Abstract
Conversion of components of the Thermobifida fusca free-enzyme system to the cellulosomal mode using the designer cellulosome approach can be employed to discover the properties and inherent advantages of the cellulosome system. In this article, we describe the conversion of the T. fusca xylanases Xyn11A and Xyn10B and their synergistic interaction in the free state or within designer cellulosome complexes in order to enhance specific degradation of hatched wheat straw as a model for a complex cellulosic substrate. Endoglucanase Cel5A from the same bacterium and its recombinant dockerin-containing chimera were also studied for their combined effect, together with the xylanases, on straw degradation. Synergism was demonstrated when Xyn11A was combined with Xyn10B and/or Cel5A, and ∼1.5-fold activity enhancements were achieved by the designer cellulosome complexes compared to the free wild-type enzymes. These improvements in activity were due to both substrate-targeting and proximity effects among the enzymes contained in the designer cellulosome complexes. The intrinsic cellulose/xylan-binding module (XBM) of Xyn11A appeared to be essential for efficient substrate degradation. Indeed, only designer cellulosomes in which the XBM was maintained as a component of Xyn11A achieved marked enhancement in activity compared to the combination of wild-type enzymes. Moreover, integration of the XBM in designer cellulosomes via a dockerin module (separate from the Xyn11A catalytic module) failed to enhance activity, suggesting a role in orienting the parent xylanase toward its preferred polysaccharide component of the complex wheat straw substrate. The results provide novel mechanistic insight into the synergistic activity of designer cellulosome components on natural plant cell wall substrates.
Thermobifida fusca is an aerobic thermophilic soil bacterium with strong cellulolytic activity (52). The T. fusca enzyme system is an extensively studied free cellulase system in which nearly all of the cellulolytic enzymes have been fully characterized, from the individual enzyme sequences to the three-dimensional structures, as well as the biochemical activities of the native and recombinant proteins. The genome sequence has been published (36), and the number and types of carbohydrate-active enzymes produced by the organism are known. This actinomycete produces six different cellulases that have been well studied (29, 31, 32, 50, 52). T. fusca also has the ability to grow on xylan and produces several enzymes involved in xylan degradation, such as xylanases, β-xylosidase, α-l-arabinofuranosidase, and acetylesterases (1, 21).
Previous research has suggested that the multienzyme cellulosome complex from Clostridium thermocellum is far more efficient than free cellulase systems that were tested in degrading polysaccharides (33). The cellulosome system is characterized by the strong bimodular interaction between the cohesin and dockerin modules that integrates the various enzymes into the complex (5, 35, 55). Scaffoldin subunits (nonenzymatic protein components) contain the cohesin modules that incorporate the enzymes into the complex via their resident dockerins. The primary scaffoldin subunit also includes a carbohydrate (cellulose)-binding module (CBM) through which the complex recognizes and binds to the cellulosic substrate (42, 46).
In order to evaluate the reasons for the apparent advantage of cellulosomes over free enzymes, it is interesting to compare the properties of the best-characterized free-enzyme systems for degradation of polysaccharides with those of the best-studied cellulosome system. We have initiated a program to convert the free-enzyme system of T. fusca into an artificial designer cellulosome (11-13). The designer cellulosome concept is based on the very high affinity (20, 44) and specific interaction (37, 43, 55) between a cohesin and a dockerin module from the same species. Since the various scaffoldin-borne cohesins of a given species essentially show the same specificity of binding for the enzyme-borne dockerins, designer cellulosomes are constructed from recombinant chimeric scaffoldins containing divergent cohesins from different species, for which matching dockerin-containing enzyme hybrids are prepared, as a platform for promoting synergistic action among enzyme components (5). Free cellulases from the T. fusca system were converted to the cellulosomal mode by replacing their native CBM with a desired dockerin module, and in some cases, the resultant “designer cellulosomes” exhibited enhanced synergistic activity on crystalline cellulosic substrates compared to that of the mixture of wild-type enzymes (11).
In this study, we incorporated xylanolytic enzymes into designer cellulosomes and investigated their hydrolytic effects on purified xylans and on a native, complex cellulosic substrate (hatched wheat straw). We focused on T. fusca xylanases 11A and 10B (Xyn11A and Xyn10B), which are the most abundant xylanases produced during growth on xylan (34). Xyn11A and Xyn10B function as endoxylanases (28, 34); Xyn11A contains a C-terminal family 2 CBM that binds both cellulose and xylan, whereas Xyn10B lacks a CBM. In some experiments, one of the previously converted (dockerin-containing) T. fusca endoglucanases, f-5A (11), was also introduced into the designer cellulosomes in order to evaluate cooperation between xylanases and cellulases in hydrolysis of a natural substrate. This study contributes primary information concerning a major feature of cellulosomes that had not been suitably addressed in earlier research: although xylanases are integral components of cellulosomes, their synergistic action in the cellulosome mode has yet to be examined experimentally. The xylan-binding CBM (termed XBM for the purposes of this report) was found to contribute to the activity of the parent Xyn11A enzyme.
MATERIALS AND METHODS
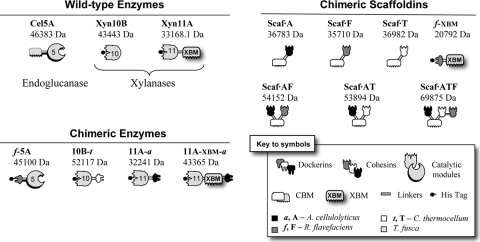
The recombinant enzymes and structural proteins (scaffoldins) designed for this study are presented schematically in Fig. 1.
FIG. 1.
Schematic representation of the recombinant proteins used in this study. The source of the representative module (see the key) is indicated. In the shorthand notation for the engineered enzymes, the numbers 5, 10, and 11 refer to the corresponding GH family (GH5, GH10, and GH11) of the catalytic module; uppercase letters (A, F, and T) indicate the source of the cohesin module, and lowercase letters (a, f, and t) indicate the source of the dockerin module.
Cloning.
Plasmids encoding the wild-type enzymes pCel5A, pXyn11A, and pXyn10B were cloned as described previously (28, 30, 34). The recombinant pf-5A was engineered as reported previously (11).
The 11A-a chimera, i.e., the catalytic module of Xyn11A attached directly to a dockerin from Acetivibrio cellulolyticus ATCC 33288 but lacking the family 2 XBM, was constructed from pXyn11A using primers 5′-CTGCCGCTAGCATGCACCATCACCATCACCACGCCGTGACCTCCAACGAGACC-3′ (NheI site in boldface) and 5′-TCCCAAGAGCTCCGTCGAACTAGTGCCACCGCCACCGGGGGGGTTGCC-3′ (SpeI and SacI sites in boldface) for amplifying the DNA encoding the catalytic module and primers 5′-ATGTATACTAGTAAATTTATATATGGTGATGT-3′ (SpeI site in boldface) and 5′-TACCAAGAGCTCTTATTCTTCTTTCTCTTCAACAG-3′ (SacI site in boldface) for amplification of the A. cellulolyticus ScaB dockerin DNA. The two modules were ligated (T4 DNA ligase: Fermentas UAB, Vilnius, Lithuania) into NheI-SacI (New England Biolabs Inc., Beverly, MA)-linearized pET21a (Novagen Inc., Madison, WI) to form p11A-a.
Primer 5′-TATCCGGAGCTCGTTGGCGCTGCAGGACACCG-3′ (SacI site in boldface) was used to clone the full-length Xyn11A DNA (together with the forward Xyn11A primer mentioned above), and primers for cloning A. cellulolyticus ScaB dockerin DNA were designed: 5′-TTATTCGAGCTCACAGCAACTACAACACCAACTACAACACCAACTACAACACCAACGCCTAAAT-3′ (SacI site in boldface) and 5′-TTATTCGAGCTCACAGCAACTACAACACCAACTACAACACCAACTACAACACCAACGCCTAAAT-3′ (XhoI site in boldface). The two PCR products were then ligated into NheI-XhoI-linearized pET21a to form p11A-XBM-a.
The DNA encoding C. thermocellum dockerin S (from Cel48S) was amplified from the genomic DNA (strain YS) using primers 5′-TTATTCACTAGTACATATAAAGTACCTGGTACTCC-3′ and 5′-TTATTCCTCGAGTTAGTTCTTGTACGGCAATGTATC-3′ (SpeI and XhoI sites in boldface). The DNA encoding the Xyn10B catalytic module was cloned from the Xyn10B plasmid using primers 5′-CATATTGCTAGCCATCACCATCACCATCACGGACCGGTCCACGACCATCATCCC-3′ and 5′-TTATTCCTCGAGTTATTAACTAGTACAGTGATCGTGCTTGGGGCCC-3′ (NheI, SpeI, and XhoI sites in boldface). The two modules were then ligated into NheI-XhoI-linearized pET21a to form p10B-t. All constructs were designed to encode an attached His tag for subsequent purification of the gene product.
Scaffoldins were assembled from modules (cohesins, dockerins, and CBM) cloned from different genomic DNAs. The following primers represent the homologous gene sequences only and were used with different restriction sites, either NcoI, KpnI, BamHI, or XhoI, depending on the desired construct. Cohesin F (DNA encoding cohesin 1 from Ruminococcus flavefaciens strain 17 scaffoldin B) was amplified using 5′-CGCCGGTGGTTTATCCGCTGTG-3′ and 5′-TTAATGGTGATGGTGATGGTGAACAATGATAGCGCCATCAGT-3′. Cohesin A (DNA encoding cohesin 3 from A. cellulolyticus scaffoldin C) was cloned from genomic DNA using 5′-ATTTACAGGTTGACATTGGAAGT-3′ and 5′-GATGCAATTACCTCAATTTTTCC-3′. DNA encoding CBM-T from C. thermocellum YS genomic DNA was amplified using 5′-GACAAACACACCGACAAACACA-3′ and 5′-CTATATCTCCAACATTTACTCCAC-3′. The different modules were assembled in the linearized pET28a plasmid to form the chimeric scaffoldins.
pScaf·T, pScaf·F, and pScaf·A were cloned as described previously (24).
To form the dockerin-containing pf-XBM, the DNA encoding XBM was amplified from the Xyn11A plasmid using the primers 5′-AAATAAGGTACCTACCAGCGGCGGTGGAAACCCC-3′ (KpnI site in boldface) and 5′-AAATTACTCGAGCTAGTTGGCGCTGCAGGACA-3′ (XhoI site in boldface) and ligated to linearized pET28a, together with the R. flavefaciens ScaB dockerin, cloned from genomic DNA using 5′-TGATCCATGGCACACCATCACCATCACCATGCACCATCACCCGGCACAAAGC-3′ (BamHI site in boldface) and 5′-ATGCTTGGTACCGCTTGAGGAAGTGTGATGAGTTCAA-3′ (KpnI site in boldface).
PCRs were performed using ABgene Reddymix ×2 (Advanced Biotechnologies Ltd., Epsom, United Kingdom), and DNA samples were purified using a HiYield Gel/PCR Fragments Extraction Kit (Real Biotech Corporation, RBC, Banqiao City, Taiwan).
Protein expression and purification.
Cel5A, Xyn11A, and Xyn10B were prepared as described previously (28, 30, 34). The f-5A chimera was expressed as reported previously (11). The 11A-a, 11A-XBM-a, 10B-t, and f-XBM plasmids were expressed in Escherichia coli BL21(λDE3) pLysS cells and purified on an Ni-nitrilotriacetic acid (NTA) column (Qiagen Inc., Valencia, CA), as reported previously (13). Scaffoldins were expressed and purified on phosphoric acid-swollen cellulose (7.5 mg ml−1, pH 7) (PASC) according to the previously described methodology (24). The purity of the recombinant proteins was tested by SDS-PAGE on 12% acrylamide gels. The concentration of each purified protein was estimated by absorbance (280 nm) based on the known amino acid composition of the protein using the Protparam tool (http://www.expasy.org/tools/protparam.html). Proteins were stored in 50% (vol/vol) glycerol at −20°C.
Affinity-based enzyme-linked immunosorbent assay (ELISA).
The matching fusion protein procedure of Barak et al. and Caspi et al. (2, 13) was followed to determine cohesin-dockerin specificity.
Binding to insoluble polysaccharides.
Insoluble xylan was prepared by boiling oat spelt xylan (Sigma Chemical Co., St. Louis, MO) for 30 min in distilled water and recovering the residue by centrifugation; this was followed by 3 washes with distilled water, and the dry weight was determined. Microcrystalline cellulose (Avicel) was purchased from FMC Biopolymer (Philadelphia, PA). The binding of each protein to insoluble polysaccharides (insoluble xylan from oat spelt and microcrystalline cellulose) was determined qualitatively using SDS-PAGE. Pure protein (10 μg for xylan-binding assays and 5 μg for cellulose-binding experiments in 50 mM citrate buffer, pH 6.0, 12 mM CaCl2, 2 mM EDTA) was mixed with 0.5 mg of insoluble xylan or 10 mg of microcrystalline cellulose in a final volume of 100 μl. The tubes were incubated on ice for 1 h with gentle mixing before being centrifuged at 14,000 rpm for 1 min, and the supernatant fluids (containing unbound protein) were carefully removed. The polysaccharide pellet was then washed once by resuspension in 100 μl of the same buffer and by centrifugation before resuspension in 60 μl of SDS-containing buffer and boiling for 10 min to dissociate any bound protein. Bovine serum albumin (BSA) was used as a negative control to ensure specificity of binding. Bound and unbound fractions were analyzed by SDS-PAGE using a 12% polyacrylamide gel.
Nondenaturating PAGE.
To check the full interaction between scaffoldin and enzymes, a differential mobility assay on nondenaturing gels was used. Into a 30-μl reaction mixture (15 μl of Tris-buffered saline, pH 7.4 [TBS], supplemented with 10 mM CaCl2 and 0.05% Tween 20), 4 to 8 μg of each protein was added in an equimolar manner. The 1.5-ml tubes were incubated for 1.5 h at 37°C. Sample buffer (7.5 μl, in the absence of SDS) was added to 15 μl of the reaction mixture, and the samples were loaded onto nondenaturating gels (4.3% stacking/9% separating phase). A parallel SDS-PAGE (10%) was performed on the remaining 15-μl sample.
Enzymatic activity.
Xylanase activity was determined quantitatively by measuring the reducing sugars released from xylan by the dinitrosalicylic acid (DNS) method (22, 39). A typical assay mixture consisted of 100 μl buffer (50 mM citrate buffer, pH 6.0, 12 mM CaCl2, 2 mM EDTA) with enzyme (0 to 10 nM). The reaction was started by adding 100 μl of 2% xylan (birchwood, beechwood, or oat spelt; Sigma Chemical Co., St. Louis, MO) suspended in 50 mM citrate buffer, pH 6.0, and the reaction was continued for 20 min at 50°C. The reaction was stopped by transferring the tubes to an ice-water bath; 100 μl of the supernatant was then added to 150 μl DNS reagent, and the tubes were boiled for 10 min, after which the absorbance was measured at 540 nm. Dockerin-containing enzymes were subjected to a 1.5-h incubation (37°C, in the absence of substrate) in the presence of equimolar concentrations of scaffoldin prior to assay.
Hatched wheat straw (0.2 to 0.8 mm), provided by Valagro (Poitiers, France), was treated as described previously (19, 48): the crude substrate was incubated in distilled water with mild stirring for 3 h at room temperature, vacuum filtered on a 2.7-μm glass filter, resuspended in water, and incubated for 16 h with mild stirring at 4°C. The suspension was filtered and washed three times with water, and a sample was dried at 100°C overnight for estimation of the dry weight. The composition of the assay mixture was the same as described above, except that hatched wheat straw was used at a concentration of 3.5 g/liter and the concentration of xylanases was set at 0.3 μM or 0.2 μM when combined with cellulases. The reaction mixtures were incubated for 17 h at 50°C. All assays were performed in triplicate.
Sugar identification and analysis.
Analysis of sugar content was performed using a high-performance anion-exchange chromatography (HPAEC) system equipped with a PA1 column (Dionex, Sunnyvale, CA). Reaction mixtures were loaded onto the column and eluted with NaOH (200 mM). Sugar concentrations were determined by integration of the chromatographic peaks, based on arabinose, xylose, xylobiose, xylotriose, and cellobiose standards. Low levels of arabinose and xylose were observed in blanks (double-distilled water); these values were deducted in all the samples.
Xylose concentrations were confirmed with a d-xylose assay kit purchased from Megazyme (Wicklow, Ireland), and glucose (absence) was determined using a Glucose Assay Kit GAGO20 (Sigma-Aldrich), according to the manufacturers' instructions.
RESULTS
Construction and expression of recombinant proteins.
The recombinant proteins designed for use in this study are shown schematically in Fig. 1. Three different T. fusca enzymes were used in the study: two xylanases, Xyn11A and Xyn10B, and the family 5 endoglucanase Cel5A. Cel5A is a typical free (noncellulosomal) enzyme that contains a family 2 cellulose-binding CBM. Xyn11A contains a CBM from the same family that shows binding specificity for both cellulose and xylan. Xyn10B lacks a CBM.
In order to convert these T. fusca enzymes into the cellulosomal mode, each was joined to a dockerin of divergent specificity. Cel5A was the topic of a previous study, and a dockerin from R. flavefaciens was used to replace the CBM of the native enzyme, generating f-5A.
Two recombinant forms of Xyn11A were designed: one, 11A-XBM-a, in which the ScaB dockerin from A. cellulolyticus was appended at the C terminus of the original Xyn11A, thus retaining the original catalytic module and XBM, and a second, 11A-a, in which XBM was replaced by the same A. cellulolyticus dockerin. The resultant fusion protein was identical to 11A-XBM-a but lacked the XBM. 11A-a was employed as a crucial control in this study in order to assay the importance of the XBM module in the enzymatic activity of the enzyme alone or within a complex. XBM alone was also examined for its contribution to activity; therefore, the dockerin of scaffoldin A from R. flavefaciens (14) was fused to the XBM module at the N terminus of the protein.
In order to integrate Xyn10B into an enzymatic complex, the dockerin from the exoglucanase Cel48S of C. thermocellum (51) was fused at its C terminus, resulting in 10B-t.
Scaf·AF has two cohesins with divergent specificities, allowing the possibility of binding two different dockerin-containing proteins selectively. The specific modules that comprise the construct are as follows: cohesin 3 from A. cellulolyticus scaffoldin C (designated A) (54); CBM3a from C. thermocellum, which binds strongly to cellulose (42); and cohesin 1 from R. flavefaciens scaffoldin B (designated F) (14). Scaf·AF allows the specific incorporation of the previously described enzymes (either 11A-XBM-a or 11A-a and f-5A) and directs the complex to the substrate via the CBM.
Scaf·AT also has 2 different cohesins and a cellulose-binding CBM. A. cellulolyticus cohesin (A, as specified above) interacts specifically with enzymes carrying the matching dockerin, i.e., 11A-XBM-a or 11A-a. At the C terminus, T—cohesin 3 from the CipA C. thermocellum scaffoldin (55)—binds with the dockerin S-containing enzyme, 10B-t.
Another scaffoldin, Scaf·ATF, was produced and included all three above-described cohesin types, together with the cellulose-binding CBM. This 3-cohesin scaffoldin enabled the integration of the two xylanases, 10B-t and 11A-XBM-a (or 11A-a), and endoglucanase f-5A. All purified recombinant proteins showed a single major band on SDS-PAGE (not shown), and in each case, their mobilities were consistent with their molecular masses.
Affinity-based ELISA.
The specificities of the cohesins for the chimeric dockerin-bearing enzymes were examined semiquantitatively by a sensitive enzyme-linked affinity assay in microtiter plates (2). All of the cohesins in each scaffoldin specifically bound their respective dockerins and did not bind (or bound very poorly) other, nonmatching dockerin-bearing molecules (data not shown). The scaffoldin-borne cohesins bound their matching dockerins just as efficiently as the individual single-cohesin scaffoldins, indicating that the binding capabilities of the scaffoldins were reliable and selective. All specific cohesin-dockerin interactions, for each scaffoldin, were of similar intensities, indicating that similar amounts of protein were bound in each well and suggesting a molar equivalent of a 1:1 scaffoldin (cohesin)/dockerin ratio.
Binding to insoluble polysaccharides.
The majority of the family 10B enzyme was able to bind to insoluble xylan (data not shown); this was due to the inherent binding capacity of the catalytic module only, since the protein does not include any XBM. The same result was obtained for 11A-a, in which the XBM module was replaced by the dockerin module; the enzyme was found in both the bound and unbound fractions. The Xyn11A, 11A-XBM-a, and f-XBM proteins were located in the bound fractions, suggesting that the dockerin module in 11A-XBM-a did not disturb the binding function of the XBM and that the XBM alone is able to bind xylan.
Upon mixing the proteins with microcrystalline cellulose, Xyn11A and 11A-XBM-a were found exclusively in the bound fractions; thus, the cellulose-binding ability of the protein is maintained in 11A-XBM-a and has not been affected by the dockerin module. f-XBM was found in approximately equal portions in both fractions, suggesting that the binding function to Avicel reflects the combined action of the entire protein (catalytic module and XBM) and that the lack of the catalytic module results in a weakened ability to bind the substrate. Indeed, 11A-a, which lacks the XBM, was also found in both fractions (the major part was found in the bound fraction), reinforcing the hypothesis that the protein needs its catalytic module, together with its XBM, to achieve full substrate-binding capacity. The Xyn10B and 10B-t enzymes bound cellulose very weakly, indicating a low but measurable cellulose-binding activity associated with the family 10 catalytic module. As expected, more than ∼95% of the BSA negative control was found in the unbound fraction. These results are in perfect accord with previous publications (28, 34) in which the binding capacities of the wild-type Xyn11A and Xyn10B enzymes were investigated. Xyn11A was found to bind strongly to insoluble xylan and cellulose, and a weak ability to bind insoluble xylan was demonstrated for Xyn10B. In previous experiments with binding to microcrystalline cellulose, Cel5A exhibited an ability to bind to cellulose, whereas f-5A failed to bind cellulose due to a lack of CBM2 (11).
Nondenaturing PAGE.
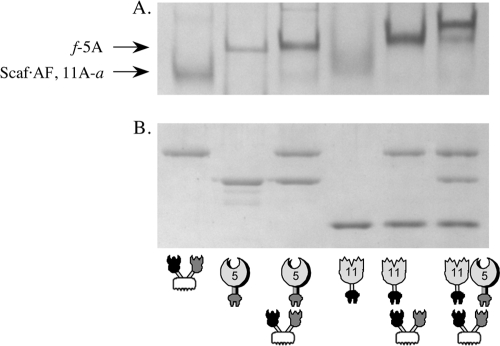
For each chimeric designer cellulosome, complex formation was tested by nondenaturing PAGE. Denaturing PAGE was used as a control for sample content verification. Stoichiometric mixtures of the enzymes and the scaffoldin resulted in a single band with altered mobility (the band strengthened and shifted), indicating that complete or near-complete complexation was achieved in all cases (an example is given in Fig. 2). The stabilities of the complexes were demonstrated after the desired incubation period at 50°C; the designer cellulosomes appeared to be stable at high temperature despite the inclusion of cohesin and dockerin modules derived from mesophilic organisms.
FIG. 2.
Electrophoretic mobility of components and assembled complexes on nondenaturing and denaturing gels. Equimolar concentrations of the chimeric enzymes (f-5A and 11A-a) and their matching scaffoldin (Scaf·AF) were combined. The single fusion proteins and the mixtures were subjected to nondenaturing PAGE (A) and SDS-PAGE (B). Analysis of the matching components by native PAGE clearly showed their complete or near-complete interaction.
Enzymatic activities on xylans.
All recombinant xylanases were tested for xylan degradation on a variety of xylan substrates. Three different substrates were used to test the degradation activities of the transformed xylanases: birchwood xylan, beechwood xylan, and oat spelt xylan. The characteristics (compositions and properties) of xylans from different origins were investigated by Hespell and Cotta in 1995 (26). Birchwood xylan is more than 90% soluble in water and is composed of a high percentage of neutral sugars (87.7% mainly xylose residues; small amounts of glucose, and traces of arabinose and galactose can be found) and 10.2% hexuronic acids. The ratio of sugars in beechwood xylan is comparable to that in birchwood xylan, but they differ in their relative contents of hexuronic acids (less than 3% is found in beechwood xylan) and in their water solubilities: beechwood xylan is approximatively 95% insoluble in water. Oat spelt xylan is a mixture of a high percentage of xylose (84%) and some arabinose, glucose, and galactose; its water solubility varies greatly depending on the temperature and extent of centrifugation. The family 11A enzymes were more effective in the degradation of xylans than the family 10B enzymes. On each substrate, the enzymatic activities of the wild-type family 11 enzyme and its derivatives were very close, suggesting that the addition of the dockerin module had very little effect on the structural conformation of the enzymes. Table 1 summarizes the specific activity values obtained for all chimeras and wild-type constructs produced in this work. Oat spelt xylan was the most efficiently degraded substrate for all the enzymes. For the 11A enzymes, 11A-a had similar specific activities on all three substrates, which were lower than those of the XBM-containing enzymes, i.e., Xyn11A and 11A-XBM-a (which had comparable specific activities). Deletion of the XBM module may have a negative impact on its activity on these substrates, suggesting that the XBM-targeting role is important even for easily degraded substrates, like purified xylan. Interestingly, the specific activities observed for 10B-t were higher than those of wild-type Xyn10B on all substrates, so the addition of the dockerin module may have allowed better access of the catalytic module to its substrate. Xyn10B exhibited the lowest activity on beechwood xylan compared to the other substrates. The solubility properties of the xylan may thus play a role in degradation by this enzyme: the more insoluble the substrate, the more difficult for the enzyme to degrade it. The enzymatic activities of Cel5A and f-5A on a variety of cellulosic substrates were reported earlier (11).
TABLE 1.
Specific activities of recombinant enzymes on various xylans
| Substrate | Sp acta |
||||
|---|---|---|---|---|---|
| Xyn10B | 10B-t | Xyn11A | 11A-a | 11A-XBM-a | |
| Birchwood xylan | 89.9 ± 4.9 | 136 ± 7.6 | 430 ± 1.9 | 337 ± 12.0 | 425 ± 7.0 |
| Oat spelt xylan | 125 ± 2.2 | 152 ± 1.8 | 449 ± 2.7 | 342 ± 6.9 | 433 ± 8.8 |
| Beechwood xylan | 40.3 ± 1.9 | 97.4 ± 6.8 | 438 ± 0.7 | 345 ± 3.8 | 445 ± 6.4 |
Katal/mol enzyme.
Assays of free enzymes on hatched wheat straw.
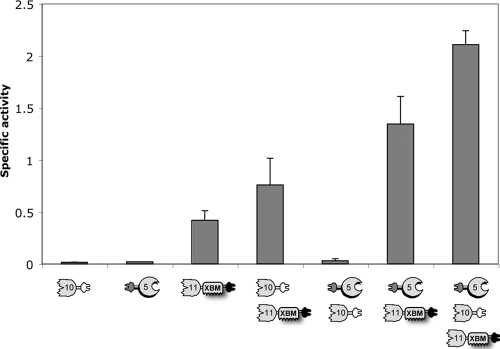
Each of the dockerin-containing enzymes and their combinations were tested for activity on hatched wheat straw (Fig. 3). 10B-t and f-5A exhibited very weak activity on straw, whereas 11A-XBM-a showed higher levels of hydrolysis.
FIG. 3.
Specific activities of single dockerin-containing enzymes or combinations thereof. The composition of the free-enzyme systems is indicated at the bottom of the graph. Each enzyme was assayed at 0.3 μM with 3.5 g/liter hatched wheat straw in a 200-μl reaction mixture. Specific activity is defined as μM reducing sugar per min per μM enzyme. Triplicate samples of each reaction mixture were employed, and standard deviations for straw hydrolysis are indicated.
Synergism was demonstrated for the following combinations: 10B-t plus 11A-XBM-a, 11A-XBM-a plus f-5A, and the three enzymes together (f-5A plus 11A-XBM-a plus 10B-t), with respective activity enhancements of 1.8-, 3.1-, and 4.6-fold (compared to the theoretical total of the individual activities). Notably, synergism was observed within all the combinations containing 11A-XBM-a, suggesting that 11A-XBM-a attacks the straw substrate in such a manner that it allows the other enzymes access to their specific sites on the complex substrate.
Xyn10B proved to be less active than Xyn11A during initial degradation of the different xylans, but the enzyme seemed to contribute to the complete conversion of xylan into xylobiose, as reported earlier (34). The same synergistic action may also occur in the degradation of a more complex substrate, like straw, which could involve divergent cleaving mechanisms and would explain why Xyn10B (or its chimeric derivative, 10B-t) cannot achieve substantial levels of degradation by itself but contributes to the reaction when combined with 11A-XBM-a.
This hypothesis could explain the synergistic activities observed for the combinations of enzymes, i.e., the two-enzyme system 11A-XBM-a plus f-5A and the three-enzyme mixture f-5A plus 11A-XBM-a plus 10B-t. Interestingly, the combination of 10B-t plus f-5A reactions did not show improved activity. Xyn10B and Cel5A (or their chimeric derivatives) may have difficulty in accessing their target substrate within the complex matrix of the straw composite and require association with an additional enzyme, such as Xyn11A, to degrade straw efficiently. The same experiment was carried out with the wild-type enzymes, and equivalent results were obtained (data not shown), indicating that the presence of dockerins in the enzymes does not substantially affect the overall actions of these enzymes on the crude substrate. Additional trials were carried out in subsequent experiments only with the enzyme combinations that showed clear synergistic activity.
Several concentrations of the enzyme combinations were tested in order to ensure that 0.3 μM enzyme provided linear reactions for the given time points (data not shown). Previous results were confirmed, as the three-enzyme system Cel5A plus Xyn11A plus Xyn10B (or f-5A plus 11A-XBM-a plus 10B-t) appeared to be more effective than the enzyme pair Cel5A plus Xyn11A (or f-5A plus 11A-XBM-a), which was more effective than Xyn11A plus Xyn10B (11A-XBM-a plus 10B-t). The same trend was evident for the entire range of experimental data.
Kinetics studies also proved that the reaction was still in the linear part of the curve after 16 to 18 h of enzymatic action (data not shown).
Enzymatic assays of designer cellulosomes on hatched wheat straw.
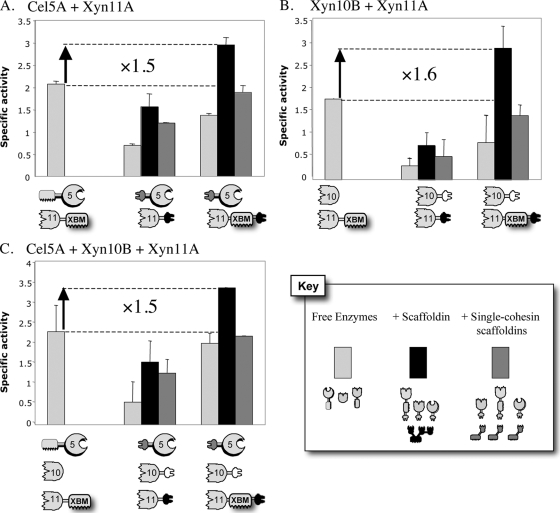
The combination of the cellulase Cel5A with the xylanase Xyn11A was examined in three different modes: (i) the free-enzyme mixture, (ii) the designer cellulosome, and (iii) the individually targeted enzyme system. Designer cellulosomes were constructed by mixing the desired dockerin-containing enzymes with the chimeric scaffoldins bearing the appropriate matching cohesins. The targeting effect was assessed by attaching the individual enzymes to a single-cohesin (CBM-containing) scaffoldin construct. The specific activities of the enzyme mixtures in each of the three modes were compared, and the results are shown in Fig. 4.
FIG. 4.
Comparative degradations of straw by the various complexes and free-enzyme systems. The compositions of the complexes and free-enzyme systems are indicated in symbolic form below each graph. Specific activity is defined as μM reducing sugar per min per μM enzyme. Each reaction was performed in triplicate, and standard deviations for straw hydrolysis are indicated.
As free enzymes (cellulase Cel5A and xylanase Xyn11A), the wild-type enzymes appeared to have better specific activities than the dockerin-bearing enzymes—both f-5A plus 11A-a and f-5A plus 11A-XBM-a (Fig. 4A). This can be explained by the cellulose-binding CBM2 in Cel5A, which targets the cellulase to the cellulose substrate, leading to more efficient degradation. The combination of f-5A plus 11A-XBM-a was also more active than f-5A plus 11A-a, suggesting that the lack of XBM has a negative influence on the capacity of the enzymes to degrade straw. The addition of the matching single-cohesin-bearing CBM to the dockerin-containing enzymes improved their specific activities by returning the cellulose-targeting feature to the enzyme. In fact, in the case of the f-5A-plus-11A-XBM-a combination, the wild-type enzyme activity was almost fully recovered by restoration of the targeting function. In both cases, incorporation of the enzymes into designer cellulosomes served to increase the activity substantially. The resultant enhancement in enzyme activity can be attributed to the proximity effect between the enzymes in the designer cellulosome. Although f-5A plus 11A-a was still less efficient than the wild-type enzymes, the combination of f-5A plus 11A-XBM-a gave a 1.5-fold enhancement compared to the wild-type T. fusca enzymes, demonstrating the impact of assembling the enzymes into a designer cellulosome complex. This also suggests that the XBM in the xylanase 11A provides a major contribution to the overall degradation of the complex cellulosic substrate.
Similar results were also obtained for the combination of the two xylanases, as well as for the three-enzyme system (Fig. 4B and C). In the free state, wild-type enzymes always degraded straw more efficiently than the dockerin-bearing chimeras, due to the fact that in most cases, the dockerin replaces the CBM, which has an important influence (substrate targeting) on the activity. Reactions involving 11A-a showed very weak activities, demonstrating once again the crucial role of the XBM.
Connecting each enzyme to its matching single-cohesin scaffoldin improved activity in each case, confirming the CBM effect. Furthermore, as observed in the combined Xyn11A-plus-Cel5A interaction, placing the enzymes in close proximity via the scaffoldin unit had a significant positive impact on the specific activity. Likewise, complexation of the xylanases (11A-XBM-a plus 10B-t) and the three-enzyme system (11A-XBM-a plus 10B-t plus f-5A) provided activity enhancements of approximately 1.6- and 1.5-fold, respectively, relative to the wild-type enzymes. Nevertheless, complexes including 11A-a showed markedly reduced levels of activity compared to the wild-type enzymes.
Interestingly, parallel experiments using soluble and insoluble xylan as substrates showed no apparent difference between free and complexed enzymes (data not shown), reinforcing the theory that designer cellulosomes are advantageous on complex substrates relative to free enzymes (19).
Sugar analysis.
Sugar concentration and identification were performed using known concentrations of standards, and the relative amounts were calculated via integration of the identified peaks in the given samples. Combinations of free and scaffoldin-borne enzymes were applied to samples of hatched wheat straw, and the degradation products were analyzed. Various quantities of arabinose, cellobiose, xylose, and xylobiose were found in the samples (Table 2).
TABLE 2.
Sugar concentrations obtained by HPLC analysis following digestion of hatched wheat straw for 17 h by various enzyme combinationsa
| Enzyme or combinationb | Sugar concn (μmol/g substrate) |
|||
|---|---|---|---|---|
| Arabinose | Xylose | Xylobiose | Cellobiose | |
| Xyn11A | 20.3 ± 0.4 | 49.3 ± 0.5 | 7.9 ± 0.9 | ND |
| Xyn10B | NDc | ND | ND | ND |
| Cel5A | ND | ND | ND | ND |
| Xyn11A + Xyn10B | 23.6 ± 0.5 | 42.7 ± 0.0 | 24.6 ± 1.2 | ND |
| Scaf (11A-XBM-a + 10B-t) | 26.7 ± 0.2 | 34.7 ± 0.2 | 33.7 ± 0.2 | ND |
| Xyn11A + Cel5A | 21.5 ± 0.4 | 41.5 ± 0.3 | 24.4 ± 1.1 | 20.6 ± 1.3 |
| Scaf (11A-XBM-a + f-5A) | 24.2 ± 0.0 | 20 ± 0.5 | 32.4 ± 2.0 | 22.2 ± 0.5 |
| Xyn11A + Xyn10B + Cel5A | 24.3 ± 0.2 | 48 ± 0.0 | 20.2 ± 1.7 | 21.8 ± 1.4 |
| Scaf (11A-XBM-a + 10B-t + f-5A) | 31 ± 0.3 | 33.3 ± 0.1 | 24.8 ± 1.3 | 27.3 ± 0.7 |
Absence of glucose was confirmed by using a glucose assay kit. Values for xylose were corroborated using a xylose assay kit. An unidentified peak, present only after enzymatic treatments, eluted at ∼3.9 min (between the xylose and xylobiose peaks), suggesting a monosaccharide, or more likely, a modified monosaccharide. The area under the unassigned peak was larger in all designer cellulosome samples, which, together with the unmeasurable xylotriose values, may account for the discrepancies of these data with the reducing sugar assays shown in Fig. 4.
Scaf indicates that the designated chimeric enzymes were complexed to the scaffoldin in the designer cellulosome format.
ND, not detected.
In accordance with the findings shown in Fig. 3, Xyn10B and Cel5A were essentially inactive on the wheat straw substrate. In contrast, Xyn11A alone produced significant amounts of xylobiose and xylose, as well as arabinose, but not cellobiose or glucose (Table 2), indicating its specificity for xylan. Many xylanases exhibit residual activity toward l-arabinose due to the structural similarities between α-l-arabinofuranoside and β-d-xylopyranoside. Relatively large amounts (41.7 ± 1.2 mmol/g substrate) of xylotriose were also produced by Xyn11A treatment of this substrate. When Xyn11A was included in combination with other enzymes, in either the free or cellulosomal mode, xylotriose was also produced but could not be quantified, owing to the presence of contaminating products that caused an erratic baseline in this region. The presence of Cel5A in the reaction mixtures resulted in production of significant quantities of cellobiose, which was absent in samples lacking the cellulase. Incorporation of the dockerin-containing enzyme derivatives into chimeric scaffoldins served to enhance the levels of disaccharides and arabinose at the expense of xylose.
Yield calculations.
Fierobe and colleagues (19) analyzed the wheat straw composition after sulfuric acid treatment. The washed straw was found to contain 3.3 mmol of acid-extractable reducing sugars/g of dry matter, using the method of Park and Johnson (45). Quantification of glucose by high-performance liquid chromatography (HPLC) analysis and the glucose oxidase method indicated that the substrate contained approximately 40% cellulose (2.3 mmol of glucose/g of dry matter). The content of xylose was found to be 0.8 mmol/g of dry matter, whereas the amount of arabinose was around 0.1 mmol/g. Accordingly, reaction yields after 17 h comprised about 8.2% and 9.6% for the bi- and trienzyme designer cellulosome systems, respectively (versus the corresponding yields, 4.9% and 6.3%, of the wild-type enzymes).
Disposition of the XBM.
In order to examine whether the importance of the XBM resides in the structural conformation of Xyn11A or reflects the mere presence of that particular CBM in the complex, we designed a scaffoldin that would include XBM together with a dockerin. The dockerin of R. flavefaciens was attached to the N-terminal end of the XBM from Xyn11A in order to effect its physical separation from the catalytic module. Three complexes were tested to examine the extrinsic contribution of the XBM to straw degradation (Fig. 5): (i) 11A-a plus 10B-t, as a negative control for an XBM-lacking system; (ii) 11A-XBM-a plus 10B-t as a positive control for an XBM-containing system; and (iii) the designer cellulosome (11A-a plus 10B-t plus f-XBM). The results clearly demonstrate that the structural conformation of 11A-XBM-a is responsible for the enhancement of activity and synergism between the two enzymes. Thus, the function of the XBM is dependent on its presence in the native enzyme, since independent addition of the dockerin-fused XBM to higher-order designer cellulosomes had little or no effect on the activity on wheat straw. Both free systems and scaffoldin-bound designer cellulosome systems remained in the same range of efficiency as the 10B-t-plus-11A-a system, which did not contain XBM.
FIG. 5.
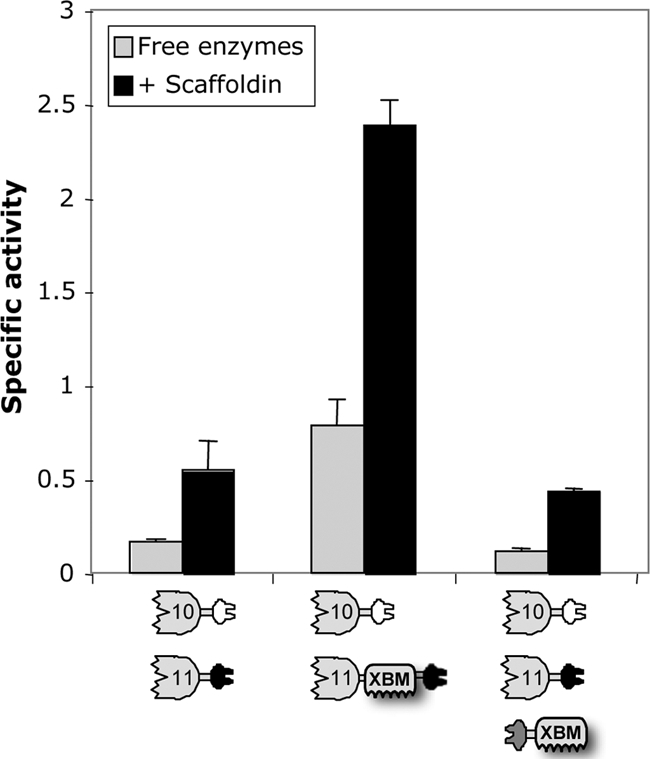
Analysis of the influence of scaffoldin-borne versus native (enzyme-borne) XBM on degradation of wheat straw. The compositions of the complexes and free-enzyme systems are indicated below each graph. Specific activity is defined as μM reducing sugar per min per μM enzyme. Triplicates of each reaction were carried out, and standard deviations for straw hydrolysis are indicated.
DISCUSSION
The original concept of designer cellulosomes was presented in 1994 (5), although many years passed before its experimental verification. During the past decade, the principle of designer cellulosomes was investigated (11-13, 17-19, 40, 41), and enhanced synergistic cooperation between the enzymes in the complex was frequently demonstrated. On the basis of these efforts, some rules have been established, such as the importance of a CBM for substrate targeting (18), the disadvantages of more than one CBM per complex, the advantage of designer cellulosomes on complex substrates (19), the influence of cellulosome geometry (41), and the significance of linker length and dockerin position in enzyme design (11).
The designer cellulosome approach is the method of choice for investigating the contributions of individual components to the overall mechanism of cellulosome action. This approach is the only confirmed way in which specific cellulosome components can be precisely incorporated into defined complexes. In previous work, designer cellulosomes were employed mainly to investigate the actions of known cellulases on different types of purified cellulosic substrates, such as amorphous cellulose, bacterial microcrystalline cellulose, and Avicel. One exception, however, involved a study of the effect of an added xylanase to the cellulase-mediated degradation of a complex cellulosic substrate (hatched wheat straw), which revealed a marked enhancement in its synergistic hydrolysis (19). In the present work, we have extended such studies by comparing the enzymatic activities, on the same natural substrate, of xylanases with and without an added endoglucanase in the free and cellulosomal modes.
In contrast to the earlier work, in which cellulosomal enzymes were used on the wheat straw substrate, we employed enzymes from the cellulolytic aerobe T. fusca, which produces only free (noncellulosomal) enzymes. This strategy provides a defined basal level of activity of the free enzymes, and our capacity to convert these enzymes to the cellulosomal mode via inclusion of a dockerin provides a genuine basis for comparison.
Here, we focused on two different T. fusca xylanases, which were converted from the free state to the cellulosomal state by integrating divergent dockerins into recombinant enzymes. The two xylanases differed in content; one (Xyn11A) contained a family 2 xylan-binding CBM (termed XBM for the purposes of the present report), whereas the other (Xyn10B) lacked a CBM. This allowed us to assess the contribution of the XBM to the degradation of the complex substrate by the designer cellulosomes. In addition, a recombinant dockerin-containing T. fusca family 5 endoglucanase (f-5A) was included in trifunctional designer cellulosomes in order to explore the mixed interaction of the cellulase with the xylanases in the hydrolysis of the complex substrate.
It is significant that Xyn11A interacted synergistically with Xyn10B and/or Cel5A, all with attached dockerins. In contrast, Xyn10B and Cel5A exhibited little or no synergism. Interestingly, enhanced synergism was observed in all Xyn11A-containing combinations; in cases where the intrinsic XBM was maintained, an ∼1.5-fold enzymatic activity enhancement was demonstrated in the designer cellulosome complex compared to the free wild-type enzymes. The observed enhancement in synergistic activity was due to both substrate targeting and enzyme proximity. The XBM within Xyn11A proved to contribute to efficient substrate degradation. Its ability to bind to cellulose, as well as xylan, may indeed be useful to the organism, since xylan is invariably associated with cellulose in plant cell walls. In addition, since cellulose is the definitive structural polysaccharide of the plant cell wall, it serves as a universal binding receptor for plant cell wall hydrolases (23). Thus, the dual binding specificity of the C/XBM of Xyn11A is particularly advantageous for the enzyme in binding to natural substrates.
Clearly, the type and disposition of CBM present on this enzyme defines its contribution to designer cellulosome action. In view of our results, not only is the presence of the particular XBM of Xyn11A important to the degradation of the complex substrate, but its attachment to the enzyme appears to be essential. Indeed, the presence of XBM in the designer cellulosome enhanced synergism only when the natural modular organization of the enzyme was maintained; when the XBM was attached separately to the complex via a dockerin module, the resultant designer cellulosome failed to act in a synergistic manner. This suggests that the XBM plays a role in positioning the parent enzyme within the designer cellulosome complex in the correct orientation with respect to its substrate to achieve efficient degradation (10, 25).
In parallel with the designer cellulosomes approach, another interesting attempt to increase enzyme synergism has been reported recently in the form of multifunctional enzyme conjugates (15, 16). These authors observed an increase in the degradation of natural substrates upon fusing two or three complementary xylan-degrading activities (xylanase, arabinofuranosidase, and xylosidase) into the same polypeptide chain. The main advantage of this strategy compared to designer cellulosomes may be the cost-reducing component in future industrial applications. However, in contrast to designer cellulosomes, strategies involving multifunctional enzymes are limited to small numbers of enzymes and restricted to suboptimal equimolar ratios of enzymes (7-9, 29, 50).
Improvement of xylanase activity has great potential for industrial applications, e.g., prebleaching of kraft pulps and recovering fermentable sugars from natural plant cell wall substrates or directly from hemicellulose (38, 47, 49, 53). Moreover, the worldwide quest to find an alternative to fossil fuels offers major challenges, and studies of designer cellulosomes may eventually provide an approach to meet this challenge (3, 4, 6). In this context, improved degradation of cellulosic biomass, the most abundant renewable source of carbon and energy on our planet, may lead to efficient processes for production of soluble sugars en route to biofuels with concomitant reduction in environmental pollution (27). Understanding and improving the synergistic actions of glycoside hydrolases by means of their incorporation into designer cellulosomes could lead to future cost-effective degradation of plant-derived biomass by overcoming cellulose recalcitrance and thus abolish a major barrier to commercialization of biofuels.
Acknowledgments
We are grateful for the critique and assistance of Ely Morag (Designer Energy, Rehovot, Israel), Dan Goldman (Technion, Haifa, Israel), and Will York and Malcolm O'Neill (The University of Georgia Complex Carbohydrate Research Center, Athens, GA) throughout the stages of preparation of the manuscript.
This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel; by the Brazilian friends of the Weizmann Institute of Science Alternative Energy Research Initiative (research grants from Charles Rothschild, Mario Fleck, and Roberto and Renata Ruhman); by the Technion-Niedersachsen Research Cooperation Program; and by the Israel Science Foundation (grant no. 966/09 and 159/07). Y.S. holds the Erwin and Rosl Pollak Chair in Biotechnology at the Technion, and E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Bachmann, S. L., and A. J. McCarthy. 1991. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microbiol. 57:2121-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, Y., T. Handelsman, D. Nakar, A. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., B. Henrissat, and R. Lamed. 2008. The cellulosome: a natural bacterial strategy to combat biomass recalcitrance, p. 407-426. In M. E. Himmel (ed.), Biomass recalcitrance. Blackwell, London, United Kingdom.
- 4.Bayer, E. A., R. Lamed, and M. E. Himmel. 2007. The potential of cellulases and cellulosomes for cellulosic waste management. Curr. Opin. Biotechnol. 18:237-245. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., Y. Shoham, and R. Lamed. 2008. Cellulosome-enhanced conversion of biomass: on the road to bioethanol, p. 75-96. In J. Wall, C. Harwood, and A. L. Demain (ed.), Bioenergy. ASM Press, Washington, DC.
- 7.Berger, E., D. Zhang, V. V. Zverlov, and W. H. Schwarz. 2007. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol. Lett. 268:194-201. [DOI] [PubMed] [Google Scholar]
- 8.Boisset, C., C. Fraschini, M. Schulein, B. Henrissat, and H. Chanzy. 2000. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 66:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisset, C., C. Petrequin, H. Chanzy, B. Henrissat, and M. Schulein. 2001. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 72:339-345. [DOI] [PubMed] [Google Scholar]
- 10.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caspi, J., Y. Barak, R. Haimovitz, D. Irwin, R. Lamed, D. B. Wilson, and E. A. Bayer. 2009. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl. Environ. Microbiol. 75:7335-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caspi, J., D. Irwin, R. Lamed, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2008. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J. Biotechnol. 135:351-357. [DOI] [PubMed] [Google Scholar]
- 13.Caspi, J., D. Irwin, R. Lamed, Y. Shoham, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatal. Biotransformation 24:3-12. [Google Scholar]
- 14.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, Z. M., K. Wagschal, W. Chen, M. D. Montross, C. C. Lee, and L. Yuan. 2009. Multimeric hemicellulases facilitate biomass conversion. Appl. Environ. Microbiol. 75:1754-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, Z. M., K. Wagschal, C. C. Lee, Q. Kong, K. A. Shen, I. B. Maiti, and L. Yuan. 2009. The construction and characterization of two xylan-degrading chimeric enzymes. Biotechnol. Bioeng. 102:684-692. [DOI] [PubMed] [Google Scholar]
- 17.Fierobe, H.-P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J.-P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras: substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 18.Fierobe, H.-P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J.-P. Belaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras: Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 19.Fierobe, H.-P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, R. Lamed, C. Tardif, J.-P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 20.Fierobe, H.-P., S. Pagès, A. Belaich, S. Champ, D. Lexa, and J.-P. Belaich. 1999. Cellulosome from Clostridium cellulolyticum: molecular study of the dockerin/cohesin interaction. Biochemistry 38:12822-12832. [DOI] [PubMed] [Google Scholar]
- 21.Ghangas, G. S., Y. J. Hu, and D. B. Wilson. 1989. Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J. Bacteriol. 171:2963-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghose, T. K. 1987. Measurements of cellulase activity. Pure Appl. Chem. 59:257-268. [Google Scholar]
- 23.Gilbert, H. J., and G. P. Hazlewood. 1993. Bacterial cellulases and xylanases. J. Gen. Microbiol. 139:187-194. [Google Scholar]
- 24.Haimovitz, R., Y. Barak, E. Morag, M. Voronov-Goldman, R. Lamed, and E. A. Bayer. 2008. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8:968-979. [DOI] [PubMed] [Google Scholar]
- 25.Hammel, M., H.-P. Fierobe, M. Czjzek, V. Kurkal, J. C. Smith, E. A. Bayer, S. Finet, and V. Receveur-Bréchot. 2005. Structural basis of cellulosome efficiency explored by small angle X-ray scattering. J. Biol. Chem. 280:38562-38568. [DOI] [PubMed] [Google Scholar]
- 26.Hespell, R. B., and M. A. Cotta. 1995. Degradation and utilization by Butyrivibrio fibrisolvens H17c of xylans with different chemical and physical properties. Appl. Environ. Microbiol. 61:3042-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himmel, M. E., S.-Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. (Erratum, 316:982.) [DOI] [PubMed] [Google Scholar]
- 28.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irwin, D., L. Walker, M. Spezio, and D. Wilson. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 30.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 31.Jeoh, T. 2004. Modeling and analysis of cellulose hydrolysis by Thermobifida fusca Cel5A, Cel6A, and Cel9A and binary mixtures of these cellulases. Ph.D. thesis. Cornell University, Ithaca, NY.
- 32.Jeoh, T., D. B. Wilson, and L. P. Walker. 2002. Cooperative and competitive binding in synergistic mixtures of Thermobifida fusca cellulases Cel5A, Cel6B, and Cel9A. Biotechnol. Prog. 18:760-769. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, E. A., M. Sakojoh, G. Halliwell, A. Madia, and A. L. Demain. 1982. Saccharification of complex cellulosic substrates by the cellulase system from Clostridium thermocellum. Appl. Environ. Microbiol. 43:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, J. H., D. Irwin, and D. B. Wilson. 2004. Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50:835-843. [DOI] [PubMed] [Google Scholar]
- 35.Lamed, R., and E. A. Bayer. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1-46. [Google Scholar]
- 36.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. DiBartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lytle, B., C. Myers, K. Kruus, and J. H. D. Wu. 1996. Interactions of the CelS binding ligand with various receptor domains of the Clostridium thermocellum cellulosomal scaffolding protein, CipA. J. Bacteriol. 178:1200-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mechaly, A., A. Teplitsky, V. Belakhov, T. Baasov, G. Shoham, and Y. Shoham. 2000. Overproduction and characterization of seleno-methionine xylanase T-6. J. Biotechnol. 78:83-86. [DOI] [PubMed] [Google Scholar]
- 39.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 31:426-428. [Google Scholar]
- 40.Mingardon, F., A. Chanal, A. M. López-Contreras, C. Dray, E. A. Bayer, and H.-P. Fierobe. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mingardon, F., A. Chanal, C. Tardif, E. A. Bayer, and H.-P. Fierobe. 2007. Exploration of new geometries in cellulosome-like chimeras. Appl. Environ. Microbiol. 73:7138-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morag, E., A. Lapidot, D. Govorko, R. Lamed, M. Wilchek, E. A. Bayer, and Y. Shoham. 1995. Expression, purification and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pagès, S., A. Belaich, J.-P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 44.Pagès, S., A. Belaich, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, J. T., and M. S. A. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 46.Poole, D. M., E. Morag, R. Lamed, E. A. Bayer, G. P. Hazlewood, and H. J. Gilbert. 1992. Identification of the cellulose binding domain of the cellulosome subunit S1 from Clostridium thermocellum. FEMS Microbiol. Lett. 99:181-186. [DOI] [PubMed] [Google Scholar]
- 47.Suurnäkki, A., M. Tenkanen, J. Buchert, and L. Viikari. 1997. Hemicellulases in the bleaching of chemical pulps. Adv. Biochem. Eng. Biotechnol. 57:261-287. [DOI] [PubMed] [Google Scholar]
- 48.Tabka, M. G., I. Herpoël-Gimbert, F. Monod, M. Asther, and J. C. Sigoillot. 2006. Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzyme Microb. Technol. 39:897-902. [Google Scholar]
- 49.Viikari, L., M. Pauna, A. Kantelinen, J. Sundquist, and M. Linko. 1986. Bleaching with enzymes, p. 67-69. In Proceedings of the 3rd International Conference on Biotechnology of the Pulp Paper Industry. Swedish Forest Products Research Laboratory, Stockholm, Sweden.
- 50.Walker, L. P., C. D. Belair, D. B. Wilson, and D. C. Irwin. 1993. Engineering cellulase mixtures by varying the mole fraction of Thermomonospora fusca E5 and E3, Trichoderma reesei CBHI, and Caldocellum saccharolyticum β-glucosidase. Biotechnol. Bioeng. 42:1019-1028. [DOI] [PubMed] [Google Scholar]
- 51.Wang, W. K., K. Kruus, and J. H. D. Wu. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, D. B. 2004. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4:72-82. [DOI] [PubMed] [Google Scholar]
- 53.Woodward, J. 1984. Xylanases: functions, properties and applications. Top. Enzyme Ferment. Biotechnol. 8:9-30. [Google Scholar]
- 54.Xu, Q., W. Gao, S.-Y. Ding, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell-surface anchoring protein. J. Bacteriol. 185:4548-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]