Abstract
Monomethylamine can be used by nonmethylotrophs as a sole nitrogen source but not as a carbon source; however, little is known about the genes and enzymes involved. The γ-glutamylmethylamide/N-methylglutamate pathway for monomethylamine utilization by methylotrophs has recently been resolved. We have identified genes encoding key enzymes of this pathway in nonmethylotrophs (e.g., Agrobacterium tumefaciens) and demonstrated that this pathway is also involved in the utilization of monomethylamine as a nitrogen source by nonmethylotrophs.
Monomethylamine (MMA) (CH3NH2) is ubiquitous in the environment and is released during the degradation of many nitrogen-containing compounds (1, 2, 6). Bacteria can use MMA as a sole carbon (C) and/or as a sole nitrogen (N) source (1). For methylotrophic bacteria that use MMA as a source of both C and N, different pathways have been elucidated, including the MMA dehydrogenase pathway and the MMA oxidase pathway and additional pathways involving methylated glutamate, i.e., γ-glutamylmethylamide (GMA) and N-methylglutamate (NMG) (1). The genes involved in the GMA/NMG-mediated MMA utilization pathway, including GMA synthetase (gmas), “NMG synthase” (mgsABC), and NMG dehydrogenase (mgdABCD), were recently identified in the methylotroph Methyloversatilis universalis (Fig. 1 A) (11). Although MMA can serve as a sole N source but not as a C source for many nonmethylotrophs (3, 5), the mechanisms involved are unclear. A search for GMA/NMG gene clusters in microbial genome sequence databases revealed that similar clusters are present in many nonmethylotrophs, including Agrobacterium tumefaciens C58, Rhizobium leguminosarum bv. viciae 3841, Mesorhizobium loti MAFF303099, and Ruegeria pomeroyi DSS-3. We tested the hypothesis that the GMA/NMG-mediated MMA utilization pathway is also involved in the metabolism of MMA as the sole N source by nonmethylotrophs.
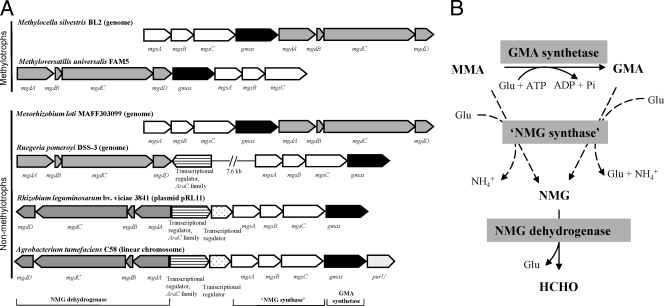
FIG. 1.
(A) Gene organization of NMG dehydrogenase (mgdABCD), GMA synthetase (gmas), and “NMG synthase” (mgsABC) in representative methylotrophs and nonmethylotrophs. purU, formyltetrahydrofolate deformylase. (B) Proposed pathway of GMA- and NMG-mediated MMA metabolism in bacteria. The substrate specificity of “NMG synthase” is not well established (shown in dashed lines), and it is proposed that both MMA and GMA can be used as a substrate for this enzyme. MMA, monomethylamine; GMA, γ-glutamylmethylamide; NMG, N-methylglutamate; Glu, glutamate.
Identification of the gene clusters involved in metabolism of MMA via the GMA/NMG pathway in nonmethylotrophs.
An eight-gene cluster which encodes three key enzymes involved in the GMA/NMG pathway for MMA oxidation in methylotrophs was identified (reference 11 and Y. Chen, unpublished data). The overall reaction for MMA oxidation in methylotrophs containing this pathway is the conversion of MMA to formaldehyde and ammonium (CH3NH2 → HCHO + NH4+), which are used as a C and an energy source and as an N source, respectively (Fig. 1B). Searches for similar gene clusters encoding the GMA/NMG pathway in other microorganisms were carried out using the integrated microbial genomes (IMG) tool (http://img.jgi.doe.gov). Similar gene clusters are present in many methylotrophs but are also present in nonmethylotrophs such as Agrobacterium tumefaciens (Fig. 1A). Therefore, the enzymes encoded by this gene cluster might be used by nonmethylotrophs to metabolize MMA as a sole N source. If so, the ammonium released from MMA oxidation would serve as the N source. The formaldehyde produced is toxic and would need to be detoxified. Indeed, a formyltetrahydrofolate deformylase (purU) gene is found immediately downstream of this gene cluster (Fig. 1A) in A. tumefaciens and an folD gene (encoding methylenetetrahydrofolate dehydrogenase/cyclohydrolase) is also present in the genome. Therefore, it is likely that formaldehyde released from this pathway in nonmethylotrophs is converted to formate via 5,10-methylenetetrahydrofolate, 5,10-methenyltetrahydrofolate, and 10-formyltetrahydrofolate (11). Formate may further be oxidized to carbon dioxide, and genes encoding formate dehydrogenase are present in the genome of A. tumefaciens. The gmas homologue (encoding GMA synthetase) found in these nonmethylotrophs is annotated as a putative glutamine synthetase; however, multiple sequence alignments of glutamine synthetases and characterized GMA synthetases, as shown in Fig. S1 and S2 in the supplemental material, indicated that they are likely to be GMA synthetases. They cluster together with characterized GMA synthetases from methylotrophs, and they lack key residues for ammonium binding but contain conserved domains for ATP and glutamate binding, as do glutamine synthetases. Furthermore, the tyrosine residue (Tyr397), which is commonly found in type I glutamine synthetases and which is subjected to adenylylation, is missing. It is interesting that in two cases (R. leguminosarum bv. viciae 3841 and Burkholderia phymatum STM815), this gene cluster is encoded on a plasmid, indicating a potential for horizontal gene transfer of this metabolic pathway in bacteria. Since the genetics of MMA metabolism in nonmethylotrophs is not established, we therefore hypothesized that the genes in this cluster are involved in the utilization of MMA as a sole N source by nonmethylotrophs such as A. tumefaciens.
MMA can be used as a sole nitrogen source for Agrobacterium tumefaciens.
We first tested whether MMA could support the growth of these bacteria as a sole N source, since this has not been previously documented. The growth media used were an N-free medium from Kanvinde and Sastry (10) for A. tumefaciens C58, a minimal medium from Brown and Dilworth (4) for R. leguminosarum bv. viciae 3841 and M. loti MAFF303099, and a modified ammonium-free marine mineral salts medium for Ruegeria pomeroyi DSS-3 (8). In all cases, glucose (5 g liter−1) was used as a C source and ammonium or MMA was added to make up a final concentration of 2 mM as a sole N source. A control for each growth experiment was set up without added N compounds to avoid the presence of contaminating N in chemicals used for making media. All growth experiments were carried out at 30°C with shaking at 150 rpm. Values for optical density at 540 nm (OD540) were recorded.
All four strains tested were able to use MMA as a sole N source (Fig. 2 A; see also Fig. S3 in the supplemental material). Control experiments set up with no added N compounds did not yield any growth for the bacteria tested. In addition, the possibility of MMA being the sole C source was ruled out, since MMA alone did not support the growth of any of these four bacteria (data not shown). Furthermore, neither A. tumefaciens nor M. loti was able to use methanol (10 mM) or formate (10 mM) as a sole carbon source (data not shown).
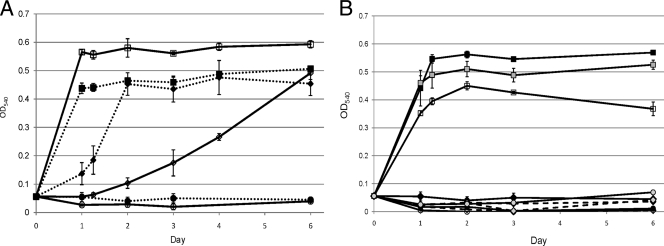
FIG. 2.
(A) Representative growth curves of wild-type A. tumefaciens (dotted lines) and a gmas::gm mutant (solid lines) grown with ammonium (squares) and MMA (diamonds) as the sole nitrogen source or with no added nitrogen (controls are indicated as circles). The means and standard deviations of the results of three replicate experiments are shown. Note that the y axis is not presented as a logarithmic scale. (B) Representative growth curves of mgdC::gm, mgsC::gm, and mgsC_gmas::gm mutants of A. tumefaciens grown with ammonium (filled, empty, and shaded squares, respectively) and MMA (filled, empty, and shaded diamonds, respectively) as the sole nitrogen source or with no added nitrogen (controls are indicated as filled, empty, and shaded circles, respectively). The means and standard deviations of the results of three replicate experiments are shown. Note that the y axis is not presented as a logarithmic scale.
The intracellular pool of amino acids was then analyzed to investigate whether GMA or NMG is involved in MMA metabolism in these nonmethylotrophs. Analyses using ion-exchange chromatography followed by ninhydrin staining were carried out at Alta Biosciences (Birmingham, United Kingdom). GMA was detected in A. tumefaciens and M. loti when they were grown on MMA but not when they were grown on ammonium as the sole N source (see Fig. S4 in the supplemental material), whereas NMG was detected in Ruegeria pomeroyi when it was grown on MMA, indicating the importance of GMA/NMG in MMA utilization in these nonmethylotrophs. The reason for the accumulation of GMA or NMG is not well understood but is probably related to the growth state, as previously shown in a study using Pseudomonas sp. MA, where GMA accumulated under low-oxygen conditions, whereas under high-oxygen conditions, MMA was primarily converted to NMG (9).
Mutation of mgdC and mgsC, but not mutation of gmas, abolishes MMA metabolism by Agrobacterium tumefaciens.
To determine whether this gene cluster is essential for MMA metabolism in A. tumefaciens, we constructed three mutants, i.e., mgdC::gm, mgsC::gm, and gmas::gm, by marker-exchange mutagenesis using a pK18mobsacB suicide vector (13). In each case, the upstream and downstream regions (∼500 bp) of the target were PCR amplified (using the primers listed in Table S1 in the supplemental material) and cloned into pK18mobsacB. A gentamicin gene cassette from p34S-Gm (7) was then inserted in between these regions. The resulting plasmids were then electroporated into A. tumefaciens. Single homologous recombination mutants were selected on LB (lysogeny broth) plates with kanamycin (50 μg ml−1). Colonies from these plates were then grown for 24 h in LB liquid medium and plated out at different dilutions (10−2 to 10−4) onto LB plates containing 10% (wt/vol) sucrose. The resulting kanamycin-sensitive colonies were then screened for double homologous mutation by PCR using primers targeting areas outside the upstream and downstream regions. Mutations of the genes were confirmed by diagnostic PCR (see Table S1 in the supplemental material) and subsequently by DNA sequencing. Mutants were checked using the medium of Kanvinde and Sastry (10) for their abilities to use MMA as a sole N source.
Mutants of mgdC and mgsC lost their capacity to use MMA as the sole N source (Fig. 2B), although they grew normally using ammonium as the sole N source. The gmas mutant, in contrast, could still grow on MMA as the N source; however, the growth rate of this mutant on MMA (0.024 h−1) was significantly reduced compared to the wild-type strain growth rate (0.043 h−1) (Fig. 2A). A fourth mutant (mgsC_gmas::gm) was therefore constructed to mutate gmas and one of the “NMG synthase”-encoding genes simultaneously. This mutant could no longer grow on MMA as the sole N source (Fig. 2B).
Our results indicated that nonmethylotrophs such as A. tumefaciens can use MMA as the sole N source via the GMA/NMG-mediated pathway, as has been demonstrated in some methylotrophs. The mgdABCD and mgsABC genes seem to be essential in A. tumefaciens, and mutation of representatives of these gene clusters completely abolished its capacity to use MMA. The presence of GMA synthetase in this bacterium is important, but not vital, since mutation of gmas caused much slower growth of this bacterium on MMA. It is likely that “NMG synthase” in A. tumefaciens (encoded by mgsABC) can use both GMA and MMA as a substrate, as shown in Fig. 1B. In fact, it has been shown that the purified “NMG synthase” is specific for glutamate but not for MMA and that a number of amines can substitute for MMA (12). The GMA/NMG-mediated pathway for MMA uptake as the N source is likely to be widespread in nature due to the potential for horizontal gene transfer of plasmids encoding this pathway. Previous studies have shown that diverse genera of bacteria can use MMA as the sole N source (3, 5), and the widespread occurrence of this physiological trait suggests that MMA might be an important N source in natural habitats. The ability to use organic nitrogen compounds such as MMA would confer some advantage to these microorganisms, considering that many environments are often limited by the availability of inorganic nitrogen. However, it remains unclear whether the GMA/NMG-mediated pathway is the only way for nonmethylotrophs to acquire N from MMA, and this issue certainly warrants further investigation.
Supplementary Material
Acknowledgments
We thank M. Challen, S. Kumar, J. Gronlund, S. Klüsener, F. Narberhaus, J. Green, P. Young, P. Poole, and H. Schäfer for providing various plasmids and bacterial strains and for advice on constructing mutants. A. Crombie and R. Boden are acknowledged for helpful discussions.
We acknowledge a vacation studentship from the Society for General Microbiology (United Kingdom) to K.L.M.
Footnotes
Published ahead of print on 16 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Barrett, E. L., and H. S. Kwan. 1985. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 39:131-149. [DOI] [PubMed] [Google Scholar]
- 3.Bicknell, B., and J. D. Owens. 1980. Utilization of methyl amines as nitrogen sources by non-methylotrophs. J. Gen. Microbiol. 117:89-96. [Google Scholar]
- 4.Brown, C. M., and M. J. Dilworth. 1975. Ammonia assimilation by Rhizobium cultures and bacteroids. J. Gen. Microbiol. 86:39-48. [DOI] [PubMed] [Google Scholar]
- 5.Budd, J. A., and C. P. Spencer. 1968. The utilisation of alkylated amines by marine bacteria. Mar. Biol. 2:92-101. [Google Scholar]
- 6.Burg, M. B., and J. D. Ferraris. 2008. Intracellular organic osmolytes: function and regulation. J. Biol. Chem. 283:7309-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin, K. D., R. K. Varner, P. M. Crill, and R. S. Oremland. 2001. Consumption of tropospheric levels of methyl bromide by C1 compound-utilizing bacteria and comparison to saturation kinetics. Appl. Environ. Microbiol. 67:5437-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, J. G., and E. Bellion. 1991. In vivo 13C and 15N NMR studies of methylamine metabolism in Pseudomonas species MA. J. Biol. Chem. 266:11705-11713. [PubMed] [Google Scholar]
- 10.Kanvinde, L., and G. R. K. Sastry. 1990. Agrobacterium tumefaciens is a diazotrophic bacterium. Appl. Environ. Microbiol. 56:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latypova, E., S. Yang, Y.-S. Wang, T. Wang, T. A. Chavkin, M. Hackett, H. Schäfer, and M. G. Kalyuzhnaya. 2010. Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 75:426-439. [DOI] [PubMed] [Google Scholar]
- 12.Pollock, R. J., and L. B. Hersh. 1971. N-Methylglutamate synthetase: purification and properties of the enzyme. J. Biol. Chem. 246:4737-4743. [PubMed] [Google Scholar]
- 13.Schäfer, A., A. Tauch, W. Jager, J. Kalinowshi, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.