peaks were identified beyond the elution range shown.
TABLE 3.
Compilation of head-to-head olefins produced by different bacteria
| Microorganism | Total hydrocarbons |
Predominant hydrocarbon |
||
|---|---|---|---|---|
| No. detected | Size range | Mass spectruma (m/z) | Formula | |
| C. aurantiacus J-10-fl | 1 | C31H58 | 430, 303 | C31H58 |
| K. rhizophila DC2201 | 12 | C24H48-C29H58 | 478, 348 | C27H54 |
| B. fuscum ATCC 15993 | 9 | C27H54-C29H58 | 406, 376 | C29H58 |
| X. campestris pv. campestris | 15b | C28H56-C31H58 | 402, 303 | C29H54 |
| S. oneidensis MR-1 | 1 | C31H46 | 418, 281 | C31H46 |
| S. putrefaciens CN-32 | 1 | C31H46 | 418, 281 | C31H46 |
| S. baltica OS185 | 1 | C31H46 | 418, 281 | C31H46 |
| S. frigidimarina NCIMB 400 | 1 | C31H46 | 418, 281 | C31H46 |
| S. amazonensis SB2B | 1 | C31H46 | 418, 281 | C31H46 |
| S. denitrificans OS217 | 1 | C31H46 | 418, 281 | C31H46 |
| C. psychrerythraea 34H | 1 | C31H46 | 418, 281 | C31H46 |
| G. bemidjiensis Bem | 1 | C31H46 | 418, 281 | C31H46 |
| O. bacterium TAV2 | 1 | C31H46 | 418, 281 | C31H46 |
| P. maris DSM 8797 | 1 | C31H46 | 418, 281 | C31H46 |
Identifying ions for the predominant long-chain olefin.
Number readily identifiable by GC-MS.
Shewanella amazonensis SB2B, isolated from the Amazon River Delta off the coast of Brazil (69), contains recognizable ole genes. It produced a single product with a carbon chain length of 31 and with nine double bonds (C31H46). The GC retention time and the mass spectrum indicated that the compound was identical to that produced by S. oneidensis strain MR-1, which had been described previously (59). The hydrocarbon in S. oneidensis MR-1 is the C31 polyolefin 3,6,9,12,15,19,22,25,28-hentriacontanonaene. Additional Shewanella strains were tested in this study, and all produced the C31 polyolefin as the only discernible hydrocarbon (Table 3).
Colwellia psychreryhtraea is an obligate psychrophile that grows at temperatures below 0°C, Geobacter bemidjiensis Bem was isolated from a petroleum-contaminated aquifer sediment, Opitutaceae TAV2 is a member of the phylum Verrucomicrobia but is not well studied (54), and Planctomyces maris DSM8797 is in the phylum Planctomycetes and was isolated from the open ocean (7). Despite the great phylogenetic and ecological diversities of these bacteria, they all produced a single hydrocarbon product with the same retention time (20.2 min) and mass spectrum, consistent with its identity as 3,6,9,12,15,19,22,25,28-hentriacontanonaene (Fig. 4 and Table 3).
A closely migrating, but clearly distinct, hydrocarbon product was produced by Chloroflexus aurantiacus strain J-10-fl (Fig. 4), a bacterium isolated from hot springs and that grows optimally at 55°C (48). The Chloroflexus hydrocarbon migrated more slowly on the GC column (20.4 min), and the mass spectrum indicated a chemical formula of C31H58, consistent with a hydrocarbon containing 31 carbon atoms and three double bonds. These data are consistent with previous reports that identified hentriaconta-9,15,22-triene (C31H58) growing in microbial mats (67) and being formed by Chloroflexus spp. in pure culture (66).
Kocuria rhizophila strain DC2201 was isolated for its ability to withstand organic solvents (23), and its complete genome sequence was reported in 2008 (60). Here it was shown to produce multiple olefinic hydrocarbon products that ranged from 24 to 29 carbon atoms (Table 3). Each identified compound contained one double bond. The clusters of compounds eluting at approximately 16 min, 16.8 min, 17.5 min, and 19 min (Fig. 4) represent isomeric clusters of C25, C26, C27, and C29 chain lengths, respectively, based on mass spectrometry. This type of hydrocarbon cluster resembled, but was not identical to, those found in species of Arthrobacter (22) and Micrococcus (2, 8, 63) that have been studied previously. The major compounds in Kocuria analyzed here contained 25 and 27 carbon atoms. Another actinobacterial strain that had not yet been tested for the presence of head-to-head hydrocarbons, Brevibacterium fuscum ATCC 15993, similarly produced isomeric clusters of hydrocarbons but in the range of 27 to 29 carbon atoms (Table 3).
The most extensive array of hydrocarbon products from those organisms tested here was observed with Xanthomonas campestris (Fig. 4 and Table 3), a bacterium that causes a range of plant diseases (11, 68). X. campestris produced hydrocarbons with chain lengths of C28, C29, C30, and C31. Based on the mass spectra, hydrocarbons containing one, two, or three double bonds could be identified. There was additional structural complexity that was likely due to isomerization, which could arise from different types of methyl branching at the hydrocarbon termini. The complexity of the mixture precluded precise structural determinations, which would require the availability of synthetic standards.
Negative controls were run to rule out artifacts that could result, for example, from hydrocarbon contamination external to the cells (72). The most telling experimental results were obtained with Geobacter sulfurreducens PCA, an organism closely related to G. bemidjiensis Bem but suggested from bioinformatics analysis, in this study, to contain an oleA homolog with a different function (Fig. 2). Olefinic hydrocarbons were not detected in G. sulfurreducens. Additionally, long-chain olefinic hydrocarbons were not detected in cultures of E. coli K-12 or Vibrio furnissii M1, both of which were determined not to contain ole genes based on the bioinformatics analysis described here.
Most previous studies had investigated bacterial head-to-head hydrocarbon biosynthesis in members of the Actinobacteria, including Micrococcus (2, 3, 8) and Arthrobacter (22). Long-chain olefinic hydrocarbons had also been demonstrated in Stenotrophomonas maltophilia (58), a member of the phylum Proteobacteria. The present study showed additional Actinobacteria (Brevibacterium) and Proteobacteria (Geobacter sp.) produce head-to-head hydrocarbons. In addition, members of the phyla Verucomicrobia, Planctomyces, and Chloroflexi were shown to contain bona fide ole genes and to produce olefinic hydrocarbons. This greatly expanded the phylogenetic diversity demonstrated experimentally to produce head-to-head olefinic hydrocarbons and revealed the type(s) of hydrocarbon produced. The latter could not be discerned from the ole gene sequences alone based on previous studies. The present study is a start in establishing a link between one of the Ole protein sequences with the hydrocarbon(s) produced as discussed in the section below.
OleA has a major role in determining the type of head-to-head products formed.
The different long-chain olefinic hydrocarbons identified in this and other studies show variable chain lengths and degrees of unsaturation. These findings could be determined largely by the fatty acid composition within the cell, by the substrate specificity of the Ole proteins, by other proteins, or by some combination of these factors. To begin to investigate this, S. oneidensis MR-1 strains with different oleA gene contents were grown identically and tested for hydrocarbon content. The S. oneidensis MR-1 strains contained, respectively (A) the native Shewanella oleA gene only, (B) the native Shewanella oleA gene plus a Stenotrophomonas oleA gene, (C) no oleA gene, (D) the Stenotrophomonas oleA gene in a Shewanella oleA deletion strain, or (E) the Stenotrophomonas oleA gene in a Shewanella oleABCD deletion strain. Each strain (A to E) was grown under the same conditions of medium, temperature, and aeration. Each strain was harvested and extracted the same way. Each extract was subjected to the same chromatographic procedures.
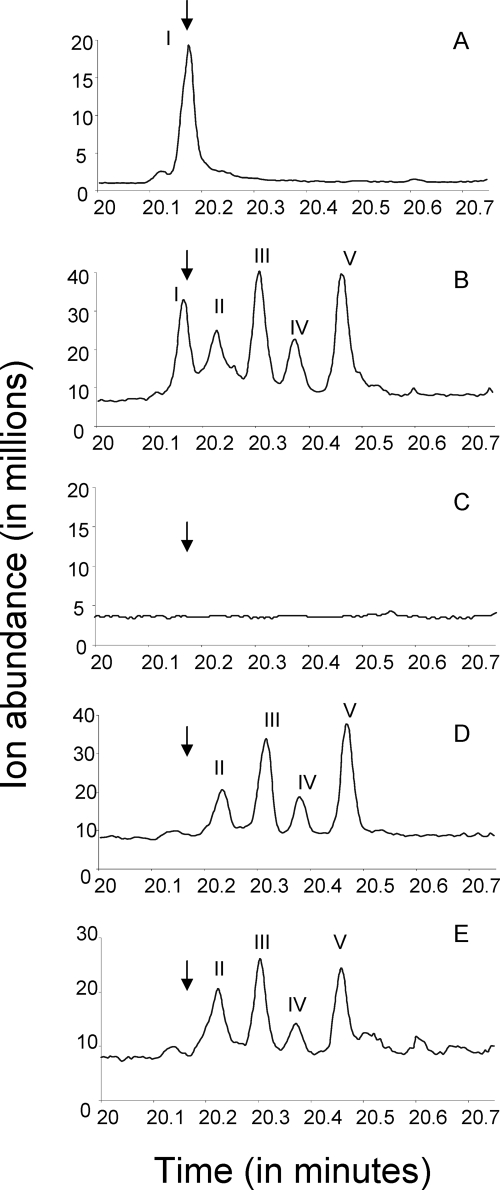
The chromatograms shown in Fig. 5 suggested that the product composition is strongly influenced by the oleA gene. In the same cell type, with cells grown under the same conditions and therefore likely having the same fatty acid precursor pools, the product distribution was completely different when oleA genes from different organisms were present. When oleA genes native to Shewanella and Stenotrophomonas were expressed in the same cell, the products were additive to what was found with either alone (Fig. 5B). Moreover, the Stenotrophomonas oleA gene, in the absence of the native oleBCD genes, was sufficient to make products of fatty acid head-to-head condensation (Fig. 5E). This has implications for the mechanism of olefin biosynthesis and will be discussed in more detail below.
FIG. 5.
Gas chromatograms of extracts from wild-type and mutant S. oneidensis strains with and without the oleA gene from S. maltophilia. Extracts are from the following strains: (A) S. oneidensis MR-1 wild type; (B) S. oneidensis MR-1 wild type with S. maltophilia oleA; (C) S. oneidensis ΔoleA; (D) S. oneidensis ΔoleA with S. maltophilia oleA; (E) S. oneidensis ΔoleABCD with S. maltophilia oleA. Products were identified as described in the text.
When the Shewanella oleA gene was present, the cells made compound I (Fig. 5A and B), which had been previously identified as a polyolefin containing nine double bonds derived from an intermediate in the polyunsaturated fatty acid biosynthetic pathway (59). The presence of the Stenotrophomonas oleA gene led to the formation of new products of fatty acid condensation. All of the later-eluting compounds, labeled II to V in Fig. 5, were ketones. This was apparent from mass spectrometry based on (i) the parent ions, (ii) prominent fragment ions, and (iii) comparison to an authentic long-chain ketone standard. From known fragmentation of alkyl ketones and the observed fragmentation with standard 14-heptacosanone, the major fragments expected were R—CH2—C=O. In the case of 14-heptacosanone, the carbonyl group is directly in the middle and fragmentation at either side on the carbonyl functionality yields a fragment of m/z 211, and this was observed experimentally using GC-MS. Compound II (Fig. 5) showed fragments of m/z 223 and 225 and a parent ion of m/z 420, consistent with a compound containing a carbonyl functionality directly in the middle of a C29 chain with 14 saturated carbon atoms on one side and a C14 chain with one double bond on the other. Compound III showed a fragment with m/z 223 and a parent ion of m/z 418. This mass spectrum is consistent with a compound containing a carbonyl functionality directly in the middle of a C29 chain flanked by two C14 chains, each containing one double bond. Compound IV showed a fragment with m/z 225 and a parent ion of m/z 422. This mass spectrum is consistent with a compound containing a carbonyl functionality directly in the middle of a C29 chain flanked by two saturated C14 chains. Compound V had a very similar mass spectrum as compound II. This suggested that it is a positional isomer of compound II and consists of hydrocarbon chains with one double bond and a saturated chain, respectively, linked together by a carbonyl functionality.
The data above are consistent with a fatty acid condensation between specific saturated and monounsaturated fatty acids. In separate experiments in which the Shewanella oleABCD deletion mutant was complemented with the Shewanella oleA gene, a compound with m/z 434 was obtained. This mass is consistent with a C31 compound containing one ketone functionality and eight carbon-carbon double bonds. The structure was confirmed by chemical modification. After hydrogenation, the compound had a parent ion of m/z 450 with a major fragment ion of m/z 239. This had the expected parent ion and major ion fragment for 16-hentriacontanone. Like the results shown in Fig. 5A to E, this result was consistent with an oleA gene product causing specific condensation of two fatty acids. The Shewanella OleA showed selectivity for polyunsaturated fatty acids, while the Stenotrophomonas OleA showed selectivity for saturated or mono- or di-unsaturated fatty acids.
A mechanism to explain the formation of ketones in the presence of oleA genes alone is proposed below. In total, these data highlight a potentially strong selectivity difference between OleA proteins from Shewanella and Stenotrophomonas. The observations here, showing that different oleA genes exert a strong influence on fatty acid condensation, have implications for the potential use of different ole genes to produce targeted hydrocarbon products commercially. Certain hydrocarbon products may be more desirable for industrial applications. In this context, a knowledge of OleA protein specificity would be critical in efforts to control product structure.
Olefin type in divergent bacteria tracks most closely with OleA sequence.
Very different types of olefin products were observed in wild-type bacteria, containing a range of from one to nine double bonds. Most bacteria in this study made exclusively the nonaene polyolefin previously identified in Shewanella. Data were presented in a previous study that indicated that the C31 nonaene compound was derived from polyunsaturated fatty acid precursors (59). However, polyunsaturated fatty acids account for 10% or less of the total fatty acids produced by Shewanella and other bacterial strains (1, 29, 31, 39). This strongly suggested that Ole enzymes must show selectivity in condensing certain fatty acids and not others. In light of the observations with oleA genes from Shewanella and Stenotrophomonas (Fig. 5), the OleABCD protein sequences were analyzed to see if, among the diverse bacteria analyzed here, Ole protein sequence relatedness correlated with the type of olefin produced by the cell.
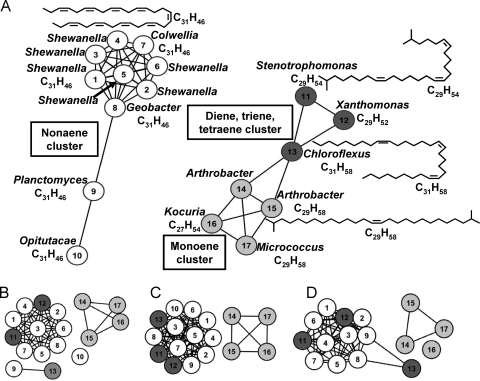
Network clustering software was used to visualize the multidimensional relatedness of different sequences, as this method has been shown to be superior to trees for visualizing protein sequence relatedness (42, 43). The method makes an all-by-all BLASTp library of a sequence set. From these data, a network diagram is created in which the nodes represent protein sequences and the edges represent a BLAST linkage that connects the two proteins. A shorter edge represents a lower e-score (greater relatedness). For example, an e-value cutoff of e−73 was used in Fig. 6. If the e-value of any pairwise comparison is lower (more related) than e−73, then the sequences (Fig. 6, circles/nodes) are connected by a line. Nodes that are not connected, or connected to fewer other nodes, are more divergent sequences. In this way, the network representation allows visualizations of connectivity more fully than protein tree analyses.
FIG. 6.
Network protein sequence clusters for OleA (A), OleB (B), OleC (C), and OleD (D). The nodes represent protein sequences, and the edges represent a BLAST linkage that connects the two proteins with an e-score better than e−73. Other methods are described in the text. The nodes are numbered to identify the organism from which each Ole protein was derived. The organism names and number identifiers for each sequence are listed in Materials and Methods. The nodes are colored to reflect the type of hydrocarbon produced by that organism: white, a C31H46 nonaene product; dark gray, diene, triene, or tetraene product; light gray, monoene product. Additional network diagrams that depict divergence of the clusters can be found in Fig. S2 of the supplemental material.
The network sequence analysis conducted for 17 each OleA, OleB, OleC, and OleD sequences is shown in Fig. 6 (see Fig. S2 in the supplemental material for results of more detailed clustering experiments). Those 17 were selected because all had been experimentally tested and shown to produce olefinic hydrocarbons, and the hydrocarbon products were identified. The top left side of Fig. 6 readily shows that 10 of the OleA proteins cluster together (having all pairwise comparisons with e-values less than e−73) and all produce the single polyolefinic hydrocarbon that is derived from polyunsaturated fatty acids. One explanation for this, which we favor based on the other data presented, is that the OleA proteins in Shewanella, Geobacter, Planctomyces, and Opitutacae specifically condense polyunsaturated fatty acids but do not condense the larger pool of more highly saturated fatty acids found in these classes of bacteria (14, 29, 31, 38).
Figure 6A also shows that the OleA proteins that make moderately saturated head-to-head olefins cluster differently than the OleA found in bacteria that produce the polyolefinic hydrocarbon. For example, Chloroflexus aurantica is known to make a C31 triene hydrocarbon (67), and that was confirmed in this study. A C31 triene would derive from the head-to-head condensation of two monounsaturated fatty acids. Chloroflexus aurantica makes predominantly C16 and C18 saturated fatty acids (66). The most obvious explanation is that the head-to-head biosynthetic pathway shows selectivity for only certain fatty acids within Chloroflexus.
Since the Ole-mediated head-to-head condensation process shows selectivity, it was investigated which Ole protein sequence networks clustered most strongly with the type of head-to-head olefin formed. Figure 6A, B, C, and D represent the clustering networks of OleA, OleB, OleC, and OleD, respectively. For OleA (Fig. 6A), sequence relatedness tracks with the type of olefinic hydrocarbon produced. For OleB, -C, and -D (Fig. 6B, C, and D), the sequences cluster differently and are less reflective of the olefinic hydrocarbon structure. This is perhaps most apparent with OleB (Fig. 6B).
With the cluster represented by the OleABCD sequences from the actinobacterial genera Arthrobacter, Kocuria, and Micrococcus, it was not possible to discern selectivity. The olefinic hydrocarbons produced are methyl branched, and the major fatty acids in Arthrobacter and Microccus are methyl branched (62, 65). The OleA proteins in the actinobacterial branch may be nonselective, or the proteins may have evolved selectivity that mirrors the major fatty acid types produced by the cell.
Potential mechanisms of OleA.
The observation that Shewanella OleA (Fig. 5), Stenotrophomonas OleA (Fig. 5), and other OleA proteins (Fig. 6) confer fatty acid substrate selectivity is consistent with OleA catalyzing the first reaction in head-to-head hydrocarbon formation. An alternative proposal has been advanced in which several β-oxidation steps precede the OleA-catalyzed condensation reaction, and the reaction is coincident with the decarboxylation step (8). That mechanism was supported by two observations, the requirement for E. coli cell extract to support in vitro olefin synthesis and sequence alignments of the Micrococcus luteus OleA with E. coli FabH. The latter enzyme catalyzes a decarboxylative fatty acyl (Claisen) condensation reaction. OleA proteins show the highest percent sequence identity with thiolase superfamily members, like FabH, that catalyze decarboxylative Claisen condensations.
The present study offers an alternative mechanism. As illustrated in Table 2, the thiolase superfamily contains several members that catalyze nondecarboxylative fatty acyl condensation reactions, for example, the biosynthetic thiolase involved in polyhydroxybutyrate (PHB) biosynthesis (17) and 3-hydroxyl-3-methylglutaryl-CoA synthase (HMG-CoA synthase) (56). The latter enzyme and other nondecarboxylative thiolase superfamily enzymes share the same highly conserved residues with those of OleA and FabH (Fig. 1). The decarboxylative and nondecarboxylative thiolase superfamily proteins use these residues in an analogous manner to acylate a cysteine and then attack the bound acyl group with an enzyme-generated carbanion (27). The differences in mechanisms are subtle. Thus, sequence arguments cannot rule in or out decarboxylative versus nondecarboxylative mechanisms for OleA proteins.
Moreover, the mechanism proposed by Beller et al. for OleA is not analogous to that catalyzed by FabH. FabH acts on condensing a fatty acyl group containing an α-carboxy group, and this activation mechanism is not shown in the proposed mechanism (8). Those authors propose a series of steps catalyzed by unidentified enzymes to generate a β-ketoacyl chain that then reacts in condensation with release of coenzyme A and carbon dioxide.
An alternative mechanism would be for OleA to catalyze a nondecarboxylative Claisen condensation directly analogous to the reaction catalyzed by biosynthetic thiolases that function in PHB (17) and steroid (27) synthesis. Both biosynthetic and catabolic thiolases show free reversibility, and dozens of enzymes in the thiolase superfamily are already known to catalyze this general reaction. While the equilibrium constant for the biosynthetic direction is typically unfavorable, subsequent steps can affect the equilibrium, as occurs in PHB and steroid biosyntheses.
The product data are also suggestive that OleA catalyzes the first step in head-to-head hydrocarbon biosynthesis. The product selectivity shown in this study to arise from the oleA gene would be unusual if the OleA protein was in the middle of the biosynthetic pathway, as proposed by Beller et al. (8). Biosynthetic pathways are typically controlled at the first committed step in the pathway (26, 41). The mechanism proposed by Beller et al. requires additional enzymes to generate the 1,3-diketone that is proposed to undergo OleA-catalyzed condensation with a second fatty acyl chain. Those putative genes were searched for in the present study. The genes would need to be present in organisms producing head-to-head hydrocarbons and they might be expected to be contiguous, at least in some organisms, to the other genes encoding enzymes in the same metabolic pathway. However, we could not identify genes contiguous to the oleABCD gene clusters encoding enzymes that act to oxidize an acyl chain and generate a β-ketoacyl chain. This suggests that the OleABCD proteins may be sufficient for ketone and olefin biosynthesis.
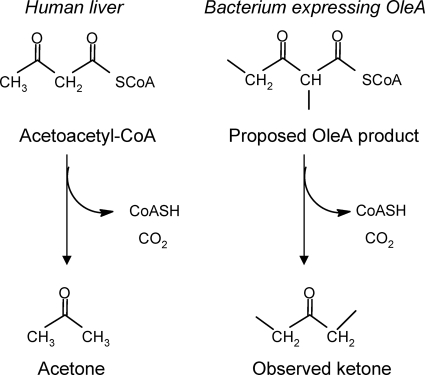
Unlike the previous study (8), a nondecarboxylative, thiolytic type of fatty acyl condensation is proposed here. The nondecarboxylative type of mechanism would explain the observed formation of ketones with OleA in vivo and in vitro (2, 8, 59; Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781; this study) and that the proposed 1,3-dione intermediate (8) has not been observed to date. Ketone formation following a direct OleA-catalyzed nondecarboxylative coupling of fatty acyl chains is chemically plausible and biochemically precedented (Fig. 7). This is reminiscent of the formation of acetone in humans via acetoacetyl-CoA (30). Acetoacetyl-CoA is a β-ketoacyl compound, as is the thiolytic product of the OleA reaction that we propose (Fig. 7). In human liver, excess acetoacetyl-CoA can give rise to acetoacetate, which is known to undergo spontaneous decarboxylation to acetone. The spontaneous decarboxylation of β-keto acids of this type has been known for more than 80 years and is quite facile (47). In a similar manner, we propose that the product(s) of the OleA reaction, if not acted upon by OleBCD, could undergo thioester hydrolysis either spontaneously (21) or enzymatically and then decarboxylation to generate a ketone(s). Note that the acyl-CoA compounds shown in Fig. 7 are directly analogous. They can both arise from thiolytic condensation of either acetyl-CoA or longer-chain acyl-CoAs, respectively. The thioester could undergo enzyme-catalyzed hydrolysis. Alternatively, spontaneous thioester hydrolysis is known to be an important step in the mammalian blood clotting cascade (21). Thioester hydrolysis and facile β-ketoacid decarboxylation offer a plausible explanation as to why monoketones have been observed whenever the oleA gene, by itself, is cloned into a heterologous host (8, 59; Friedman and Rude, WO2008/113041; Friedman and Da Costa, WO2008/147781; this study). Moreover, ketones were observed in this study when exogenous oleA genes were placed into the S. oneidensis MR-1 background.
FIG. 7.
Parallel biological reaction sequences. At left is the known reaction sequence leading to ketones in humans. At right is the proposed reaction sequence leading to ketones in bacteria expressing OleA.
At this time, there are two alternative mechanisms proposed for the initiation of head-to-head hydrocarbon biosynthesis and the specific role of OleA. Further studies will be required to discern between the mechanism proposed previously (8) and the nondecarboxylative Claisen condensation favored here.
Supplementary Material
Acknowledgments
We thank Derek Wilson from the Medical Research Council Laboratory of Molecular Biology at Cambridge for determining the number of proteins in each superfamily within the Superfamily database.
This work was supported by grant LG-B13 from the Initiative for Renewable Energy and the Environment (to L.P.W.) and a Watson Fellowship (to D.J.S.).
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abboud, R., R. Popa, V. Souza-Egipsy, C. S. Giometti, S. Tollaksen, J. J. Mosher, R. H. Findlay, and K. H. Nealson. 2005. Low-temperature growth of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 71:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albro, P. W., and J. C. Dittmer. 1969. Biochemistry of long-chain, nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry 8:394-405. [DOI] [PubMed] [Google Scholar]
- 3.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. III. The metabolic relationship of long-chain fatty acids and hydrocarbons and other aspects of hydrocarbon metabolism in Sarcina lutea. Biochemistry 8:1913-1918. [DOI] [PubMed] [Google Scholar]
- 4.Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain, nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry 8:3317-3324. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Baron, D., E. LaBelle, D. Coursolle, J. A. Gralnick, and D. R. Bond. 2009. Electrochemical measurement of electron transfer kinetics by Shewanella oneidensis MR-1. J. Biol. Chem. 284:28865-28873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauld, J., and J. T. Staley. 1976. Planctomyces maris sp. nov.: a marine isolate of the Planctomyces-Blastocaulis group of budding bacteria. J. Gen. Microbiol. 97:45-55. [Google Scholar]
- 8.Beller, H. R., E. B. Goh, and J. D. Keasling. 2010. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman, J. P., S. A. McCammon, D. S. Nichols, J. H. Skerratt, S. M. Rea, P. D. Nichols, and T. A. McMeekin. 1997. Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5 omega 3) and grow anaerobically by dissimilatory Fe(III) reduction. Int. J. Syst. Bacteriol. 47:1040-1047. [DOI] [PubMed] [Google Scholar]
- 10.Brettar, I., R. Christen, and M. G. Hofle. 2002. Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 52:2211-2217. [DOI] [PubMed] [Google Scholar]
- 11.Burnes, T. A., R. A. Blanchette, and R. L. Farrell. 2000. Bacterial biodegradation of extractives and patterns of bordered pit membrane attack in pine wood. Appl. Environ. Microbiol. 66:5201-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caccavo, F., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:497-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choo, Y. J., K. Lee, J. Song, and J. C. Cho. 2007. Puniceicoccus vermicola gen. nov., sp. nov., a novel marine bacterium, and description of Puniceicoccaceae fam. nov., Puniceicoccales ord. nov., Opitutaceae fam. nov., Opitutales ord. nov. and Opitutae classis nov. in the phylum ‘Verrucomicrobia’. Int. J. Syst. Evol. Microbiol. 57:532-537. [DOI] [PubMed] [Google Scholar]
- 15.Conti, E., N. P. Franks, and P. Brick. 1996. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4:287-298. [DOI] [PubMed] [Google Scholar]
- 16.Daum, M., S. Herrmann, B. Wilkinson, and A. Bechthold. 2009. Genes and enzymes involved in bacterial isoprenoid biosynthesis. Curr. Opin. Chem. Biol. 13:180-188. [DOI] [PubMed] [Google Scholar]
- 17.Davis, J. T., R. N. Moore, B. Imperiali, A. J. Pratt, K. Kobayashi, S. Masamune, A. J. Sinskey, C. T. Walsh, T. Fukui, and K. Tomita. 1987. Biosynthetic thiolase from zoogloea ramigera. I. Preliminary characterization and analysis of proton transfer reaction. J. Biol. Chem. 262:82-89. [PubMed] [Google Scholar]
- 18.Deming, J. W., L. K. Somers, W. L. Straube, D. G. Swartz, and M. T. MacDonnell. 1988. Isolation of an obligatory barophilic bacterium and description of a new genus, Colwellia gen. nov. Syst. Appl. Microbiol. 10:152-160. [Google Scholar]
- 19.Dennis, M., and P. E. Kolattukudy. 1992. A cobalt-porphyrin enzyme converts a fatty aldehyde to a hydrocarbon and CO. Proc. Natl. Acad. Sci. U. S. A. 89:5306-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fredrickson, J. K., M. F. Romine, A. S. Beliaev, J. M. Auchtung, M. E. Driscoll, T. S. Gardner, K. H. Nealson, A. L. Osterman, G. Pinchuk, J. L. Reed, D. A. Rodionov, J. L. Rodrigues, D. A. Saffarini, M. H. Serres, A. M. Spormann, I. B. Zhulin, and J. M. Tiedje. 2008. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6:592-603. [DOI] [PubMed] [Google Scholar]
- 21.Fredslund, F., L. Jenner, L. B. Husted, J. Nyborg, G. R. Andersen, and L. Sottrup-Jensen. 2006. The structure of bovine complement component 3 reveals the basis for thioester function. J. Mol. Biol. 361:115-127. [DOI] [PubMed] [Google Scholar]
- 22.Frias, J. A., J. E. Richman, and L. P. Wackett. 2009. C29 olefinic hydrocarbons biosynthesized by Arthrobacter species. Appl. Environ. Microbiol. 75:1774-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita, K., T. Hagishita, S. Kurita, Y. Kawakura, Y. Kobayashi, A. Matsuyama, and H. Iwahashi. 2006. The cell structural properties of Kocuria rhizophila for aliphatic alcohol exposure. Enzyme Microb. Technol. 39:511-518. [Google Scholar]
- 24.Gough, J., K. Karplus, R. Hughey, and C. Crothia. 2001. Assessment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 3131:903-919. [DOI] [PubMed] [Google Scholar]
- 25.Gulick, A. M., X. Lu, and D. Dunaway-Mariano. 2004. Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states. Biochemistry 43:8670-8679. [DOI] [PubMed] [Google Scholar]
- 26.Gunnarsson, N., A. Eliasson, and J. Nielsen. 2004. Control of fluxes toward antibiotics and the role of primary metabolism in the production of antibiotics. Adv. Biochem. Eng. Biotechnol. 88:137-178. [DOI] [PubMed] [Google Scholar]
- 27.Haapalainen, A. M., G. Meriläinen, and R. K. Wierenga. 2006. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem. Sci. 31:64-71. [DOI] [PubMed] [Google Scholar]
- 28.Heath, R. J., and C. O. Rock. 2002. The Claisen condensation in biology. Nat. Prod. Rep. 19:581-596. [DOI] [PubMed] [Google Scholar]
- 29.Hedrick, D. B., A. D. Peacock, D. R. Lovley, T. L. Woodard, K. P. Nevin, P. E. Long, and D. C. White. 2009. Polar lipid fatty acids, LPS-hydroxy fatty acids, and respiratory quinones of three Geobacter strains, and variation with electron acceptor. J. Ind. Microbiol. Biotechnol. 36:205-209. [DOI] [PubMed] [Google Scholar]
- 30.Hird, F. J. R., and R. H. Symons. 1962. The mechanism of ketone-body formation from butyrate in rat liver. Biochem. J. 84:212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanova, E. P., S. Flavier, and R. Christen. 2004. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1773-1788. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, C., S. Y. Kim, and D. Y. Suh. 2008. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 49:691-701. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen, B. R., and H. H. Huss. 1989. Growth and activity of Shewanella putrefaciens isolated from spoiling fish. Int. J. Food Microbiol. 9:51. [DOI] [PubMed] [Google Scholar]
- 34.Jörnvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzàlez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita, S., K. Kadota, T. Inoue, H. Sawada, and H. Taguchi. 1988. Purification and properties of three kinds of a-hydroxysteroid dehydrogenases from Brevibacterium fuscum DC33. J. Ferment. Technol. 66:145-152. [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Kovacs, G., J. Burghardt, S. Pradella, P. Schumann, E. Stackebrandt, and K. Marialigeti. 1999. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int. J. Syst. Bacteriol. 49:167-173. [DOI] [PubMed] [Google Scholar]
- 38.Kulichevskaya, I. S., A. O. Ivanova, S. E. Belova, C. T. Baulina, P. L. Bodelier, W. I. Rijpstra, J. S. Sinninghe Damsté, G. A. Zavarzin, and S. N. Dedysh. 2007. Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from sphagnum-dominated boreal wetlands. Int. J. Syst. Evol. Microbiol. 57:2680-2687. [DOI] [PubMed] [Google Scholar]
- 39.Lee, W. H., K. W. Cho, S. Y. Park, K. S. Shin, D. S. Lee, S. K. Hwang, S. J. Seo, J. M. Kim, S. Y. Ghim, B. H. Song, S. H. Lee, and J. G. Kim. 2008. Identification of psychrophile Shewanella sp. KMG427 as an eicosapentaenoic acid producer. J. Microbiol. Biotechnol. 18:1869-1873. [PubMed] [Google Scholar]
- 40.Lee, S. K., H. Chou, T. S. Ham, T. S. Lee, and J. D. Keasling. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556-563. [DOI] [PubMed] [Google Scholar]
- 41.Lehninger, A. L. 1978. Biochemistry: the molecular basis of cell structure and function. Worth Publishers, New York, NY.
- 42.Meng, E. C., E. F. Pettersen, G. S. Couch, C. C. Huang, and T. E. Ferrin. 2006. Tools for integrated sequence-structure analysis with UCSF Chimer. BMC Bioinformatics 7:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris, J. H., C. C. Huang, P. C. Babbitt, and T. E. Ferrin. 2007. StructureViz: linking Cytoscape and UCSF Chimera. Bioinformatics 23:2345-2347. [DOI] [PubMed] [Google Scholar]
- 44.Nardini, M., and B. W. Dijkstra. 1999. α/β-Hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 45.Nevin, K. P., D. E. Holmes, T. L. Woodard, E. S. Hinlein, D. W. Ostendorf, and D. R. Lovley. 2005. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int. J. Syst. Evol. Microbiol. 55:1667-1674. [DOI] [PubMed] [Google Scholar]
- 46.Park, M. O., M. Tanabe, K. Hirata, and K. Miyamoto. 2001. Isolation and characterization of a bacterium that produces hydrocarbons extracellularly which are equivalent to light oil. Appl. Microbiol. Biotechnol. 56:448-452. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen, K. J. 1929. The ketonic decomposition of beta-keto carboxylic acids. J. Am. Chem. Soc. 51:2098-2107. [Google Scholar]
- 48.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 49.Qian, Z., C. J. Fields, Y. Yu, and S. Lutz. 2007. Recent progress in engineering alpha/beta hydrolase-fold family members. Biotechnol. J. 2:192-200. [DOI] [PubMed] [Google Scholar]
- 50.Rollefson, J. B., C. E. Levar, and D. R. Bond. 2009. Identification of genes involved in biofilm formation and respiration via mini-himar transposon mutagenesis of Geobacter sulfurreducens. J. Bacteriol. 191:4207-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito, N., and S. Sugiyama. 1964. Production of dihydroxyacetone and other ketoses by Brevibacterium fuscum. Agric. Biol. Chem. 28:48-55. [Google Scholar]
- 52.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 19:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayers, E. W., T. Barrett, D. A. Benson, E. Bolton, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. Dicuccio, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, Z. Lu, T. L. Madden, T. Madej, D. R. Maglott, A. Marchler-Bauer, V. Miller, I. Mizrachi, J. Ostell, A. Panchenko, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, D. Slotta, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, Y. Wang, W. J. Wilbur, E. Yaschenko, and J. Ye. 2010. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 38:D5-D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt, T. D. Accessed 23 November 2009. Genome Project: Opitutaceae bacterium TAV2. DOE Joint Genome Institute, Walnut Creek, CA. http://genome.jgi-psf.org/opiba/opiba.info.html.
- 55.Seffernick, J. L., S. K. Samanta, T. M. Louie, L. P. Wackett, and M. Subramanian. 2009. Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J. Biotechnol. 143:7-26. [DOI] [PubMed] [Google Scholar]
- 56.Steussy, C. N., A. D. Robison, A. M. Tetrick, J. T. Knight, V. W. Rodwell, C. V. Stauffacher, and A. L. Sutherlin. 2006. A structural limitation on enzyme activity: the case of HMG-CoA synthase. Biochemistry 45:14407-14414. [DOI] [PubMed] [Google Scholar]
- 57.Stevenson, B., S. Elchhorst, J. Wertz, T. Schmidt, and J. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suen, Y., G. U. Holzer, J. S. Hubbard, and T. G. Tornabene. 1988. Biosynthesis of acyclic methyl branched poly-unsaturated hydrocarbons in Pseudomonas maltophilia. J. Ind. Microbiol. 2:337-348. [Google Scholar]
- 59.Sukovich, D. J., J. L. Seffernick, J. E. Richman, K. A. Hunt, J. A. Gralnick, J. A. Gralnick, and L. P. Wackett. 2010. Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1. Appl. Environ. Microbiol. 76:3842-3849. [DOI] [PMC free article] [PubMed]
- 60.Takarada, H., M. Sekine, H. Kosugi, Y. Matsuo, T. Fujisawa, S. Omata, E. Kishi, A. Shimizu, N. Tsukatani, S. Tanikawa, N. Fujita, and S. Harayama. 2008.Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 190:4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thoden, J. B., A. M. Gulick, and H. M. Holden. 1997. Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 36:10685-10695. [DOI] [PubMed] [Google Scholar]
- 62.Tornabene, T. G., E. Gelphi, and J. Oro. 1967. Identification of fatty acids and aliphatic hydrocarbons in Sarcina lutea by gas chromatography and combined gas chromatography-mass spectrometry. J. Bacteriol. 94:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tornabene, T. G., S. J. Morrison, and W. E. Kloos. 1970. Aliphatic hydrocarbon contents of various members of the family Micrococcaceae. Lipids 5:929-934. [DOI] [PubMed] [Google Scholar]
- 64.Tornabene, T. G., and S. P. Markey. 1971. Characterization of branched monounsaturated hydrocarbons of Sarcina lutea and Sarcina flava. Lipids 3:190-195. [DOI] [PubMed] [Google Scholar]
- 65.Unell, M., N. Kabelitz, J. K. Jansson, and H. J. Heipieper. 2007. Adaptation of the psychrotroph Arthrobacter chlorophenolicus A6 to growth temperature and the presence of phenols by changes in the anteiso/iso ratio of branched fatty acids. FEMS Microbiol. Lett. 266:138-143. [DOI] [PubMed] [Google Scholar]
- 66.van der Meer, M. T., S. Schouten, B. E. van Dongen, W. I. Rijpstra, G. Fuchs, J. S. Damste, J. W. de Leeuw, and D. M. Ward. 2001. Biosynthetic controls on the 13C contents of organic components in the photoautotrophic bacterium Chloroflexus aurantiacus. J. Biol. Chem. 276:10971-10976. [PubMed] [Google Scholar]
- 67.van der Meer, M. T., S. Schouten, D. M. Ward, J. A. Geenevasen, and J. S. Sinninghe Damste. 1999. All-cis hentriaconta-9,15,22-triene in microbial mats formed by the phototrophic prokaryote Chloroflexus. Org. Geochem. 30:1585-1587. [DOI] [PubMed] [Google Scholar]
- 68.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 69.Venkateswaran, K., M. E. Dollhopf, R. Aller, E. Stackebrandt, and K. H. Nealson. 1998. Shewanella amazonensis sp. nov., a novel metal-reducing facultative anaerobe from Amazonian shelf muds. Int. J. Syst. Bacteriol. 48:965-972. [DOI] [PubMed] [Google Scholar]
- 70.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 71.Verschueren, K. H., F. Seljée, H. J. Rozeboom, K. H. Kalk, and B. W. Dijkstra. 1993. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363:693-698. [DOI] [PubMed] [Google Scholar]
- 72.Wackett, L. P., J. A. Frias, J. L. Seffernick, D. J. Sukovich, and S. M. Cameron. 2007. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl. Environ. Microbiol. 73:7192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ward-Rainey, N., F. A. Rainey, and E. Stackebrandt. 1997. The presence of a dnaK (HSP70) multigene family in members of the orders Planctomycetales and Verrucomicrobiales. J. Bacteriol. 179:6360-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu, R., J. Cao, X. Lu, A. S. Reger, A. M. Gulick, and D. Dunaway-Mariano. 2008. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry 47:8026-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yumoto, I., K. Kawasaki, H. Iwata, H. Matasuyama, and H. Okuyama. 1998. Assignment of Vibrio sp. strain ABE-1 to Colwellia maris sp. nov., a new psychrophilic bacterium. Int. J. Syst. Bacteriol. 48:1357-1362. [DOI] [PubMed] [Google Scholar]
- 76.Ziemke, F., M. G. Höfle, J. Lalucat, and R. Rossellö-Mora. 1998. Reclassification of Shewanella putrefaciens Owen's genomic group II as Shewanella baltica sp. nov. Int. J. Syst. Bacteriol. 48:179-186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.