Abstract
Clavibacter michiganensis subsp. michiganensis is a Gram-positive bacterium that causes wilting and cankers, leading to severe economic losses in commercial tomato production worldwide. The disease is transmitted from infected seeds to seedlings and mechanically from plant to plant during seedling production, grafting, pruning, and harvesting. Because of the lack of tools for genetic manipulation, very little is known regarding the mechanisms of seed and seedling infection and movement of C. michiganensis subsp. michiganensis in grafted plants, two focal points for application of bacterial canker control measures in tomato. To facilitate studies on the C. michiganensis subsp. michiganensis movement in tomato seed and grafted plants, we isolated a bioluminescent C. michiganensis subsp. michiganensis strain using the modified Tn1409 containing a promoterless lux reporter. A total of 19 bioluminescent C. michiganensis subsp. michiganensis mutants were obtained. All mutants tested induced a hypersensitive response in Mirabilis jalapa and caused wilting of tomato plants. Real-time colonization studies of germinating seeds using a virulent, stable, constitutively bioluminescent strain, BL-Cmm17, showed that C. michiganensis subsp. michiganensis aggregated on hypocotyls and cotyledons at an early stage of germination. In grafted seedlings in which either the rootstock or scion was exposed to BL-Cmm17 via a contaminated grafting knife, bacteria were translocated in both directions from the graft union at higher inoculum doses. These results emphasize the use of bioluminescent C. michiganensis subsp. michiganensis to help better elucidate the C. michiganensis subsp. michiganensis-tomato plant interactions. Further, we demonstrated the broader applicability of this tool by successful transformation of C. michiganensis subsp. nebraskensis with Tn1409::lux. Thus, our approach would be highly useful to understand the pathogenesis of diseases caused by other subspecies of the agriculturally important C. michiganensis.
Clavibacter michiganensis subsp. michiganensis is a Gram-positive, aerobic bacterium that belongs to a group of plant-pathogenic actinomycetes (37). Infections by C. michiganensis subsp. michiganensis cause bacterial canker and wilt in tomato, which is considered one of the most destructive and economically significant diseases of this crop. Severe epidemics can cause up to 80% yield loss, mainly due to wilting and death of plants and lesions on fruit. Bacterial canker was first discovered in Michigan greenhouses in 1909 and has now been reported to occur in most tomato production areas around the world (11, 40).
Plant wounds facilitate but are not required for infection by C. michiganensis subsp. michiganensis, which invades the xylem vessels and causes vascular disease with high titers (109 bacteria/g of plant tissue) (2, 29), impairing water transport and leading to plant wilting, canker stem lesions, and death (17, 23). Alternatively, asymptomatic infections can be induced by C. michiganensis subsp. michiganensis during late stages of plant development, resulting in the production of contaminated seeds, a major source of outbreaks of C. michiganensis subsp. michiganensis infections in tomato production (13, 34). Traditional bacterial-disease management measures, such as applications of antibiotics and copper bactericides, have not been successful against this disease, and canker-resistant tomato cultivars are not available. As a result, C. michiganensis subsp. michiganensis has been included under international quarantine regulation (10, 11). Consequently, seed testing and maintaining pathogen-free seeds and transplants is currently the most appropriate approach to minimize the spread of disease (23). However, even a low C. michiganensis subsp. michiganensis transmission rate (0.01%) from seed to seedling can cause a disease epidemic under favorable conditions (5). Due to overcrowding of seedlings during transplant production, the pathogen can easily spread through splashing of irrigation water and leaf contact. Despite its apparent significance in C. michiganensis subsp. michiganensis epidemiology, the mechanism of seed-to-seedling transmission of C. michiganensis subsp. michiganensis is not well understood.
Another critical point for disease spread is the grafting process, which is now a common practice for the majority of plants used in production greenhouses. Desirable tomato cultivars (scions) are grafted onto rootstocks that provide greater vigor, longevity, or, in some cases, disease resistance (26). Grafting requires cutting both rootstock and scion, providing a quick way for C. michiganensis subsp. michiganensis to spread from plant to plant. However, grafting is a relatively recent innovation in tomato production, and little is known about how grafting affects the dynamics of C. michiganensis subsp. michiganensis infection. Developing adequate control measures for C. michiganensis subsp. michiganensis is complicated by the complexity of genetic manipulation of Gram-positive bacteria, which impairs analysis and characterization of pathogenesis mechanisms (23). Consequently, there is a need to develop molecular techniques that would allow a better understanding of C. michiganensis subsp. michiganensis infections.
One method of interest is using engineered bioluminescent bacteria to monitor plant-pathogen interactions in real time. By exploiting natural light-emitting reactions that are encoded by the luxCDABE genes, bioluminescent bacteria have been used to assess gene expression and to monitor the internalization and distribution of bacteria in hosts (3, 6, 7, 8, 9, 12, 15, 24, 31, 35, 36). In particular, bioluminescent phytopathogenic Xanthomonas campestris pathovars and Pseudomonas spp. have been used to track bacterial movement and distribution in host plants (7, 8, 15, 31, 36), as well as to assess host susceptibility quantitatively (15). Likewise, the lux genes have also been transferred to beneficial bacteria, such as Rhizobium leguminosarum and Pseudomonas spp. to visualize colonization patterns in rhizospheres (3, 9).
The genes that carry the function of light emission are luxAB, which express luciferase enzymes that catalyze the bioluminescent reaction, while luxCDE encode the enzymes required for biosynthesis of a fatty aldehyde substrate necessary for the reaction (28, 39). Bioluminescence involves an intracellular oxidation of the reduced form of flavin mononucleotide and the fatty aldehyde by luciferase in the presence of molecular oxygen; therefore, bacterial bioluminescence also requires oxygen, a source of energy (38). Cells that express the lux operon spontaneously emit photons that can be captured by a sensitive charge-coupled-device (CCD) camera, enabling imaging and visualization of bacterial cells (22). Luciferase activity depends on the metabolic integrity of the cell, while the number of photons emitted correlates with the biomass of living bacteria (12, 31). Furthermore, since the half-life of luciferase binding to its substrate is several seconds (28), captured light events reflect processes in real time and are not artifacts of accumulated signals. Consequently, live imaging of bioluminescence provides a sensitive means of visualizing bacterial colonization and invasion of hosts and allows real-time representation and examination of pathogen-plant interactions (24, 36).
Very little information is available about the mechanisms of C. michiganensis subsp. michiganensis pathogenesis and its colonization of seeds and subsequent transmission to seedlings. This is largely attributable to a lack of tools and difficulties in genetically manipulating this Gram-positive bacterium (30). However, recent development of an insertion sequence element IS1409 (Tn1409)-based efficient transposon mutagenesis system for C. michiganensis subsp. michiganensis has increased our knowledge of the pathogenesis of tomato canker (16, 25). To better understand the dynamics of seed-to-seedling transmission of C. michiganensis subsp. michiganensis, as well as movement of C. michiganensis subsp. michiganensis in grafted plants, we constructed a bioluminescent C. michiganensis subsp. michiganensis strain using the Tn1409 transposon mutagenesis system. Our results demonstrated the utility of using a bioluminescent C. michiganensis subsp. michiganensis strain as a novel approach to elucidate the interaction of plants with this economically important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. C. michiganensis subsp. michiganensis strains A300 and C290 were isolated from infected plants in Ohio and belong to BOX-PCR fingerprint types A and C, respectively (27). C. michiganensis subsp. nebraskensis strain LexIIC was obtained from David L. Coplin, Ohio State University. Depending on the assay, Clavibacter strains were grown in yeast-dextrose-calcium carbonate (YDC) medium (43), nutrient broth-yeast extract broth (NBY) (41), tryptone broth with yeast (TBY), or SB medium (25). Escherichia coli strains (DH5α and ER2925) were grown on LB at 37°C. The growth medium was supplemented with the antibiotics chloramphenicol (20 μg/ml for E. coli and 10 μg/ml for C. michiganensis subsp. michiganensis and C. michiganensis subsp. nebraskensis), kanamycin (50 μg/ml for C. michiganensis subsp. michiganensis), and ampicillin (150 μg/ml for E. coli) where necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| C. michiganensis subsp. michiganensis | ||
| C290 | Wild-type virulent strain isolated in Ohio | Laboratory collection |
| A300 | Wild-type virulent strain isolated in Ohio | Laboratory collection |
| C. michiganensis subsp. nebraskensis LexIIC | Wild-type virulent strain isolated in Nebraska | D. L. Coplin |
| E. coli | ||
| DH5α | Used in cloning experiments | Invitrogen |
| ER2925 | dam and dcm methylation-negative strain | New England Biotechnology |
| Plasmids | ||
| pXen5 | Source for promoterless lux operon optimized for expression in Gram-positive bacteria; Ery Kan; 18.3 kb | Xenogen; 14 |
| pXen13 | Source for Gram-negative lux operon from P. luminescens; Amp; 8.7 kb | Xenogen |
| pMod3 | Contains EZ::TN transposon; 2.8 kb | Epicentre |
| pKGT452Cβ | Tn1409 Cβ; tnpA cmx Amp Cm; 6.4 kb | 25 |
| pUC4K | Source of kanamycin resistance marker; Kan Amp | Amersham |
| pUWGR4 | pMod3 containing EZ::TN transposon and lux operon from pXen13; Kan; 10 kb | 33 |
| pXX1 | pMod3 containing promoterless lux operon from pXen5; Kan; 9.3 kb | This study |
| pXX2 | pKGT452Cβ containing promoterless lux operon from pXen5; Amp Cm; 12.2 kb | This study |
| pXX3 | pKGT452Cβ containing promoterless lux operon from pXen13; Amp Cm; 12.3 kb | This study |
Kan, kanamycin resistance; Amp, ampicillin resistance; Cm, chloramphenicol resistance; Ery, erythromycin resistance.
Construction of plasmids pXX1, pXX2, and pXX3.
Both pXen5 and pXen13 (Xenogen Corporation, Alameda, CA) carry the promoterless lux operon originally isolated from Photorhabdus luminescens. The lux operon (luxABCDE) in pXen5 had been modified by adding a Gram-positive bacterial ribosome binding site (AGGAGG) upstream of each lux gene for optimal expression in Gram-positive bacteria (14). To construct pXX1, the lux operon and kanamycin resistance gene from pXen5 were amplified by a long-range, high-fidelity PCR using Herculase Polymerase (Stratagene, La Jolla, CA) with LuxpXen5F/LuxpXen5R primers. The oligonucleotides were designed using Vector NTI software (Invitrogen) and commercially synthesized by Integrated DNA Technologies (Skokie, IL). All the oligonucleotides used in this study are listed in Table 2. Appropriate restriction sites were included in the primers to facilitate cloning. The amplified PCR product was digested and ligated to a similarly digested Tn5 transposon construction vector, pMod 3 (Epicentre, Madison, WI). pXX1 carries an EZ::TN<lux-kan> cassette, which is flanked by the Tn5 transposon mosaic ends.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea | Purpose |
|---|---|---|
| LuxpXen5F | 5′ ATAAAAGAATTCGACTCCTGTGAAATGATC | Amplification of the lux operon and kanamycin gene from pXen5 |
| LuxpXen5R | 5′ AAAAAACTCGACCGGATGTACTTCAGAAAAGA | |
| pKGT1F | 5′ AAAAAAGCGGCCGCATCACGCCGCTAGAGCTT | Inverse PCR amplification of pKGT452Cβ to clone lux operon from pXen5 |
| pKGT1R | 5′ AAAAAACTCGAGTCAGAGAAGGTGAGGGCCTC | |
| KanF | 5′ GAAGCGTTTGATAGTTAAGT | For confirmation of EZ::TN<lux-kan> integration into chromosome |
| KanR | 5′ GGTACTAAAACAATTCATCC | |
| LuxpXen5F2 | 5′ ATAAAAGCGGCCGCGAAACAGCTATGACCATGAT | Amplification of the lux operon from pXen5 |
| LuxpXen5R2 | 5′ AAAAAACTCGAGTTATTATTTCCCTCCTCGAC | |
| pKGT2F | 5′ AAAAAACTCGAGATCACGCCGCTAGAGCTTGG | Inverse PCR amplification of pKGT452Cβ to clone lux operon from pXen13 |
| pKGT2R | 5′ AAAAAGCGGCCGCTCAGACAAGGTGAGGGCCTC | |
| CmxF | 5′ AGAGTACTGCCGACGCCGA | Confirmation of Tn1409-lux integration into chromosome |
| CmxR | 5′ ACTGTCGATCCTGCTCTCCG | |
| LuxCF | 5′ GTTGATGAATATCCACCTCT | Confirmation of Tn1409-lux integration into chromosome |
| LuxCR | 5′ GGACATACATTCGTGACTTA | |
| Xu3F | 5′ GGATTTGTCGGGGTGTTTCG | Mapping of insertion site of Tn1409-lux in C. michiganensis subsp. michiganensis genome |
| Xu3R | 5′ CGGCCCCACAGAAGCAATTA |
Restriction sites added to the 5′ end of each primer are underlined.
pXX2 was constructed by amplifying the lux operon from pXen-5 with primers LuxpXen5F2/LuxpXen5R2. Inverse PCR was performed on the Tn1409 transposon vector pKGT452Cβ (25) with pKGT1F/pKGT1R primers that were designed to amplify all but the region of lux insertion. Appropriate restriction sites (NotI and XhoI) were added to the PCR primers to facilitate cloning. The inverse-PCR product was digested and ligated to the similarly digested lux operon from pXen5, generating pXX2 (Cmx resistance gene::luxABCDE::Tn1409).
To construct pXX3, the lux operon (luxCDABE) was obtained by digesting pXen13 with NotI and XhoI and ligated to the similarly digested inverse-PCR product of the transposon vector pKGT452Cβ amplified using the primers pKGT2F/pKGT2R. pXX3 carries Cmx resistance gene::luxCDABE::Tn1409. The recombinant plasmids pXX1, pXX2, and pXX3 were transformed to E. coli DH5α and subsequently propagated in the dam- and dcm-deficient E. coli strain ER2925, since the use of unmethylated DNA improves transformation efficiency in C. michiganensis subsp. michiganensis (25).
Mutagenesis and isolation of bioluminescent C. michiganensis subsp. michiganensis.
Bioluminescent C. michiganensis subsp. michiganensis strains were generated either by electroporation of the modified EZ::TN transposomes from pXX1 or pUWGR4 (33) or by directly electroporating the suicide vectors containing the modified transposon Tn1409 (pXX2 and pXX3). Before electroporation, the EZ::TN transposome was prepared as described by the manufacturer (Epicentre), pUWGR4 and pXX1 were digested with PvuII, and the resulting approximately 7-kb linear DNA fragments containing the EZ::TN-lux, and Kanr genes were gel purified using a Qiagen gel purification kit (Qiagen, Valencia, CA). Two microliters of each purified DNA (100 ng/μl) was mixed separately with 4 μl of EZ::TN transposase (Epicentre) and 2 μl of 100% glycerol in an 8-μl reaction mixture. Following incubation for 30 min at room temperature, the transposome complexes were stored at −20°C until further use.
Electrocompetent C. michiganensis subsp. michiganensis aliquots were prepared as described previously (25). Briefly, the bacteria were grown to an optical density at 600 nm (OD600) of 0.5 to 0.7 in TBY broth at 25°C. The cells were then pretreated with 2.5% glycine for 2 h and harvested by centrifugation at 9,000 × g for 10 min, followed by washing with sterile ice-cold distilled water three times and with 10% glycerol twice. The cell pellet was then resuspended in 15% glycerol to 1/250 of the original volume. Transformation was achieved by mixing 100 μl of electrocompetent C. michiganensis subsp. michiganensis cells with 2 μl of the EZ::TN transposome complex or 1 μl of the suicide plasmid pXX2 or pXX3 (1 μg each), and electroporation was performed in a 0.2-cm cuvette using a Gene Pulser Xcell electroporator (Bio-Rad, Hercules, CA) with the following settings: voltage, 2.5 kV; capacitance, 25 μF; resistance, 600 Ω. Immediately after electroporation, the cells were mixed with 0.4 ml of SB medium, transferred into a 10-ml sterile disposable tube, and incubated with shaking (140 rpm) for 3 h at 28°C. Finally, the cells were spread on SB agar plates containing kanamycin or chloramphenicol and incubated for 4 to 7 days at 28°C. The Kanr or Cmxr colonies were recovered and streaked onto NBY plates containing appropriate antibiotics and screened for bioluminescence using a CCD camera in an in vivo imaging system (IVIS) (Xenogen). The bioluminescent colonies were purified and stored at −80°C for further analysis.
For electroporation of C. michiganensis subsp. nebraskensis, electrocompetent cells were prepared and transformed with 1 μg of pXX2 as described above. Transformants were selected on SB agar containing chloramphenicol and screened for bioluminescence using a CCD camera after 4 days of incubation at 28°C.
PCR analysis of bioluminescent C. michiganensis subsp. michiganensis strains.
To confirm the integration of the lux operon into the C. michiganensis subsp. michiganensis chromosome, PCR analysis was performed on all recovered colonies. After electroporation with pXX1, the colonies were tested using the KanF/KanR primers, which target part of the kanamycin resistance gene. Additionally, colonies recovered after electroporation with pXX2 or pXX3 were tested for the presence of the chloramphenicol resistance (cmx) and luxC genes using the primers CmxF/CmxR and LuxCF/LuxCR, respectively. PCR was performed on genomic DNA obtained from bioluminescent strains using a Taq PCR Master Mix kit (Qiagen).
Analysis of bioluminescence.
To quantify the amount of bioluminescence emitted by the transformed C. michiganensis subsp. michiganensis, isolates were grown in NBY broth containing the appropriate antibiotics with aeration at 28°C for 48 h. Three replicates of 100 μl of each culture (1 × 109 CFU) were then transferred to a black 96-well plate, and the bioluminescence was quantified using an IVIS. The photon output (radiance) of each well (photons per second) was normalized for background luminescence by subtracting the value of a control well containing NBY broth only. The OD600 was measured, and the radiance was divided by the OD to normalize for the culture cell density. Highly bioluminescent clones were selected for further studies.
HR and pathogenicity assays.
The bioluminescent C. michiganensis subsp. michiganensis strains were tested in the four o'clock plant (Mirabilis jalapa) for hypersensitive response (HR) and by tomato seedling inoculation for pathogenicity. Briefly, for the HR test, four o'clock plants were grown in autoclaved soil mix (50% silt loam-50% muck was mixed and sieved; then, 30.3 liters of peat was added to every 151.5 liters of soil mixture and supplemented with 350 g of lime and 150 g of Peters 4-45-15 fertilizer). Bacterial suspensions (108 CFU/ml) were prepared in sterile water and injected into fully expanded leaves of four o'clock plants using syringes without needles, ensuring that the bacteria would infiltrate into the intercellular spaces. Typically, the wild-type virulent C. michiganensis subsp. michiganensis induces HR-characteristic necrosis in the infiltrated areas within 36 to 48 h (18). For each strain, four injections were applied on one leaf, and the inoculated plants were placed for up to 48 h in a greenhouse with a 14-h light/10-h dark photoperiod. The wild-type C. michiganensis subsp. michiganensis strain C290 and sterile water were used as positive and negative controls, respectively.
For the pathogenicity assay, the inoculation of tomato seedlings with bioluminescent C. michiganensis subsp. michiganensis strains was performed as previously described (32). The cotyledons of tomato seedlings were clipped using dissecting scissors that had been dipped in a C. michiganensis subsp. michiganensis suspension (108 CFU/ml). Tomato seedlings (n = 3) were inoculated with each strain and monitored for symptoms for 3 weeks. Furthermore, virulence was assessed by determining the number of plants that exhibited wilting symptoms within 3 weeks after inoculation. To quantify the bacterial populations in infected tomato plants, stem samples from three plants were taken from 3 cm above the cotyledons. Samples were ground in 3 ml phosphate buffer (pH 7.4; 10 mM) and centrifuged at 400 × g for 3 min. The supernatant was serially diluted and plated onto NBY agar plates that were supplemented with chloramphenicol. For samples inoculated with C290, serial dilutions were spread onto D2ANX (a semiselective medium for C. michiganensis subsp. michiganensis) (20). Colony counts were recorded on the fifth day. The number of bacteria present in tomato tissue (CFU/g) was calculated as follows: number of colonies × dilution factor × 3 ml/weight of sample tissue. The HR and pathogenicity assays were conducted twice.
Assessing in vitro growth, constitutive expression of bioluminescence, and stability of Tn1409-lux insertion in a virulent bioluminescent C. michiganensis subsp. michiganensis strain.
To compare the in vitro growth of BL-Cmm17 to that of its parent strain, C290, the growth rates of BL-Cmm17 and C290 were compared in a rich medium, NBY broth, as well as in C. michiganensis subsp. michiganensis minimal medium (1). BL-Cmm17 growing on NBY plates for 48 h at 28°C was inoculated into a 200-ml flask containing 50 ml NBY broth or C. michiganensis subsp. michiganensis minimal medium and incubated at 28°C with shaking at 160 rpm for 50 h (three replications per strain). Bacterial growth was monitored by measuring the OD600 from 1 ml culture every 4 h. To assess constitutive lux expression, at each time point, the light intensity from 100 μl of culture was detected using an IVIS. Relative light units were determined by log transformation of the average radiance.
The stability of the transposon insertion was verified by growing BL-Cmm17 without antibiotic selection in 5 ml NBY broth at 28°C for 72 h, for a total of five rounds (with 5 μl for each reinoculation). After the fifth round, the culture was diluted in NBY broth, plated on NBY agar plates without antibiotics, and incubated at 28°C for 72 h. To assess the stability of insertion, 50 colonies from each of the three replicates were randomly selected and plated onto NBY agar medium with chloramphenicol. The growth on the antibiotic plate and the bioluminescence of each colony was assessed to determine the stability of insertion.
Mapping the Tn1409 insertion site.
Genomic DNA was extracted from BL-Cmm17 using a MasterPure Gram-positive DNA Purification Kit (Epicentre), and the insertion site was determined by bidirectional sequencing using the primers Xu3F/Xu3R. Sequencing was performed using dye terminators at a DNA-sequencing core facility (the Plant-Microbe Genomic Facility, Ohio State University). Sequences were compared to the C. michiganensis subsp. michiganensis genome to determine the site of insertion.
Seed inoculation with bioluminescent C. michiganensis subsp. michiganensis.
Healthy tomato seeds (cv. OH9242) were inoculated by soaking them in BL-Cmm17 suspension (108 CFU/ml) in a 100-ml sterilized beaker. The beaker was placed in a Nucerite Desiccator (Nalge Sybron Corporation, Rochester, NY), and a vacuum was applied for 5 min using an Air Cadet pump (Barnant, Barrington, IL) with a maximum of 18 lb/in2 pressure. Seeds inoculated with sterilized water were used as controls. After inoculation, the seeds were air dried and placed on water agar (1%) in a square petri dish (10 by 10 cm, with a 13-mm grid) and incubated at 25°C in the dark. Bioluminescent C. michiganensis subsp. michiganensis colonization of germinating seeds was monitored daily using the IVIS. Six seeds were sampled each day for 5 days to estimate the bacterial population on each seed. On the first and second days of germination, seeds were ground individually in 0.5 ml potassium phosphate buffer, and serial dilutions were plated on YDC medium supplemented with chloramphenicol. From the third to fifth days, seedlings were carefully dissected, and the seed coat, cotyledons, hypocotyls, and radicals were collected aseptically with sterile forceps for bacterial isolation. The experiment was conducted twice.
Grafted plant inoculation with bioluminescent C. michiganensis subsp. michiganensis.
A BL-Cmm17 suspension adjusted to 102, 104, 106, or 108 CFU/ml was tested on tomato “Maxifort” (Johnny's Selected Seeds, ME) rootstock and Celebrity F1 (Johnny's Selected Seeds, ME) scion seedlings for bacterial movement during grafting. Rootstock and scion seeds were sanitized by hot-water treatment (a 10-min presoak at 38°C, followed by treatment for 25 min at 50°C) prior to sowing. The grafting tool, a Menno Knife with a presterilized number 11 blade (Royal Brinkman, Netherlands), was exposed to different concentrations of bacterial suspensions by cutting a 1-cm-diameter cotton roll that had been submerged in the bacterial suspension. For rootstock inoculation, 21-day-old rootstock seedlings were decapitated with one horizontal cut 5 mm below the cotyledons with a blade exposed to BL-Cmm17 suspensions, and 18-day-old scion seedlings were derooted by a single cut with a sterile blade exposed to sterile water. For scion inoculation, rootstock seedlings were decapitated as described above with a sterile-water-exposed sterile blade, and scion seedlings were derooted by a single cut with a blade exposed to a BL-Cmm17 suspension. Plants cut with a blade exposed to sterile water served as controls. At the time of grafting, the cut ends of scions and rootstocks were tissue printed onto D2ANX medium for the day zero evaluation. Three replicate samples with three plants per replicate were evaluated at day zero and each subsequent sampling time. Following grafting, exterior portions of the cut stems (grafted area) were surface sterilized with 70% ethanol. The grafted plants were placed in a growth chamber at 22°C in darkness under 80 to 90% relative humidity for 7 days (three plants/replication; three replications). The movement of the bioluminescent bacteria up and down the stem from the graft union was determined by tissue imprinting the cut surfaces of both scion and rootstock on D2ANX medium at a specific distance from the graft union (0, 3, 6, 9, 12, and 15 mm), and the tissue-imprinted plates were imaged using the IVIS after 7 days of incubation. Further, to enumerate the bacteria, 0.1-cm pieces of stem tissues of both scion and rootstock were ground and cultured on YDC agar medium after serial dilution, and the bacterial numbers (CFU/g) were recorded on the fifth day. The experiment was conducted twice.
RESULTS
Isolation of bioluminescent C. michiganensis subsp. michiganensis strains.
In order to generate a bioluminescent C. michiganensis subsp. michiganensis strain, we desired stable chromosomal insertion of the lux operon, as well as constitutive expression of bioluminescence. Unlike the expression of lux from a plasmid, chromosomal integration leads to stable expression of lux even in the absence of antibiotic selection, which is necessary for real-time studies of plants, where there is no antibiotic selection. Because of the lack of tools for genetic manipulation of C. michiganensis subsp. michiganensis, we tried several approaches to integrate lux genes into the C. michiganensis subsp. michiganensis chromosome. Since the lux operon in pXen13 was originally derived from the Gram-negative bacterium P. luminescens, it might not be expressed efficiently in C. michiganensis subsp. michiganensis, a high-G+C-content Gram-positive bacterium. Consequently, the ribosome binding sites of each lux gene in the luxABCDE operon in pXen5 were optimized for expression in Gram-positive bacteria. Initially, we tried the Tn5-based transposon mutagenesis approach that has been successfully used in Corynebacterium matruchotii, an actinomycete related to C. michiganensis subsp. michiganensis (42). After electroporation of the Tn5::lux transposome complexes prepared from pUWGR4 or pXX1 into C. michiganensis subsp. michiganensis strains A300 and C290, a total of 84 kanamycin-resistant colonies were obtained; however, none were bioluminescent. PCR analysis targeting the kanamycin gene indicated that they lacked EZ::TN insertion. Further, C. michiganensis subsp. michiganensis electroporated without an EZ::TN<lux-Kan> cassette (negative control) also resulted in kanamycin-resistant colonies, which suggested that the C. michiganensis subsp. michiganensis strains used in this study can inherently develop kanamycin resistance or that Tn5 might not work in C. michiganensis subsp. michiganensis.
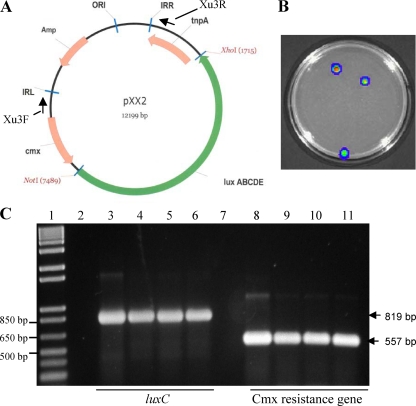
Recently, Kirchner et al. (25) developed an efficient mutagenesis system for C. michiganensis subsp. michiganensis using Tn1409. Therefore, we modified the Tn1409-containing vector pKGT452 Cβ to contain either a lux operon optimal for expression in Gram-positive bacteria (pXX2) (Fig. 1) or a lux operon from a Gram-negative bacterium (pXX3). After electroporation of pXX3 into C. michiganensis subsp. michiganensis strains, three colonies were recovered, which were found to harbor the chloramphenicol resistance gene analyzed by PCR (data not shown), indicating the presence of the transposon in their genomes. However, none of these strains were bioluminescent, which is likely due to the lack of expression of Gram-negative lux in Gram-positive bacteria, as previously suggested (14). When transposon vector pXX2 was electroporated into C. michiganensis subsp. michiganensis, bioluminescent colonies were obtained successfully. The transformation efficiency of C. michiganensis subsp. michiganensis (C290 or A300) with pXX2 was on average about 2 transformants/μg of vector DNA. Twenty-six chloramphenicol-resistant colonies were obtained; however, only 19 colonies (BL-Cmm1 to -19) were bioluminescent. Upon further analysis using PCR, both the Cmx resistance gene and the luxC gene were detected in the genomes of the bioluminescent strains (Fig. 1). In addition, all 7 nonbioluminescent strains also contained the Cmx resistance gene (data not shown).
FIG. 1.
(A) Physical map of the Tn1409 transposon vector pXX2 carrying the lux operon. Amp, ampicillin resistance; cmx, chloramphenicol resistance; tnpA, transposase IS1409; IRR, right inverted repeat; IRL, left inverted repeat; ORI, origin of replication. The arrows indicate locations of the primers used for sequencing. (B) Representative image of bioluminescent mutants recovered on an SB plate screened by IVIS. (C) PCR analysis with CmxF/CmxR and LuxCF/R primers of the colonies recovered after pXX2 electroporation. Lane 1, 1-kb-plus ladder; lanes 2 and 7, wild-type C290 as a negative control; lanes 3 and 8, pXX2 plasmid as a positive control; lanes 4 to 6 and 9 to 11, bioluminescent transposon mutants. Only results from representative strains are shown.
To evaluate the amounts of bioluminescence in these 19 strains, the light intensity from each strain was measured from late-log-phase cultures. BL-Cmm17 exhibited significantly higher bioluminescence than other strains (P ≤ 0.05), which differed by up to 2 log units (data not shown). Since integration of Tn1409 in the C. michiganensis subsp. michiganensis genome is nonspecific (25), the lux operon may be integrated downstream of promoters that are expressed at different levels. Consequently, variations in the lux operon expression are expected, leading to the observed differences in bioluminescence emitted by different bioluminescent strains.
Impact of the lux insertion on the virulence of bioluminescent C. michiganensis subsp. michiganensis strains.
Since Tn1409 is inserted nonspecifically into the C. michiganensis subsp. michiganensis genome (25), the lux operon might affect genes that are essential for virulence. Nine bioluminescent strains (BL-Cmm1 to -8 and -17) that emitted a different range of light intensities were selected for the HR assay. All of the strains induced strong HR in four o'clock plants, indicating that their virulences were similar to that of the wild type (Table 3 and Fig. 2). The same nine BL-Cmm strains were inoculated into tomato seedlings, and all isolates caused plant wilting similar to that caused by wild-type C. michiganensis subsp. michiganensis C290. In addition, the numbers of bacteria retrieved from the stem tissue of plants inoculated with any of the bioluminescent strains or the wild-type strain were similar, suggesting that the abilities of the bioluminescent strains to colonize the vascular tissue were not affected by the lux insertion. Since the bacteria recovered from the stem tissues were strongly bioluminescent, it was concluded that the lux insertion was stable in these strains.
TABLE 3.
Induction of HR in four o'clock plants and virulence to tomato plants by bioluminescent C. michiganensis subsp. michiganensis strains
| Strain | HRa | Virulenceb | Log CFU/g in plantac |
|---|---|---|---|
| BL-Cmm1 | + | 6/6 | 9.57 ± 0.18 |
| BL-Cmm2 | + | 5/6 | 10.63 ± 0.14 |
| BL-Cmm3 | + | 6/6 | 10.55 ± 0.18 |
| BL-Cmm4 | + | 6/6 | 8.77 ± 1.03 |
| BL-Cmm5 | + | 5/6 | 9.88 ± 1.04 |
| BL-Cmm6 | + | 6/6 | 9.11 ± 0.57 |
| BL-Cmm7 | + | 6/6 | 9.23 ± 0.9 |
| BL-Cmm8 | + | 6/6 | 8.77 ± 0.94 |
| BL-Cmm17 | + | 6/6 | 9.43 ± 0.28 |
| C290 | + | 6/6 | 9.08 ± 0.60 |
| Water | − | 0/6 | 0.00 |
+, positive for HR reaction; −, negative for HR reaction.
Number of wilting plants/total number of inoculated plants.
Mean population of bacteria ± standard deviation (SD) recovered from BL-Cmm-inoculated stem tissue; no significant differences in population counts from inoculated plants were observed (P ≤ 0.05).
FIG. 2.
(A) Hypersensitive response in a four o'clock leaf induced by bioluminescent C. michiganensis subsp. michiganensis strain BL-Cmm17. Bacterial suspensions (108 CFU/ml) were injected into fully expanded leaves of four o'clock plants using syringes without needles, and HR reactions were recorded after 24 h. WT, wild type. (B) Virulence test on 4-week-old tomato seedlings. The seedlings were monitored for wilting for 3 weeks after inoculation.
Characterization of a bioluminescent C. michiganensis subsp. michiganensis strain.
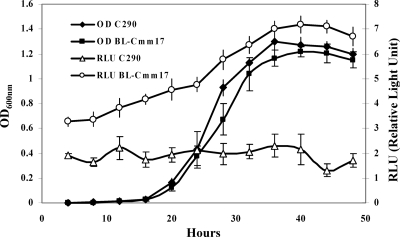
Since BL-Cmm17 exhibited strong bioluminescence and was virulent in both HR and tomato pathogenicity assays, similar to its parental C290, we used this strain for future studies. The in vitro growth of BL-Cmm17 was similar to that of C290: both displayed a generation time of 4 h (Fig. 3). Bioluminescence increased as BL-Cmm17 grew in log phase and started to decrease when its growth reached the stationary phase, suggesting a constitutive expression of Lux (Fig. 3). Furthermore, the similar growth patterns of BL-Cmm17 and C290 were observed in minimal medium, indicating that the lux insertion in BL-Cmm17 had no auxotrophic effect on the growth and survival of BL-Cmm17 (data not shown). The lux insertion was stable in BL-Cmm17; after five rounds of nonselective growth for approximately 50 generations, 150 independent BL-Cmm17 colonies from three replicate experiments were resistant to chloramphenicol and were bioluminescent.
FIG. 3.
Bioluminescence and growth curves of C. michiganensis subsp. michiganensis wild-type C290 and BL-Cmm17 strains. Both strains were inoculated into NBY broth, and the optical density at 600 nm and photon emissions were measured from 4 h to 48 h. The relative light units are log-transformed values of the average radiance (photons/s); the error bars indicate standard deviations.
To identify the insertion site of the lux operon in the BL-Cmm17 genome, chromosomal DNA was isolated and sequenced bidirectionally using the outward primers Xu3F and Xu3R, which bind just inside the inverted repeats of Tn1409 (Fig. 1 shows the primer locations). Sequence analysis suggested a whole-vector integration, since the nucleotide sequences obtained were highly aligned (90% identity) with pXX2 sequence flanking the inverted repeats. However, this is not surprising and has been observed previously when using this mutagenesis system in C. michiganensis subsp. michiganensis (K. H. Gartemann, unpublished data). The plasmid profile from BL-Cmm17 was identical to that of wild-type C290, further confirming the plasmid integration. In addition, restriction analysis of plasmids extracted from BL-Cmm17 and C290 with XhoI/NotI enzymes suggested a chromosomal integration of Tn1409::lux (data not shown). Therefore, further analysis is needed to identify the precise insertion site. Since the insertion of the lux operon did not influence the growth of BL-Cmm17 and the expression of the lux operon was stable and constitutive, it can be assumed that the lux operon was inserted downstream of a strong promoter without disturbing any housekeeping or essential virulence genes in the BL-Cmm17 genome.
Monitoring the colonization of bioluminescent C. michiganensis subsp. michiganensis in germinating tomato seeds.
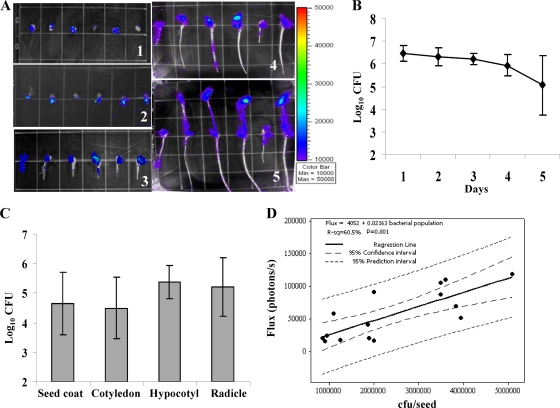
In total, 100 seeds were imaged using the IVIS, and representative results are shown in Fig. 4 A. Luminescent signals were detected on the seed coat throughout 5 days of germination. On the third day, the radicle punctured the seed coat, and luminescence was also observed on some of the radicles. On the fourth and fifth days, cotyledons emerged from the seed coat and hypocotyls and radicles elongated. Luminescent signals were usually stronger on the hypocotyls, cotyledons, and seed coats than on the radicles. Consistent with the bioluminescence imaging, up to 6 log units of bacteria was detected on the seed coat during the first 3 days postinfection and up to 5 log units of bacteria was detected on day 5 postinfection (Fig. 4B). Further, hypocotyls, cotyledons, and radicles contained on average 4.5 to 5.4 log units of bacteria at day 5 (Fig. 4C). Light intensity was positively correlated with the number of bacteria on individual seedling components, with the highest determination coefficient (R2 = 60.5%) for seed coats tested from days 1 to 3 (Fig. 4D).
FIG. 4.
(A) Bioluminescence of strain BL-Cmm17 inoculated onto tomato seeds. Images were acquired daily by using an IVIS. Bacteria were observed on seed coats throughout the germination period and colonized other seedling components as they emerged. The number at the bottom of each image indicates the day of germination. The rainbow scale represents photon counts (photons/s). (B) Population dynamics of the bioluminescent BL-Cmm17 strain on tomato seed coats during germination. Bacterial counts were estimated from serial-dilution plating, and the total CFU were log transformed. Each point represents the mean bacterial count for 12-seed (days 1 to 3) and seed coat (days 4 and 5) isolations; the error bars indicate standard deviations. (C) Distribution of the bioluminescent BL-Cmm17 strain on tomato seedlings on the fifth day of germination. Each bar represents the mean bacterial count for 12 of each seedling component; the error bars indicate standard deviations. (D) Regression analysis of light intensity versus populations of strain BL-Cmm17 on tomato seed coats during days 1, 2, and 3 of germination.
Visualization of bioluminescent C. michiganensis subsp. michiganensis colonization of grafted plants.
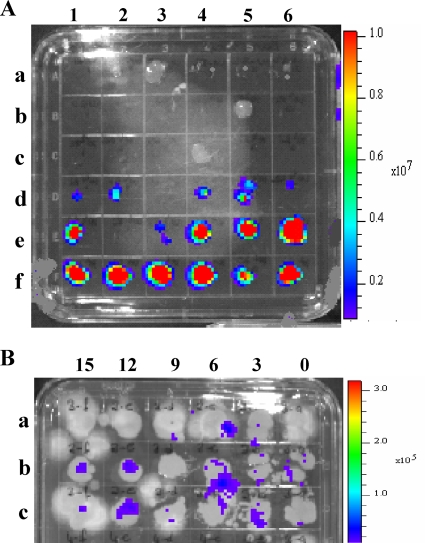
Cut surfaces of tomato rootstock and scion stems became contaminated with BL-Cmm17 when exposed to inoculum doses of 104, 106, and 108 CFU/ml, shown by tissue imprinting on D2ANX medium on day zero, followed by IVIS imaging of recovered colonies (Table 4 and Fig. 5). These results were confirmed by colony counting (Table 4). Seven days after the grafting, only plants exposed to these inoculum doses were colonized by BL-Cmm17 (Table 5). In rootstock-inoculated plants, colonization was detected in the rootstock up to 15 mm from the graft union for all three BL-Cmm17 doses, whereas scions of the same plants were colonized only at doses of 106 and 108 CFU/ml. When scions were exposed to BL-Cmm17, colonization of rootstocks and scions was not observed at doses below 106 CFU/ml. At the 106-CFU/ml dose, BL-Cmm17 was detected only up to 9 mm beyond the graft union in the rootstock. Similar to tissue imprint imaging, colony counts of tissues taken from either the rootstock or scion after grafting at specified distances from the graft union also contained an average 105 to 108 CFU/g of tissue, depending on the inoculation dose, with most of the tissue sections from the 108-CFU/ml scion-inoculated group containing more than 108 bacteria/g of tissue (data not shown).
TABLE 4.
Isolation of BL-Cmm17 from tomato rootstock and scion seedling tissues at time of exposure to the pathogen (day zero) via a contaminated grafting tool
| Inoculum concn (CFU/ml) | BL-Cmm17 detected by tissue imprinting and luminescence imaginga |
BL-Cmm17 population in planta (log CFU/g ±SD) |
||
|---|---|---|---|---|
| Rootstock inoculated | Scion inoculated | Rootstock inoculated | Scion inoculated | |
| Water control | − | − | ||
| 102 | − | − | ||
| 104 | + (7) | + (4) | 4.31 ± 0.04 | 4.31 ± 0.08 |
| 106 | + (9) | + (9) | 6.22 ± 0.38 | 6.32 ± 0.11 |
| 108 | + (9) | + (9) | 8.27 ± 0.27 | 8.31 ± 0.05 |
+, detected; −, not detected. The values in parentheses are the number of tissue imprints from which BL-Cmm17 was cultured from nine exposed plants.
FIG. 5.
Recovery of a bioluminescent BL-Cmm17 strain from tomato tissue imprinted on D2ANX semiselective medium and identified by an IVIS. The images show representative plates of tissue imprints after exposure of cut stems to BL-Cmm17 at day zero (A) and 7 days postgrafting (B). Each grid was imprinted with one tissue cut. (A) Three plants were imprinted per inoculum dose, and each dose exposure was replicated three times. Sections a1 to b3, rootstock cut with a tool exposed to water; sections b4 to c6, d1 to e3, and e4 to f6, rootstock cut with a tool exposed to 102, 104, or 106 CFU/ml of BL-Cmm17, respectively. (B) Sections of grafted seedlings 0, 3, 6, 9, 12, and 15 mm below the graft union. Each row represents one replicate rootstock seedling that was inoculated with the grafting tool exposed to 104 CFU/ml of BL-Cmm17. BL-Cmm17 colonies were visible even in the presence of fungal contamination.
TABLE 5.
Colonization of tomato seedlings 7 days postgrafting after rootstocks or scions were exposed to different doses of BL-Cmm17
| Grafted plant part imprinted | Distance (mm) from graft union | Presence of BL-Cmm17a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rootstock inoculated at (CFU/ml): |
Scion inoculated at (CFU/ml): |
||||||||||
| 0 | 102 | 104 | 106 | 108 | 0 | 102 | 104 | 106 | 108 | ||
| Scion | 15 | − | − | − | − | + | − | − | − | + | + |
| Scion | 12 | − | − | − | + | ++ | − | − | − | +++ | ++ |
| Scion | 9 | − | − | − | + | + | − | − | − | +++ | ++ |
| Scion | 6 | − | − | − | + | +++ | − | − | − | + | ++ |
| Scion | 3 | − | − | − | + | ++ | − | − | − | +++ | + |
| Graft union | 0 | − | − | − | + | + | − | − | − | ++ | + |
| Graft union | 0 | − | − | + | ++ | + | − | − | − | + | +++ |
| Rootstock | −3 | − | − | ++ | ++ | + | − | − | − | ++ | +++ |
| Rootstock | −6 | − | − | ++ | − | +++ | − | − | − | + | +++ |
| Rootstock | −9 | − | − | ++ | + | ++ | − | − | − | + | +++ |
| Rootstock | −12 | − | − | ++ | + | ++ | − | − | − | − | +++ |
| Rootstock | −15 | − | − | ++ | ++ | ++ | − | − | − | − | +++ |
Three plants were sampled for each inoculum dose, and the presence of BL-Cmm17 was determined by tissue imprinting the cut surface on D2ANX medium at the indicated distance from the graft union. The number of pluses indicates the number of tissue sections from which BL-Cmm17 was cultured and identified by luminescence imaging. −, not present.
Isolation of bioluminescent C. michiganensis subsp. nebraskensis.
Tn1409 transposon mutagenesis is effective in other subspecies of C. michiganensis (25). Therefore, to demonstrate the utility of our modified Tn1409-lux for the generation of bioluminescent bacteria in a closely related subspecies of C. michiganensis, we transformed pXX2 into C. michiganensis subsp. nebraskensis, the causal agent of wilt and leaf blight in maize. Electroporation of pXX2 resulted in a total of 22 chloramphenicol-resistant colonies, 8 of which were bioluminescent. The transformation frequency was very similar to that observed with C. michiganensis subsp. michiganensis. These results highlight the usefulness of our approach to elucidate, in real time, the plant-pathogen interactions in diseases caused by other agriculturally important subspecies of C. michiganensis.
DISCUSSION
Despite the importance of C. michiganensis subsp. michiganensis as a debilitating crop pathogen with a significant economic impact on agricultural production, little is known about its ecology in and on tomato plants under modern propagation and production practices. This lack of knowledge is partly attributable to the difficulty of genetically manipulating this Gram-positive bacterium. Consequently, molecular approaches that would facilitate the study of C. michiganensis subsp. michiganensis interactions with plant hosts, colonization of seeds, and transmission to seedlings and the spread of the pathogen among tomato crops are needed. Of particular interest is the lux operon, which has been successfully used as a bioluminescent reporter in bacterial pathogens, facilitating the characterization of bacterial distribution, growth, and infection in plant hosts and allowing the assessment of plant responses to bacterial infection (6, 7, 8, 9, 12, 15, 31). Therefore, we generated a bioluminescent C. michiganensis subsp. michiganensis strain that retained its virulence and growth properties and enabled visualization of the colonization dynamics of the pathogen in planta in real time.
The construction of a bioluminescent C. michiganensis subsp. michiganensis strain required testing different mutagenesis approaches, among which the use of a modified Tn1409 vector, pKGT452Cβ, carrying the lux operon optimized for expression in Gram-positive bacteria proved successful. Insertion of the lux operon allowed the isolation of a strain that constitutively expressed strong bioluminescence. We desired a promoter that was active both in planta and in vitro and a bioluminescent strain that was not compromised in virulence to use for in planta real-time infection. Initially, we constructed a Tn5-based transposon-containing lux operon; however, we were not successful in generating bioluminescent C. michiganensis subsp. michiganensis strains using this approach, though Tn5 transposon mutagenesis has been used successfully in the related Corynebacterium (42). Alternatively, use of vectors carrying the Tn1409 transposon (pKGT452Cβ) effectively resulted in transformed bioluminescent C. michiganensis subsp. michiganensis that harbored the lux genes. The pKGT452Cβ vector was of particular interest, since transposon Tn1409 was shown to integrate nonspecifically in the Arthrobacter chromosome (a genus closely related to Clavibacter) and later proved to be efficient in C. michiganensis subsp. michiganensis mutagenesis (16, 25). Specifically, electroporation with the suicide vector-carrying transposon Tn1409 yielded approximately 1 × 103 mutants per μg of DNA and resulted in random insertion of the transposon in the C. michiganensis subsp. michiganensis chromosome (25). However, C. michiganensis subsp. michiganensis electroporation with pXX3 carrying Tn1409::lux resulted in a very small number of transformants, which carried the lux operon but were not bioluminescent. Although the low transformation efficiency was likely due to the large size of the lux insert, the lack of bioluminescence in the transposon mutants suggests a lack of expression of the Gram-negative bacterium-derived lux operon in C. michiganensis subsp. michiganensis. The luxCDABE operon originated from the Gram-negative bacterium P. luminescens, which is relatively low in GC (42%) (NCBI genome database). The C. michiganensis subsp. michiganensis genome has a high GC content (∼73%), which possibly explains the lack or inefficient expression of lux genes. Therefore, we used pXX2, which has a lux operon optimized for expression in Gram-positive bacteria. Similar to pXX3, the transformation frequency with pXX2 was very low, which again could be attributed to the large size of the lux operon; however, we cannot exclude the possibility of genetic variation among C. michiganensis subsp. michiganensis strains contributing to observed differences in the transformation frequencies (25).
For the bioluminescent C. michiganensis subsp. michiganensis strains to constitute a suitable tool for studying colonization and plant invasion, it was important to test if the lux insertion affected their growth and virulence. Our results showed that strain BL-Cmm17, which was strongly bioluminescent, exhibited growth similar to that of the wild-type strain (Fig. 3), carried a stable lux operon that was not lost after successive growth cycles in nonselective medium, and possessed virulence properties comparable to those of the wild type (Fig. 2 and Table 3). These observations showed that although the precise lux operon insertion site within BL-Cmm17 could not be determined, insertion of the lux operon did not impede essential functions involved in bacterial growth and virulence. Furthermore, the bioluminescence could be detected in BL-Cmm17 at different growth stages. Specifically, the bioluminescence of BL-Cmm17 reached its highest level in late stages of exponential growth. This could be the result of efficient activation of the promoter controlling the lux expression or a reflection of bacterial numbers during this stage of growth. The decrease in BL-Cmm17 bioluminescence after entering the stationary growth phase might have been the result of downregulation of cell metabolism, as it has been shown that bioluminescence requires energy derived from cell metabolism (21). These observations suggest that this strain is suitable for experiments that mimic the plant colonization processes of wild-type bacteria.
Few studies have addressed C. michiganensis subsp. michiganensis seedling infections initiated by contaminated seeds in general, and data are almost completely lacking concerning C. michiganensis subsp. michiganensis transmission from seeds, a major route for long-distance spread of the pathogen. Since it has been shown that naturally deposited C. michiganensis subsp. michiganensis on the surfaces of seeds during seed extraction precedes colonization (11), it was assumed that tomato seeds artificially inoculated with BL-Cmm17 simulated naturally infected seeds. Subsequently, real-time imaging showed that BL-Cmm17 aggregated on the hypocotyls and cotyledons (Fig. 4) of the seedlings, possibly advancing colonization and prompting the onset of future infections. These observations and conclusions were supported by previous reports showing that early canker symptoms appeared on the cotyledons and stems and that epiphytic C. michiganensis subsp. michiganensis populations were associated with tomato leaves (2, 19, 40). Perhaps one of the most important features of using a bioluminescent C. michiganensis subsp. michiganensis strain is the ability to determine the number of pathogens colonizing the plant in real time. Emitted light can be correlated to bacterial abundance using predetermined growth curves. Therefore, we were able to monitor C. michiganensis subsp. michiganensis colonization and assess the abundance of the population across time (Fig. 4). Our seed inoculation experiments were primarily designed to highlight the value of using bioluminescence in studying C. michiganensis subsp. michiganensis interactions with the plant host. Our results corroborated this value and shed new light on early C. michiganensis subsp. michiganensis colonization of tomato seedlings. The correlation between radiance and bacterial colony counts was highest for the seed coat and lowest for radicles. It is possible that C. michiganensis subsp. michiganensis was less aggregated on radicles than on the seed coat, resulting in lower photon emissions per unit area. Additionally, electrolytes diffused through the seed coat may promote bacterial growth in the early stages of germination (4). Further analysis and longer observation intervals are needed to fully characterize the epiphytic stage of C. michiganensis subsp. michiganensis on seeds and its transmission to seedlings and mature plants.
Grafting is a widely applied technique in greenhouse tomato production used to improve plant vigor and yield and to provide resistance to soilborne pathogens and nematodes (26). For tomato bacterial canker, grafting plays an important role in its transmission by directly exposing C. michiganensis subsp. michiganensis to vascular tissue. However, the dynamics of C. michiganensis subsp. michiganensis colonization of tomato seedlings during grafting has not been studied. Such studies have been hindered by the lack of truly selective media to specifically isolate C. michiganensis subsp. michiganensis without interference from other epiphytic microorganisms. The imaging system used in this study is highly sensitive and detected phosphorescence (autobioluminescence) from green plant tissue, which interfered with detection of bioluminescent C. michiganensis subsp. michiganensis in grafted plants (which was not observed in seedlings germinated for up to 5 days in the dark). Therefore, we used tissue imprinting to monitor bacterial colonization of grafted rootstocks and scions. Our results have shown that grafting tools readily transmit C. michiganensis subsp. michiganensis, but an inoculum threshold (>104 CFU/ml) may be required. At higher inoculum concentrations, C. michiganensis subsp. michiganensis moved past the graft union at least 12 to 15 mm within 7 days, even though vascular connections were forming during that time (Table 5 and Fig. 5). Our study provides visualization of C. michiganensis subsp. michiganensis movement in grafted tomato plants and may serve as an important tool for the development of effective grafting procedures to restrict the spread of C. michiganensis subsp. michiganensis in seedling propagation units.
In summary, we have developed a novel approach to characterize C. michiganensis subsp. michiganensis plant infection and colonization. Incorporation of a bioluminescent marker in the C. michiganensis subsp. michiganensis chromosome has allowed us to monitor the infection temporally. Significantly for C. michiganensis subsp. michiganensis pathogenesis, our results reveal (i) the colonization dynamics of C. michiganensis subsp. michiganensis infection of hypocotyls and cotyledons of the seedlings that promote the onset of future infections and (ii) visualization of the translocation of C. michiganensis subsp. michiganensis in grafted plants, which may have a significant impact on the development of grafting procedures to limit the transmission of C. michiganensis subsp. michiganensis. Therefore, our approach provides a novel visual representation of the disease process and accelerates understanding of C. michiganensis subsp. michiganensis ecology. Furthermore, these bioluminescent strains will have a wide scope of application, including determining plant factors that affect C. michiganensis subsp. michiganensis growth and metabolism and screening of antibiotics and bactericides for canker control in an effective real-time approach. Equally important, our results indicated that Tn1409-lux can be used to generate bioluminescent C. michiganensis subsp. nebraskensis. Since Tn1409 is also an effective mutagenesis system for other important subspecies of C. michiganensis, including subsp. insidiosus (which causes wilt and stunt on alfalfa) and subsp. sepedonicus (which causes ring rot on potato) (25), the vector pXX2 constructed in this study could also be used for the transformation of the lux operon into these important pathogens, facilitating real-time transmission, control, and plant interaction studies.
Acknowledgments
We thank Larry Madden and Pierce Paul for their assistance with statistical analyses, David L. Coplin for providing the C. michiganensis subsp. nebraskensis strain and critical comments on virulence tests and growth assays, and Issmat Kassem for critical review of the manuscript.
The research in the Rajashekara and Miller laboratories is supported by a USDA-NRI grant and by the Ohio Agricultural Research and Development Center, Ohio State University.
Footnotes
Published ahead of print on 23 April 2010.
REFERENCES
- 1.Alarcón, C., J. Castro, F. Muñoz, P. Arce-Johnson, and J. Delgado. 1998. Protein(s) from the gram-positive bacterium Clavibacter michiganensis subsp. michiganensis induces a hypersensitive response in plants. Phytopathology 88:306-310. [DOI] [PubMed] [Google Scholar]
- 2.Carlton, W. M., E. J. Braun, and M. L. Gleason. 1998. Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology 88:525-529. [DOI] [PubMed] [Google Scholar]
- 3.Chabot, R., H. Antoun, J. W. Kloepper, and C. J. Beauchamp. 1996. Root colonization of maize and lettuce by bioluminescent Rhizobium leguminosarum biovar phaseoli. Appl. Environ. Microbiol. 62:2767-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chachalis, D., E. M. Khah, and M. K. Darawsheh. 2008. Effects of initial seed moisture content, imbibition temperature and seed vigour on germination, electrolyte leakage and seedling growth in plum tomatoes. J. Food Agri. Environ. 6:299-304. [Google Scholar]
- 5.Chang, R. J., S. M. Ries, and J. K. Pataky. 1991. Dissemination of Clavibacter michiganensis subsp. michiganensis by practices used to produce tomato transplants. Phytopathology 81:1276-1281. [Google Scholar]
- 6.Cirvilleri, G., and S. E. Lindow. 1994. Differential expression of genes of Pseudomonas syringae on leaves and in culture evaluated with random genomic lux fusions. Mol. Ecol. 3:249-257. [Google Scholar]
- 7.Cirvilleri, G., P. Bella, and V. Catara. 2000. Luciferase genes as a marker for Pseudomonas corrugata. J. Plant Pathol. 82:237-241. [Google Scholar]
- 8.Cirvilleri, G., P. Bella, R. La Rosa, and V. Catara. 2008. Internalization and survival of Pseudomonas corrugata from flowers to fruits and seeds of tomato plants, p. 73-79. In M. Fatmi, A. Collmer, N. S. Lacobellis, J. Mansfield, J. Murillo, and N. W. Schaad (ed.), Pseudomonas syringae pathovars and related pathogens—identification, epidemiology and genomics. Springer Netherlands, Dordrecht, Netherlands.
- 9.de Weger, L. A., P. Dunbar, W. F. Mahafee, J. J. Lugtenberg, and G. S. Sayler. 1991. Use of bioluminescent markers to detect Pseudomonas spp. in the rhizosphere. Appl. Environ. Microbiol. 57:3641-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichenlaub, R., K. H. Gartemann, and A. Burger. 2006. Clavibacter michiganensis, a group of Gram-positive phytopathogenic bacteria, p. 385-421. In S. S. Gnanamanickam (ed.), Plant-associated bacteria. Springer Netherlands, Dordrecht, Netherlands.
- 11.European and Mediterranean Plant Protection Organization. 2005. Clavibacter michiganensis subsp. michiganensis. EPPO Bull. 35:275-283. [Google Scholar]
- 12.Fan, J., C. Crooks, and C. Lamb. 2008. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 53:393-399. [DOI] [PubMed] [Google Scholar]
- 13.Fatmi, M., N. W. Schaad, and H. A. Bolkan. 1991. Seed treatments for eradicating Clavibacter michiganensis subsp. michiganensis from naturally infected tomato seeds. Plant Dis. 75:383-385. [Google Scholar]
- 14.Francis, K. P., D. Joe, C. Bellinger-Kawahara. M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construction. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui, R., H. Fukui, R. McElhaney, S. C. Nelson, and A. M. Alvarez. 1996. Relationship between symptom development and actual sites of infection in leaves of anthurium inoculated with a bioluminescent strain of Xanthomonas campestris pv. dieffenbachiae. Appl. Environ. Microbiol. 62:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartemann, K.-H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J. Bacteriol. 183:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartemann, K.-H., O. Kirchner, J. Engemann, I. Gräfen, R. Eichenlaub, and A. Burger. 2003. Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J. Biotechnol. 106:179-191. [DOI] [PubMed] [Google Scholar]
- 18.Gitaitis, R. D. 1990. Induction of a hypersensitive-like reaction in four o'clock by Clavibacter michiganensis subsp. michiganensis. Plant Dis. 74:58-60. [Google Scholar]
- 19.Gleason, M. L., R. D. Gitaitis, and M. D. Ricker. 1993. Recent progress in understanding and controlling bacterial canker of tomato in Eastern North America. Plant Dis. 77:1069-1076. [Google Scholar]
- 20.Hadas, R., G. Kritzman, F. Klietman, T. Gefen, and S. Manulis. 2005. Comparison of extraction procedures and determination of the detection threshold for Clavibacter michiganensis ssp. michiganensis in tomato seeds. Plant Pathol. 54:643-649. [Google Scholar]
- 21.Hakkila, K., M. Maksimow, M. Karp, and M. Virta. 2002. Reporter genes lucFF, luxCDABE, gfp and dsRed have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 301:235-242. [DOI] [PubMed] [Google Scholar]
- 22.Hutchens, M., and G. D. Luker. 2007. Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 9:2315-2322. [DOI] [PubMed] [Google Scholar]
- 23.Jahr, H., R. Bahro, A. Burger, J. Ahlemeyer, and R. Eichenlaub. 1999. Interactions between Clavibacter michiganensis and its host plants. Environ. Microbiol. 1:113-118. [DOI] [PubMed] [Google Scholar]
- 24.Kamoun, S., and C. I. Kado. 1990. A plant-inducible gene of Xanthomonas campestris pv. campestris encodes an exocellular component required for growth in the host and hypersensitivity on nonhosts. J. Bacteriol. 172:5165-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchner, O., K.-H. Gartemann, E.-M. Zellermann, R. Eichenlaub, and A. Burger. 2001. A highly efficient transposon mutagenesis system for the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 14:1312-1318. [DOI] [PubMed] [Google Scholar]
- 26.Kubota, C., M. A. McClure, N. Kokalis-Burelle, M. G. Bausher, and E. N. Rosskopf. 2008. Vegetable grafting: history, use and current technology status in North America. HortScience 43:1664-1669. [Google Scholar]
- 27.Louws, F., J. Bell, C. Medina-Mora, C. D. Smart, D. Opgenorth, C. Ishimaru, F. R. deBruijin, M. Hausbeck, and D. W. Fulbright. 1998. rep-PCR-mediated genomic finger-printing: a rapid and effective method to identify Clavibacter michiganensis subsp. michiganensis. Phytopathology 88:862-868. [DOI] [PubMed] [Google Scholar]
- 28.Meighen, E. A. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7:1016-1022. [DOI] [PubMed] [Google Scholar]
- 29.Meletzus, D., A. Bermpohl, J. Dreier, and R. Eichenlaub. 1993. Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175:2131-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzler, M. C., M. J. Laine, and S. H. De Boer. 1997. The status of molecular biological research on the plant pathogenic genus Clavibacter. FEMS Microbiol. Lett. 150:1-8. [Google Scholar]
- 31.Paynter, C. D., V. C. Salisbury, D. L. Arnold, and R. W. Jackson. 2006. The use of bioluminescence for monitoring in planta growth dynamics of a Pseudomonas syringae plant pathogen. Eur. J. Plant Pathol. 115:363-366. [Google Scholar]
- 32.Poysa, V. 1993. Evaluation of tomato breeding lines resistant to bacterial canker. Can. J. Plant Pathol. 15:301-304. [Google Scholar]
- 33.Rajashekara, G., D. A. Glover, M. Krepps, and G. A. Splitter. 2005. Temporal analysis of pathogenic events in virulent and avirulent Brucella melitensis infections. Cell. Microbiol. 7:1459-1473. [DOI] [PubMed] [Google Scholar]
- 34.Ricker, M. D., and R. M. Riedel. 1993. Effect of secondary spread of Clavibacter michiganensis subsp. michiganensis on yield of northern processing tomatoes. Plant Dis. 77:364-366. [Google Scholar]
- 35.Shaw, J. J., L. G. Settles, and C. I. Kado. 1988. Transposon Tn4431 mutagenesis of Xanthomonas campestris pv. campestris: characterization of a nonpathogenic mutant and cloning of a locus for pathogenicity. Mol. Plant-Microbe Interact. 1:39-45. [Google Scholar]
- 36.Shaw, J. J., F. Dane, D. Geiger, and J. W. Kloepper. 1992. Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl. Environ. Microbiol. 58:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stackebrandt, E., A. F. Rainey, and L. N. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 38.Stewart, G. S., and P. Williams. 1992. lux genes and the applications of bacterial bioluminescence. J. Gen. Microbiol. 138:289-300. [DOI] [PubMed] [Google Scholar]
- 39.Szittner, R., and E. Meighen. 1990. Nucleotide sequence, expression, and properties of luciferase coded by lux genes from a terrestrial bacterium. J. Biol. Chem. 265:16581-16587. [PubMed] [Google Scholar]
- 40.Strider, D. L. 1969. Bacterial canker of tomato, a literature review and bibliography. N.C. Agric. Exp. Stn. Tech. Bull. 193:1-80. [Google Scholar]
- 41.Vidaver, A. K. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, C., B. Hayes, M. M. Vestling, and K. Takayama. 2006. Transposome mutagenesis of an integral membrane transporter in Corynebacterium matruchotii. Biochem. Biophy. Res. Commun. 340:953-960. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, E. E., F. M. Zeitoun, and D. L. Fredrickson. 1967. Bacterial phloem canker, a new disease of Persian walnut trees. Phytopathology 57:618-621. [Google Scholar]