Abstract
Insect endosymbiont genomes reflect massive gene loss. Two Blattabacterium genomes display colinearity and similar gene contents, despite high orthologous gene divergence, reflecting over 140 million years of independent evolution in separate cockroach lineages. We speculate that distant homologs may replace the functions of some eliminated genes through broadened substrate specificity.
Obligate symbionts of insects exhibit extreme patterns of genome evolution and include the smallest known bacterial genomes (10, 11, 14). Two recently published sequences of Blattabacterium, the obligate symbiont of cockroaches (7, 16), present the opportunity to analyze genome evolution in an additional symbiont lineage with extreme genome reduction.
General features.
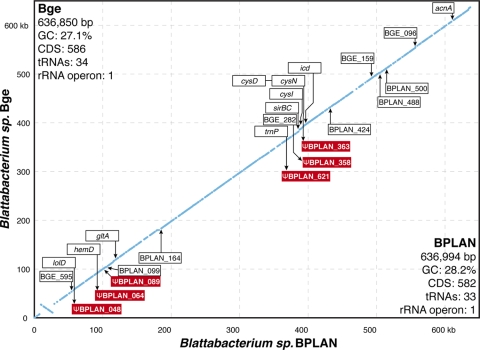
Genomes of the Blattabacterium species (Bacteroidetes) of Periplaneta americana (the BPLAN bacterium) and Blattella germanica (the BGE bacterium) are similarly sized at 637 kb and are nearly identical in numbers of protein-encoding sequences (BPLAN, 582; BGE, 586) and RNAs (BPLAN, 37; BGE, 40). Despite differential gene loss, >96% of the genes in each genome are shared (BPLAN, 609/619; BGE, 607/627). Neither the 3.4-kb plasmid harbored by BPLAN nor the genes encoded on the plasmid were reported for the BGE sequenced strain (7). However, we de-tected a plasmid bearing at least one of these genes (nrdF) in BGE residing within B. germanica specimens obtained from North Carolina and maintained in Arizona, indicating either that the plasmid was overlooked or that the strains differ in its presence (for details, see the supplemental material). Nevertheless, the main chromosomes show considerable conservation of genome colinearity with the exception of a single ∼19-kb inversion (Fig. 1). This overall structural stability resembles that observed in three other obligate endosymbiont clades for which multiple genomes are available, namely, Buchnera aphidicola, “Candidatus Blochmannia” sp., and “Candidatus Sulcia muelleri” (3, 9, 17, 18).
FIG. 1.
Gene order comparison of shared Blattabacterium orthologs. BGE gene coordinates are plotted on the BPLAN genome. Locus names for orthologs present in one genome and absent in the other are indicated in open boxes, and the arrows point to their approximate locations. Locus names for Blattabacterium sp. BGE have been shortened from “BLBBGE” to “BGE.” Arrows that cross the slope indicate the transition from an intact open reading frame (ORF) to a pseudogenized ORF. Annotated pseudogenes are shown as red boxes.
Blattabacterium forms a distinct clade within the phylum Bacteroidetes, which is relatively distant from free-living members. “Ca. Sulcia muelleri,” the obligate nutritional endosymbiont of cicadas, spittlebugs, and leafhoppers, is their closest relative (Fig. 2). Orthologous pairs of intact Blattabacterium protein coding sequences exhibit elevated rates of nonsynonymous substitutions per nonsynonymous sites (dN) compared to free-living relatives (Table 1). This genome-wide trend has affected all loci, although genes encoding proteins with poorly defined functions have slightly higher dN estimates than do others (see Table S1 in the supplemental material). Elevated dN is also observed in “Ca. Sulcia muelleri” and the sequenced gammaproteobacterial endosymbionts (Table 1). Obligate nutritional endosymbionts encounter dramatic reductions in effective population sizes and selective regimens, which contribute to both gene loss and the elevated accumulation of nonsynonymous substitutions (2, 3, 9, 13, 18).
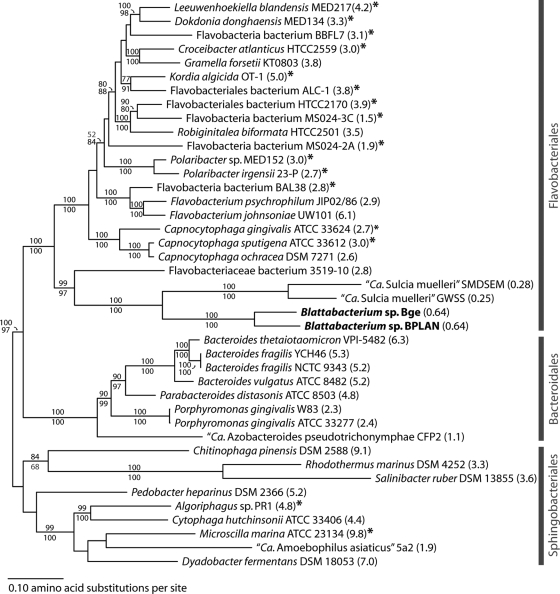
FIG. 2.
Core gene phylogeny of the Bacteroidetes. Maximum likelihood phylogeny of the Bacteroidetes was estimated using a concatenation of 38 conserved proteins identified in 41 complete and in-progress genomes (see Table S1 in the supplemental material). Consistent with previous phylogenies, Blattabacterium (in bold) is sister to “Ca. Sulcia muelleri,” endosymbiont of cicadas, leafhoppers, and spittlebugs, and falls just inside the Flavobacteriales. Both insect endosymbionts exhibit long branches, indicative of genome-wide elevation in the rate of fixation of slightly deleterious mutations (including nonsynonymous mutations), which is common among obligate endosymbionts. Genome sizes in megabase pairs are indicated in parentheses. Size estimates of unfinished genomes marked with asterisks are based on total base pairs in finished contigs. Support values from RAxML (above) and PhyML (below) were generated from 100 bootstrap replicates; nodes with support of <70% by both methods are not shown.
TABLE 1.
Comparison of nonsynonymous divergences among pairs of obligate insect endosymbionts and related free-living bacteria
| Genome paira | Phylum | Host | Genome wide |
Conserved core |
Estimated time since divergence (MY)e | ||
|---|---|---|---|---|---|---|---|
| n | Mean dN ± SD | n | Mean dN ± SD | ||||
| Blattabacterium (BPLAN-BGE) | Bacteroidetes | Blattaria (cockroaches) | 570 | 0.130 ± 0.073 | 125 | 0.113 ± 0.062 | 150-300 |
| “Ca. Sulcia” (DSEM-GWSS) | Bacteroidetes | Auchenorrhyncha (cicadas, leafhoppers, spittlebugs, etc.) | 202 | 0.141 ± 0.080 | 126b | 0.131 ± 0.075 | 200 |
| Flavobacterium johnsoniae-Flavobacterium psychrophilum | Bacteroidetes | Free living | 1,268 | 0.148 ± 0.075 | 119b,c | 0.097 ± 0.096 | NA |
| Bacteroides thetaiotaomicron-Bacteroides fragilis | Bacteroidetes | Free living | 1,962 | 0.115 ± 0.073 | 123b,d | 0.047 ± 0.043 | NA |
| “Ca. Blochmannia” (BPEN-BFL) | Proteobacteria | Camponotini (ants) | 577 | 0.225 ± 0.106 | 126b | 0.186 ± 0.088 | 16-20 |
| Buchnera (APS-SG) | Proteobacteria | Aphidoidea (aphids) | 458 | 0.149 ± 0.085 | 125 | 0.112 ± 0.066 | 50-70 |
| Buchnera (APS-BP) | Proteobacteria | Aphidoidea (aphids) | 458 | 0.315 ± 0.173 | 125 | 0.238 ± 0.130 | 150-200 |
| E. coli-Salmonella enterica | Proteobacteria | Free living | 2,940 | 0.067 ± 0.058 | 125 | 0.020 ± 0.023 | 100-150 |
Accession numbers for genomes used in this analysis can be found in the supplemental material.
TrpD is not fused with TrpG.
ThrABC, LeuABCD, and LysA are absent in F. psychrophilum, and TrpC is a split gene in F. johnsoniae and F. psychrophilum.
DapD, SucA, and AceF are absent in B. thetaiotamoicron and B. fragilis.
Dates are based on insect fossils except for that for E. coli-S. enterica. MY, million years; NA, not available.
These two Blattabacterium genomes are estimated to have diverged about 140 million years ago based on the insect fossil record (19) and the observation of codiversification of Blattabacterium and their hosts (1, 6, 8). Although the sequence similarity is rather low (∼85% nucleotide sequence identity), suggesting an ancient divergence, the conservation of gene content and colinearity is quite high. This pattern is echoed in all of the other obligate symbionts of insects for which multiple genomes are available (3, 9, 12, 15, 17, 18); in each case, despite many millions of years of divergence, almost no chromosome rearrangements or gene acquisitions are observed. The absence of recA has been suggested as a mechanism contributing to genome stability in obligate endosymbionts (17); however, recA is present in both Blattabacterium species.
Given the large set of shared genes in these anciently diverged endosymbionts, much of the genome reduction likely occurred early during the association of primitive cockroaches and the ancestor of Blattabacterium. Evidence of ongoing gene loss is present in the form of pseudogenes. Of the six BPLAN pseudogenes (Fig. 1), five are intact in BGE and are involved in a variety of pathways, including translation (trnP/proL) and amino acid (cysND), cofactor (hemD), and lipid (lolD) biosynthesis. Five additional intact open reading frames unique to BPLAN were also observed.
Gene loss shapes endosymbiont metabolisms.
Both genomes encode biosynthetic pathways capable of producing nearly all of the amino acids, except asparagine and glutamine, from waste nitrogen (e.g., urea and ammonia). Interestingly, BGE possesses most of the genes involved in sulfur assimilation (cysNDHIJ) and they are either completely absent (cysI) or riddled with deletions (cysND) in BPLAN. This loss of function has occurred through gradual gene decay and has not disrupted genome colinearity (Fig. 1). A similar pattern of lineage-specific loss of the cys genes was observed in Buchnera aphidicola strains in host aphids with different diets (17). B. aphidicola of Acythrosiphon pisum has intact cysIHGDN genes while in B. aphidicola of Schizaphis graminum these genes show multiple deletions and appear nonfunctional, potentially reflecting relaxed selection on sulfur assimilation genes due to the availability of organic sulfur in the S. graminum diet (17). Both B. germanica and P. americana are generalist detritivores, and it is not clear that host diet has shaped the loss/retention of these genes in Blattabacterium strains.
In addition to three conserved hypothetical proteins and a precorrin 2 dehydrogenase (sirBC), BGE encodes three tricarboxylic acid (TCA) cycle genes (gltA, acnA, and icd) that are completely missing from the BPLAN genome. However, BPLAN does encode 3-isopropylmalate dehydratase (leuCD) and 3-isopropylmalate dehydrogenase (leuB), which share functional domains with aconitate hydratase (acnA) and isocitrate dehydrogenase (icd), respectively. Phylogenetic reconstructions of aconitase family enzymes (e.g., LeuCD and AcnA) from Bacteria, Archaea, and Eukarya indicate that these enzymes are derived from a common ancestor, which underwent subsequent ancient duplications (5). Site-directed mutagenesis at amino acid positions 113, 115, and 116 in Icd of Escherichia coli resulted in broader substrate specificity and increased preference for isopropylmalate, the native substrate of LeuB, over isocitrate (4). Given the overlapping functional motifs and relatively few amino acid substitutions that could impact substrate specificity, it is possible that the losses of acnA and icd were largely neutral because of complementation by leuCD and leuB, respectively. Additionally, most insect obligate endosymbionts with completely sequenced genomes lack intact acnA or icd genes but do encode intact leuCD and leuB (Table 2), highlighting the potential dispensability of acnA and icd genes in insect endosymbionts and suggesting that endosymbiont enzymes may evolve broader substrate specificity (20). Because most of these symbionts provision their hosts with essential amino acids, retention of leuBCD genes is likely favored for their role in leucine biosynthesis. Remaining to be determined is the biological significance for acnA and icd retention by BGE.
TABLE 2.
Loss of acn and icd and retention of leuCD and leuB is widespread among nutritional insect endosymbiontsa
| Obligate insect endosymbiont | Mb | GenBank accession no. | Presence of gene: |
|||
|---|---|---|---|---|---|---|
| acn | leuCD | icd | leuB | |||
| Baumannia cicadellinicola HC | 0.69 | NC_007984 | ||||
| Blattabacterium sp. BGE | 0.64 | NC_013454 | + | + | + | + |
| Blattabacterium sp. BPLAN | 0.64 | NC_013418 | + | + | ||
| Buchnera aphidicola APS | 0.64 | NC_002528 | + | + | ||
| Buchnera aphidicola BP | 0.62 | NC_004545 | + | + | ||
| Buchnera aphidicola CC | 0.42 | NC_008513 | + | + | ||
| Buchnera aphidicola SG | 0.64 | NC_004061 | + | + | ||
| “Candidatus Azobacteroides pseudotrichonymphae” | 1.11 | NC_011565 | + | + | ||
| “Candidatus Blochmannia floridanus” | 0.71 | NC_005061 | + | + | ||
| “Candidatus Blochmannia pennsylvanicus” | 0.79 | NC_007292 | + | + | ||
| “Candidatus Carsonella ruddii PV” | 0.16 | NC_008512 | + | + | ||
| “Candidatus Hodgkinia cicadicola DSEM” | 0.14 | NC_012960 | ||||
| “Candidatus Sulcia muelleri” GWSS | 0.25 | NC_010118 | + | + | ||
| “Candidatus Sulcia muelleri” SMDSEM | 0.28 | NC_013123 | + | + | ||
| Wigglesworthia glossinidia | 0.70 | NC_004344 | ||||
Endosymbionts with completely sequenced genomes were queried with orthologous proteins from closely related organisms using the blastp program.
The comparison of the Blattabacterium genomes provides an additional case of extreme stability of gene order and ongoing gene loss in reduced bacterial genomes. Further investigation of the substrate specificity of endosymbiont enzymes, such as the aconitases, could give insight into the kind of changes that allow these bacteria to function with so few genes or with particular genes missing from pathways that are mostly intact.
Supplementary Material
Acknowledgments
Z.L.S. was supported by the University of Arizona Center for Insect Science through National Institutes of Health training grant 2 K12 GM000708. National Science Foundation Award 0626716, to N.A.M., supported this work. P.H.D. received funding from National Science Foundation Awards 0313737 and 0723472 to N.A.M.
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Clark, J. W., S. Hossain, C. A. Burnside, and S. Kambhampati. 2001. Coevolution between a cockroach and its bacterial endosymbiont: a biogeographical perspective. Proc. R. Soc. Lond. B Biol. Sci. 268:393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark, M. A., N. A. Moran, and P. Baumann. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16:1586-1598. [DOI] [PubMed] [Google Scholar]
- 3.Degnan, P. H., A. B. Lazarus, and J. J. Wernegreen. 2005. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 15:1023-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle, S. A., S. Y. Fung, and D. E. Koshland, Jr. 2000. Redesigning the substrate specificity of an enzyme: isocitrate dehydrogenase. Biochemistry 39:14348-14355. [DOI] [PubMed] [Google Scholar]
- 5.Irvin, S. D., and J. K. Bhattacharjee. 1998. A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. J. Mol. Evol. 46:401-408. [DOI] [PubMed] [Google Scholar]
- 6.Lo, N., C. Bandi, H. Watanabe, C. Nalepa, and T. Beninati. 2003. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol. Biol. Evol. 20:907-913. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Sanchez, M. J., A. Neef, J. Pereto, R. Patino-Navarrete, M. Pignatelli, A. Latorre, and A. Moya. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 5:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maekawa, K., Y. C. Park, and N. Lo. 2005. Phylogeny of endosymbiont bacteria harbored by the woodroach Cryptocercus spp. (Cryptocercidae: Blattaria): molecular clock evidence for a late Cretaceous-early Tertiary split of Asian and American lineages. Mol. Phylogenet. Evol. 36:728-733. [DOI] [PubMed] [Google Scholar]
- 9.McCutcheon, J. P., B. R. McDonald, and N. A. Moran. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 106:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCutcheon, J. P., B. R. McDonald, and N. A. Moran. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCutcheon, J. P., and N. A. Moran. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392-19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran, N. A. 2003. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6:512-518. [DOI] [PubMed] [Google Scholar]
- 13.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakabachi, A., A. Yamashita, H. Toh, H. Ishikawa, H. E. Dunbar, N. A. Moran, and M. Hattori. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Brocal, V., R. Gil, S. Ramos, A. Lamelas, M. Postigo, J. M. Michelena, F. J. Silva, A. Moya, and A. Latorre. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312-313. [DOI] [PubMed] [Google Scholar]
- 16.Sabree, Z. L., S. Kambhampati, and N. A. Moran. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. U. S. A. 106:19521-19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 18.van Ham, R. C., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernandez, L. Jimenez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Moran, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. U. S. A. 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrsansky, P., V. N. Vishniakova, and A. P. Rasnitsyn. 2002. Order Blattida Latreille, 1810. The cockroaches, p. 263-270. In A. P. Rasnitsyn and D. L. J. Quicke (ed.), History of insects. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 20.Zientz, E., T. Dandekar, and R. Gross. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68:745-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.