Abstract
The order Rickettsiales (Alphaproteobacteria) is a well-known group containing obligate endocellular prokaryotes. The order encompasses three families (Rickettsiaceae, Anaplasmataceae, and Holosporaceae) and a fourth, family-level cluster, which includes only one candidate species, “Candidatus Midichloria mitochondrii,” as well as several unnamed bacterial symbionts. The broad host range exhibited by the members of the “Candidatus Midichloria” clade suggests their eventual relevance for a better understanding of the evolution of symbiosis and host specificity of Rickettsiales. In this paper, two new bacteria belonging to the “Candidatus Midichloria” clade, hosted by two different strains of the ciliate protist Euplotes harpa, are described on the basis of ultrastructural observations, comparative 16S rRNA gene sequence analysis, and an estimation of the percentage of infection. Ultrastructure of these bacteria shows some unusual features: one has an electron-dense cytoplasm, and the other one lacks a symbiosomal membrane. The latter was up to now considered an exclusive feature of bacteria belonging to the family Rickettsiaceae. 16S rRNA gene phylogenetic analysis unambiguously places the new bacteria in the “Candidatus Midichloria” clade, although their phylogenetic relationships with other members of the clade are not clearly resolved. This is the first report of a ciliate-borne bacterium belonging to the “Candidatus Midichloria” clade. On the basis of the data obtained, the two bacteria are proposed as two new candidate genera and species, “Candidatus Anadelfobacter veles” and “Candidatus Cyrtobacter comes.”
The order Rickettsiales belongs to the Alphaproteobacteria class and contains bacteria with an obligate intracellular lifestyle and, often, parasitic habits (7). This group of microorganisms is best known for its medically important genus Rickettsia, whose species can cause mild to severe human diseases (34). Because of its clinical relevance, this genus has been well studied from both the clinical and phylogenetic point of view (for an example, see reference 55). In contrast, little is known about the phylogenetic relationships among and within the less-studied Rickettsiales taxa. This information, however, would be of great importance to shed light on the origin of some relevant features of Rickettsiales, like the parasitic habit and the genesis of their eventual virulence.
According to Dumler et al. (6), the order Rickettsiales encompasses two families, namely, Rickettsiaceae and Anaplasmataceae. The monophyly of both the families is strongly supported by phylogenetic analysis (6). A third family, Holosporaceae, has been proposed by Görtz and Schmidt (16), although deeper phylogenetic investigations are needed to verify the robustness of this clade (21). Recent studies, mainly based on 16S rRNA gene sequence characterization and analysis, showed the existence of a fourth independent clade within the order. Many authors agree that this clade should be considered a new family of Rickettsiales (for example, see references 2 and 15). However, a formal description is still lacking for the clade that, at present, contains only one “Candidatus” species, namely, “Candidatus Midichloria mitochondrii” (41), an intracellular bacterium inhabiting the perimitochondrial space of the tick Ixodes ricinus, the European vector of Lyme disease (2, 23), and of many other hard ticks (8). For this reason, we will refer to this family-level cluster as the “Candidatus Midichloria” clade. Comparative analysis of 16S rRNA gene sequences demonstrated that the “Candidatus Midichloria” clade is highly diversified, showing similarity values among sequences ranging from 82.4% to 99.5%. The broad host range showed by the members of the “Candidatus Midichloria” clade suggests their relevance for a better understanding of the evolution of symbiosis and host specificity among Rickettsiales. Within this clade, several sequences have been obtained from prokaryotes associated with ticks (33, 40, 42, 45, 53, 54) and with bed bugs (38) but also to rather different hosts like amoebae (15), marine sponges (24) (EMBL accession number EU236349; D. Sipkema, unpublished data), hydrae (14), and corals (EMBL accession number FJ425643; W. R. Johnson, unpublished data). Additionally, it has to be mentioned that this novel phylogenetic group also comprises bacteria involved in outbreaks of human and animal diseases. Indeed, a bacterium tentatively named “Montezuma” was detected both in human blood samples from patients with disease with an acute fever and in the blood-feeding ticks Ixodes persulcatus and Haemophysalis concinnae, which likely act as vectors (29). Moreover, Lloyd and colleagues (22) showed a correlation between the presence of a microorganism belonging to the same clade and the occurrence of the so-called strawberry disease in rainbow trout. Although published data strongly suggest that this bacterium plays a role in strawberry disease, its natural reservoir or vector has not yet been discovered. On account of these considerations, every new finding of “Candidatus Midichloria” clade could provide new insights about phylogenesis, host range, and possible new emerging pathogens of this group.
In the present paper, two new bacteria belonging to the “Candidatus Midichloria” clade, hosted by two different strains of the ciliate protist Euplotes harpa, are described on the basis of ultrastructural observations, comparative 16S rRNA gene sequence analysis, and an estimation of the percentage of infection. In addition, three new specific oligonucleotide probes, targeting these prokaryotes, were designed and validated. Identification of host ciliates has been obtained by 18S rRNA gene characterization. As the cultivation of these new bacteria has not been achieved, we propose to classify them as “Candidatus Anadelfobacter veles” and “Candidatus Cyrtobacter comes.”
MATERIALS AND METHODS
Cell cultures.
Strains BOD18 and HS11/7 of ciliates were established by single-cell isolation from environmental samples collected from Bodherne (Bornholm, Baltic Sea, Denmark) during the summer of 1999 and from a coastal pond (termed “Stagno 1”) near the mouth of the Serchio River (Pisa, Tuscany, Italy) during the autumn of 2006, respectively. Both strains were maintained at 19°C in artificial seawater (Red Sea salt; Foster and Smith Aquatics, Rhinelander, WI) with 5 ppt of salt and fed with the photosynthetic flagellate Dunaliella tertiolecta, grown under the same culture conditions.
DNA extraction and small-subunit (SSU) rRNA gene characterization.
As sufficient biomass was obtained only for strain BOD18, not for strain HS11/7, the following steps were performed in different ways for the two strains. Ciliate cells were concentrated by centrifugation at 200 × g for 10 min (BOD18) or by picking them up with a micropipette (HS11/7) and storing them in 70% ethanol. The DNA from both the hosts and symbionts was extracted by the method of Wisotzkey et al. (57) for strain BOD18 or by using the NucleoSpin plant DNA extraction kit (Macherey-Nagel GmbH & Co., Dueren NRW, Germany), following the protocol for mycelium DNA extraction, for strain HS11/7. PCRs were carried out with a Primus 96 Plus thermocycler (MWG Biotech, Ebersberg, Germany). The 18S rRNA gene of host ciliates was amplified using the forward primer 18S F9 (5′-CTG GTT GAT CCT GCC AG-3′ [30]) together with the reverse primer 18S R1513 Hypo (5′-TGA TCC TTC YGC AGG TTC-3′ [35]). The reaction was performed with annealing taking place at 57°C for 35 cycles. PCR products were then sequenced using appropriate internal primers (39) by MWG sequencing customer service (MWG Biotech). 16S rRNA genes of prokaryotic alphaproteobacterial symbionts were amplified using primers 16S alfa F19a (5′-CCT GGC TCA GAA CGA ACG-3′ [52]) and 16S alfa R1517 (5′-TGA TCC AGC CGC AGG TTC-3′ [52]). The reactions were performed using a “touchdown” PCR (5), with annealing taking place at 63°C, 57°C, and 50°C. For the symbionts of strain HS11/7, PCR product was directly sequenced using internal primers (52). For the BOD18 strain, the PCR product was cloned in a plasmid vector (pCR2.1-TOPO and TOPO TA cloning kit from Invitrogen [Carlsbad, CA]) and inserted into chemically competent cells (One Shot TOP10; Invitrogen). Inserted fragments from a representative number of clones were then amplified by control PCR with vector-specific primers M13F and M13R (Invitrogen). The 16S rRNA gene-sized fragments were digested with restriction endonuclease BsuRI (UAB Fermentas, Vilnius, Lithuania). Digested fragments were visualized by electrophoresis on 2% agarose gels and subsequent ethidium bromide staining. Fragments showing an identical electrophoresis pattern were grouped together by restriction fragment length polymorphism (RFLP) analysis. The dominant pattern was characterized by sequencing three cloned inserts via plasmid DNA extraction (NucleoSpin plasmid; Macherey Nagel) and sequencing with primers M13F and M13R.
Phylogenetic analysis.
For phylogenetic reconstructions, a selection of sequences, encompassing 47 sequences of bacteria belonging to the Rickettsiales and to other alphaproteobacterial orders, was used. Phylogenetic trees were built using different analytical methods and different filter sets from the ARB program package (27). The FastDNAml program (32), the TREEPUZZLE program (43), and the PHYML program (17) were applied for maximum likelihood reconstructions. Neighbor joining and maximum parsimony analysis were performed by the Distance program with Kimura correction and DNAPARS program from the PHYLIP package for phylogeny inference (10). DNAPARS and PHYML were performed with a bootstrap analysis of 1,000 pseudoreplicates. Tree topologies were finally compared to recognize stable nodes (26).
Probe design and FISH experiments.
Some preliminary fluorescence in situ hybridization (FISH) experiments were performed on ciliate cells of strains BOD18 and HS11/7 in order to verify the presence of bacterial endosymbionts and to identify the class they belonged to. These preliminary experiments were performed with probes targeting Eubacteria (EUB338 [1]), Archaea (ARCH915 [46]), and the main classes of Proteobacteria (ALF1b, BET42a, and GAM42a [28]). A FISH experiment, with probe Poly_862 (5′-GGC TGA CTT CAC GCG TTA-3′ [50]) specific for bacteria belonging to the genus Polynucleobacter (Betaproteobacteria), was also performed. On the basis of the sequences obtained, three probes, Ana_434 (5′-AAT TTT CCC CAC TAA AAG AAC-3′), Cyrt_1438 (5′-TTG CGA GGT TAG CGC ACC-3′), and EUB338_V (5′-GCT GCC CCC CGT AGG AGT-3′) were designed and synthesized as described elsewhere (36). The Ana_434 probe targets the alphaproteobacterial symbiont of E. harpa HS11/7, while the Cyrt_1438 and EUB338_V probes target the alphaproteobacterial symbiont of E. harpa BOD18. The specificity of the probes was tested on both the ARB database (37) and the RDP database (4). FISH experiments were performed by the method of Manz et al. (28), using different concentrations of formamide. Probe EUB338_V was also used in competition with probe EUB338 (1). The use of the new specific probes also allowed estimation of the percentage of infected ciliate cells for both strains.
Electron microscopy observations.
For transmission electron microscopy (TEM), ciliate cells were fixed in 2.5% glutaraldehyde and 1% OsO4 in cacodylate buffer (0.05 M) (pH 7.4). The cells were then dehydrated in ethanol and embedded in Epon 812 resin. Thin sections were stained with uranyl acetate and lead citrate. Finally, observations were made with a JEOL 100S microscope.
Nucleotide sequence accession numbers.
The EMBL accession numbers of the 18S rRNA gene sequences of strain BOD18 and strain HS11/7 are FN552693 and FN552694, respectively. The EMBL accession numbers of 16S rRNA gene sequence of the alphaproteobacterial symbiont from strain HS11/7 is FN552695. The accession numbers of the 16S rRNA gene clone sequences of the alphaproteobacterial symbiont from strain BOD18 are FN552696 to FN552698.
RESULTS
Characterization of the host ciliate.
Analysis of morphological features, such as the size, shape, number of dorsal kineties, and cirral pattern, indicated that both strains HS11/7 and BOD18 belong to the ciliate species Euplotes harpa.
For both strains, a nearly full-length 18S rRNA gene sequence (1,886 bp) (EMBL accession numbers FN552693 and FN552694 for strains BOD18 and HS11/7, respectively) was obtained. Molecular data confirmed the morphological observations. The two sequences are almost identical: there is only one difference at a single nucleotide position (nucleotide 1675, which is an “A” in HS11/7 and a “T” in BOD18). The 18S rRNA gene sequence of strain HS11/7 is identical to the sequence of Euplotes harpa FSP1.4 that had been collected some years before in the same pond (EMBL accession number AJ811015F [50]) and present one-nucleotide difference with respect to the sequences of E. harpa FC1 (EMBL accession number AJ811016 [50]) and BOD2 (EMBL accession number AJ305252 [35]). In turn, this last sequence is identical to that of BOD18; indeed, both strains were collected at the same time in the same locality, and they differ by two nucleotides respect to the 18S rRNA sequence of strain FC1.
Preliminary FISH experiments.
Preliminary FISH experiments showed the presence in both ciliate strains of two different species of bacterial symbionts. Positive, overlapping signals obtained from probe BET42a, specific for Betaproteobacteria (28), and probe Poly_862 demonstrated the presence of bacteria belonging to the genus Polynucleobacter (Betaproteobacteria). Different bacterial cells occurring in the cytoplasm of the host were positively targeted by the probe ALF1b, specific for Alphaproteobacteria (28), showing the presence of at least another (second) bacterial symbiont belonging to the class Alphaproteobacteria.
Characterization of an alphaproteobacterial symbiont from strain HS11/7.
TEM observations demonstrated the presence of two different bacteria in the cytoplasm of E. harpa HS11/7, showing different morphological features (Fig. 1). Clearly, there were bacteria belonging to the genus Polynucleobacter (Betaproteobacteria), as seen by the long rod-shaped cell with electron-dense peripheral cytoplasm and the presence of nucleoids (19). Another rod-shaped bacterium with a length of 5 to 6 μm and width of 1 to 2 μm and probably the alphaproteobacterial symbiont, was also seen. This bacterium shows an electron-dense cytoplasm and is often surrounded by a clear halo, and in some cases it was possible to see the presence of a symbiosomal membrane from the host.
FIG. 1.
Transmission electron micrographs of Euplotes harpa strain BOD18 (A) and strain HS11/7 (B). Polynucleobacter bacteria are visible (arrows), together with secondary alphaproteobacterial symbionts (arrowheads). Bars, 0.5 μm.
A nearly full-length 16S rRNA gene sequence (1,438 bp; EMBL accession number FN552695) was obtained from strain HS11/7. This sequence has a similarity value of 89.5% with the sequence of “Candidatus Midichloria mitochondrii,” and similarity values ranging from 83.8% to 89.7% with other sequences of bacteria belonging to the “Candidatus Midichloria” clade.
Characterization of an alphaproteobacterial symbiont from strain BOD18.
Polynucleobacter-like bacteria were also observed in the cytoplasm of E. harpa BOD18 by TEM. In addition, morphologically different bacteria could be detected (Fig. 1). These organisms frequently show an irregular shape, their size is about 4 to 5 μm in length and 2 to 3 μm in width, and they are not surrounded by any host membrane, although an adjacent clear zone is often visible.
A nearly full-length 16S rRNA gene sequence was obtained (1,441 bp) as a consensus sequence from three different gene clone sequences (EMBL accession numbers FN552696 for clone 1m, FN552697 for clone 8m, and FN552698 for clone 12l). This sequence has a similarity value of 88.6% with the sequence of “Candidatus Midichloria mitochondrii,” and similarity values ranging from 83.5% to 88.7% with other 16S rRNA gene sequences of bacteria belonging to the “Candidatus Midichloria” clade.
FISH experiments with specific probes and percentage of infection.
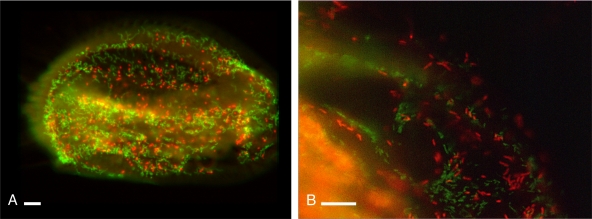
Probe Ana_434 was shown to be highly specific. Indeed, in silico, it matched only the sequence obtained from the HS11/7 symbiont, as no other matches were found screening both the ARB database and the RDP database. In FISH experiments, probe Ana_434 never cross-reacted with the symbionts belonging to the genus Polynucleobacter that were recognized by the probe Poly_862. Probe Ana_434 gave a strong and clear signal in FISH experiments with bacteria found in the cytoplasm of E. harpa strain HS11/7, confirming that the obtained sequence belongs to the symbiont (Fig. 2). FISH experiments were performed on 63 ciliate cells, and all of these cells contained the symbiont.
FIG. 2.
Results of FISH on Euplotes harpa strain HS11/7 (A) and BOD18 (B). (A) FISH performed with green-labeled probe Poly_862, targeting the primary symbionts of the genus Polynucleobacter, and red-labeled probe Ana_434, targeting “Candidatus Anadelfobacter veles.” (B) FISH performed with green-labeled probe Poly_862 and red-labeled probe Cyrt_1438, targeting “Candidatus Cyrtobacter comes.” Note that the signal from the two different probes on the same cell never overlaps. Bars, 10 μm.
In silico, probe Cyrt_1438 matched 10 and 107 alphaproteobacterial sequences in the ARB and RDP databases, respectively. None of these alphaproteobacteria belong to the Rickettsiales order. Results of FISH experiments showed that this probe binds perfectly to its target organism (Fig. 2) and never cross-reacts with symbionts belonging to the Polynucleobacter genus. The presence of a mutation in a highly conserved region allowed us to design a second probe, targeting position 338 (Escherichia coli). This probe (EUB338_V) matches 2,983 sequences in the RDP database (most of them in the group “Bacteria incertae sedis”), including 32 sequences of Rickettsiales bacteria. Although less specific, FISH experiments showed that probe EUB338_V binds perfectly to the target organism (the alphaproteobacterial symbiont of E. harpa BOD18) and discriminates between this bacterium and different prokaryotes (Polynucleobacter symbionts or engulfed bacteria), when used under appropriate stringency conditions (30% formamide and in competition with probe EUB338 [1]). If not used simultaneously, probes EUB338 and EUB338_V recognize the same organisms (cross-react). Positive results of FISH experiments with both probes on ciliate cells showed that the alphaproteobacterial symbiont of E. harpa BOD18 has a prevalence of 88% (50 ciliate cells observed).
Double hybridization with the new specific probes and alphaproteobacterial probe ALF1b excluded the presence of additional alphaproteobacterial symbionts in both ciliate strains. The sequences of the newly designed probes were deposited at probeBase (25).
Phylogenetic analysis.
Phylogenetic analysis showed that sequences belonging to organisms of the same family grouped together in strongly supported clades. Indeed, all the family-level groups, including the “Candidatus Midichloria” clade, are supported by high bootstrap values (>90%) regardless of the algorithm and filter set. Sequences from bacteria belonging to the Holospora and Caedibacter genera, together with sequences from an Acanthamoeba polyphaga symbiont and from a bacterium associated with fleas, form an independent clade, which is supported by lower bootstrap values. Relationships among different families were constant in all calculated trees, although not always supported by high bootstrap values. Sequences from bacteria belonging to the Holospora and Caedibacter genera occupy a basal position within the order, and the Anaplasmataceae family is always the sister group of the “Candidatus Midichloria” clade. Within this clade, relationships among different subclades are not stable. While the association among the sequences of “Candidatus Midichloria mitochondrii,” Candidatus Nicolleia massiliensis,” and other bacteria associated with ticks as well as a bacterium associated with the strawberry disease of trout resulted in a well-defined subclade, association of the sequence from E. harpa HS11/7 symbiont with this subclade was supported only by very low bootstrap values (under 46%) in maximum parsimony (MP)- and maximum likelihood (ML)-based trees. The sequence retrieved from the symbiont of E. harpa BOD18 occupies a deep branching position with respect to the whole “Candidatus Midichloria” clade (Fig. 3).
FIG. 3.
Maximum likelihood phylogenetic tree of the order Rickettsiales, inferred from 16S rRNA gene sequences. Numbers at bifurcations represent bootstrap values on 1,000 pseudoreplicates (values below 70% are not shown). Accession numbers are shown in parentheses. Bar, 10 nucleotide substitutions in 100 nucleotides.
DISCUSSION
The 18S rRNA sequence data unambiguously assign the two ciliate strains to the Euplotes harpa species. In the cytoplasm of each strain, two different bacterial symbionts were detected by TEM observations and FISH experiments. The presence of bacteria belonging to the Polynucleobacter genus (Betaproteobacteria) was actually expected. These prokaryotes have already been reported as obligate symbionts of the ciliate species E. harpa (47, 50). The presence of a second bacterial symbiont in Polynucleobacter-bearing host ciliates has previously been detected in Euplotes eurystomus (20), Euplotes patella (20), Euplotes octocarinatus (12, 20), and E. harpa (35, 48, 50). There may even be a connection between the presence of the primary symbiont and the colonization of the cytoplasm by a second infective species, as previously described for the ciliate symbiont Holospora (13). The importance of Polynucleobacter bacteria for Euplotes spp. is well documented (18, 20, 49, 50), but nothing is known about the nature of the relationship between the secondary symbionts and their hosts. Our data show a very high infection rate for both symbionts in the strains studied (88% in E. harpa BOD18 and 100% in E. harpa HS11/7). Moreover, the results were observed over the course of a year. Therefore, it seems possible that the association of these bacteria with their host ciliates could not be only an opportunistic and episodic event, although this hypothesis needs to be confirmed by research on a broader sample collection. The presence of secondary symbionts in Euplotes has been known for many years (for example, see reference 20). Despite this, little is known about their phylogenetic relationship. The present paper reports for the first time a complete characterization of two secondary symbionts of Euplotes.
Besides defining size and shape, ultrastructural observations highlighted some peculiar morphological features of the two alphaproteobacteria under investigation. Indeed, it must be mentioned that each of the two organisms shows morphological features that are typical of many bacteria belonging to the order Rickettsiales. Symbionts of E. harpa strain HS11/7 possess an electron-dense cytoplasm. While a symbiosomal vacuole was visible only in some cases for the symbionts of E. harpa strain HS11/7, symbionts of E. harpa strain BOD18 always live free in the host cytoplasm, not surrounded by any symbiosomal membrane. Both strains generally present an adjacent clear zone. The absence of a symbiosomal membrane in many Rickettsiales has already been reported. In fact, to date, the absence of a symbiosomal membrane was considered a distinctive feature of members of the family Rickettsiaceae (6), while members of the Anaplasmataceae family are included in a vacuole in the host cell. Among organisms belonging to the “Candidatus Midichloria” clade, an ultrastructural description is available only for bacteria associated with Hydra (14), for “Candidatus Midichloria mitochondrii” (2, 3, 8), and for Acanthamoeba symbionts (15). While the presence of a host-derived membrane is not discussed for bacteria of Acanthamoeba, both in the case of symbionts of Hydra and “Candidatus Midichloria mitochondrii” such a membrane has been reported. As bacteria associated with strain BOD18 occupy a deep branching position in our phylogenetic reconstructions, it could be speculated that the lack of a symbiosome is an ancestral feature for members of the “Candidatus Midichloria” clade, though some members of closely related clades (i.e., Anaplasmataceae) do not show such a feature. Morphotypes with an electron-dense cytoplasm were previously reported for two species of the Rickettsia genus (44). This appearance is also very similar to that of “Candidatus Midichloria mitochondrii,” while bacteria of Acanthamoeba and of Hydra look quite different from this point of view.
Molecular characterization of the two alphaproteobacterial symbionts clearly shows that these microorganisms belong to the order Rickettsiales. As stated before, this order comprises obligately intracellular bacteria, hosted by a wide range of organisms, including ciliated protists. Well-known Rickettsiales bacteria living inside ciliates belong to the genera Caedibacter and Holospora, which are classified as “incertae sedis” and in the family Holosporaceae (16), respectively, and placed in a phylogenetic basal position within the order. Recently, a novel member of the family Holosporaceae, “Candidatus Paraholospora nucleivisitans,” has been found in Paramecium sexaurelia and characterized (9), and two more bacteria belonging to the family Rickettsiaceae were described as symbionts of ciliates of the genera Pseudomicrothorax (11) and Diophrys (51). Molecular phylogenetic analysis shows that the alphaproteobacterial symbionts of E. harpa strains HS11/7 and BOD18 are always included in the “Candidatus Midichloria” clade. This is the first report of ciliate-borne Rickettsiales species belonging to this group.
At this time, the data available do not allow a clear reconstruction of phylogenetic relationships within the “Candidatus Midichloria” clade; hence, it is not yet possible to make direct inferences about the evolutionary history of this group and the identity of its ancestral host. Recent studies hypothesize, with some caution, that arthropods could represent the ancestral hosts for at least three groups of the order Rickettsiales, namely, the Rickettsiaceae and Anaplasmataceae families and the “Candidatus Midichloria” clade (56). The present work accounts for the fact that the “protist-hosted” condition is present in three out of the four major lineages of Rickettsiales (except Anaplasmataceae) including the early branching Holosporaceae. Hence, the most parsimonious interpretation of our data points to a reevaluation of the protist-hosted status as the ancestral condition of the whole order with protists acting as an evolutionary “training ground” for intracellular bacteria before they achieved the ability to infect higher eukaryotes (31).
The phylogenetic cluster of “Candidatus Midichloria mitochondrii,” which includes the symbionts studied, comprises bacteria hosted by organisms living in a wide range of different habitats, such as terrestrial, marine, freshwater, and brackish water environments. Some of these bacteria were also associated with pathological conditions (22, 29). In the case of the so-called strawberry disease of trout, the possible etiologic agent has been identified, but nothing is known about its natural reservoir in the environment (22). Finding phylogenetically related bacteria in protist ciliates, living in freshwater and brackish water habitats, may help to settle this matter.
Description of “Candidatus Anadelfobacter veles” gen. nov., sp. nov.
Anadelfobacter veles (A.na.de.lfo.bac′ter. N.L. masc. n. bacter, a rod; N.L. masc. n., Anadelfobacter; ve′les. L. n. veles, a light-armed forefront soldier [since its description precedes, as a vanguard, those of the bulk of “Candidatus Midichloria” clade species, which will follow later]).
Rod-shaped bacterium, up to 2 μm wide and up to 6 μm long. Electron-dense cytoplasm, no visible inclusions. Gram-negative cell wall organization. Surrounded by a host membrane, frequently also by a clear zone. Lives in the cytoplasm of the protist ciliate Euplotes harpa; identified in strain HS11/7. Basis of assignment: 16S rRNA gene sequence (EMBL accession number FN552695) and positive matching with the 16S rRNA-targeting oligonucleotide probe Ana_434 (5′-AAT TTT CCC CAC TAA AAG AAC-3′). Uncultured thus far.
Description of “Candidatus Cyrtobacter comes” gen. nov., sp. nov.
Cyrtobacter comes (Cyr.to.bac′ter. G. adj. kyrtòs, humped [because of cell shape], N.L. masc. n. bacter, a rod; N.L. masc. n. Cyrtobacter; co′mes. L. n. còmes, companion [of Polynucleobacter bacteria within Euplotes cell]).
Rod-shaped bacterium, often presenting an irregular shape. Up to 3 μm wide and up to 5 μm long. No visible inclusions, Gram-negative cell wall organization. Lives free in the cytoplasm of the protist ciliate E. harpa, frequently surrounded by a clear zone. Identified in strain BOD18. Basis of assignment: 16S rRNA gene sequence (EMBL accession numbers of clone sequences FN552696 to FN552698), and positive matching both with the 16S rRNA-targeting oligonucleotide probes Cyrt_1438 (5′-TTG CGA GGT TAG CGC ACC-3′) and EUB338_V (5′-GCT GCC CCC CGT AGG AGT-3′). Uncultured thus far.
Acknowledgments
This work was supported in part by a fellowship of the Bayerische Forschungsstiftung and by an EMBO short-term fellowship.
We thank S. Galati and H. Mancini for their help in performing 16S rRNA gene characterization and FISH experiments and J. Euzéby for revision of etymology. S. Gabrielli is gratefully acknowledged for photographic artwork.
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beninati, T., N. Lo, L. Sacchi, C. Genchi, H. Noda, and C. Bandi. 2004. A novel alpha-proteobacterium invades the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl. Environ. Microbiol. 70:2596-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beninati, T., M. Riegler, I. M. E. Vilcins, L. Sacchi, R. McFadyen, M. Krockenberger, C. Bandi, S. L. O'Neill, and N. Lo. 2009. Absence of the symbiont Candidatus Midichloria mitochondrii in the mitochondria of the tick Ixodes holocyclus. FEMS Microbiol. Lett. 299:241-247. [DOI] [PubMed] [Google Scholar]
- 4.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 7.Dumler, J. S., and D. H. Walker. 2005. Order II. Rickettsiales Gieszczykiewicz 1939, 25AL emend. Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa 2001, 2156, p. 96-160. In G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. II, part C. Springer-Verlag, New York, NY. [Google Scholar]
- 8.Epis, S., D. Sassera, T. Beninati, N. Lo, L. Beati, J. Piesman, L. Rinaldi, K. D. McCoy, A. Torina, L. Sacchi, E. Clementi, M. Genchi, S. Magnino, and C. Bandi. 2008. Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 135:485-494. [DOI] [PubMed] [Google Scholar]
- 9.Eschbach, E., M. Pfannkuchen, M. Schweikert, D. Drutschmann, F. Brümmer, S. I. Fokin, W. Ludwig, and H. D. Görtz. 2009. “Candidatus Paraholospora nucleivisitans,” an intracellular bacterium in Paramecium sexaurelia shuttles between the cytoplasm and the nucleus of its host. Syst. Appl. Microbiol. 32:490-500. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP-Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 11.Ferrantini, F., S. I. Fokin, L. Modeo, I. Andreoli, F. Dini, H. D. Goertz, F. Verni, and G. Petroni. 2009. “Candidatus Cryptoprodotis polytropus,” a novel Rickettsia-like organism in the ciliated protist Pseudomicrothorax dubius (Ciliophora, Nassophorea). J. Eukaryot. Microbiol. 56:119-129. [DOI] [PubMed] [Google Scholar]
- 12.Ferrantini, F., S. I. Fokin, C. Vannini, L. Modeo, F. Verni, and G. Petroni. 2007. Ciliates as natural hosts of many novel Rickettsia-like bacteria. Abstr. Fifth Eur. Cong. Protistol. and Sixth Eur. Conf. Ciliate Biol., 23 to 28 July 2007, St. Petersburg, Russia. Protistology 5:28-29. http://protistology.ifmo.ru/num5_1/abstracts.pdf. [Google Scholar]
- 13.Fokin, S. O., I. N. Skovorodkin, M. Schweikert, and H. D. Goertz. 2004. Co-infection of the macronucleus of Paramecium caudatum by free-living bacteria together with the infectious Holospora obtusa. J. Eukaryot. Microbiol. 51:417-424. [DOI] [PubMed] [Google Scholar]
- 14.Fraune, S., and T. C. G. Bosch. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U. S. A. 104:13146-13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K.-H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Görtz, H. D., and H. J. Schmidt. 2005. Family III. Holosporaceae fam. nov., p. 146-160. In G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. II, part C. Springer-Verlag, New York, NY. [Google Scholar]
- 17.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 18.Heckmann, K. 1975. Omikron, ein essentieller Endosymbiont von Euplotes aediculatus. J. Protozool. 22:97-104. [Google Scholar]
- 19.Heckmann, K., and H. J. Schmidt. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37:456-457. [Google Scholar]
- 20.Heckmann, K., R. Ten Hagen, and H. D. Goertz. 1983. Freshwater Euplotes species with a 9 type 1 cirrus pattern depend upon endosymbionts. J. Protozool. 30:284-289. [Google Scholar]
- 21.Lee, K. B., C. T. Liu, Y. Anzai, H. Kim, T. Aono, and H. Oyaizu. 2005. The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erithrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55:1907-1919. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd, S. J., S. E. LaPatra, K. R. Snekvik, S. St.-Hilaire, K. D. Cain, and D. R. Call. 2008. Strawberry disease lesions in rainbow trout from southern Idaho are associated with DNA from a Rickettsia-like organism. Dis. Aquat. Org. 82:111-118. [DOI] [PubMed] [Google Scholar]
- 23.Lo, N., T. Beninati, D. Sassera, E. A. P. Bouman, S. Santagati, L. Gern, V. Sambri, T. Masuzawa, J. S. Gray, T. G. T. Jaenson, A. Bouattour, M. J. Kenny, E. S. Guner, I. G. Kharitonenkov, I. Bitam, and C. Bandi. 2006. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ. Microbiol. 8:1280-1287. [DOI] [PubMed] [Google Scholar]
- 24.Longford, S. R., N. A. Tujula, G. R. Crocetti, A. J. Holmes, C. Holmström, S. Kjelleberg, P. D. Steinberg, and M. W. Taylor. 2007. Comparison of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 48:217-229. [Google Scholar]
- 25.Loy, A., F. Maixner, M. Wagner, and M. Horn. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:D800-D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K.-H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Y. Kumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manz, W., R. I. Amann, W., M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 29.Mediannikov, O. I., L. I. Ivanov, M. Nishikawa, R. Saito, I. N. Sidel'nikov, N. I. Zdanovskaia, E. V. Mokretsova, I. V. Tarasevich, and H. Suzuki. 2004. Microorganism “Montezuma” of the order Rickettsiales: the potential causative agent of tick-borne disease in the far east of Russia. Zh. Mikrobiol. Epidemiol. Immunobiol. January-February:7-13. (In Russian.) [PubMed]
- 30.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified 16S-like rRNA coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 31.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 33.Parola, P., J. P. Cornet, Y. O. Sanogo, R. S. Miller, H. Van Thien, J. P. Gonzalez, D. Raoult, S. R. Telford III, and C. Wongsrichanalai. 2003. Detection of Ehrlichia spp., Anaplasma spp., Rickettsia spp., and other eubacteria in ticks from the Thai-Myanmar border and Vietnam. J. Clin. Microbiol. 41:1600-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parola, P., and D. Raoult. 2001. Tick-borne bacterial diseases emerging in Europe. Clin. Microbiol. Infect. 7:80-83. [DOI] [PubMed] [Google Scholar]
- 35.Petroni, G., F. Dini, F. Verni, and G. Rosati. 2002. A molecular approach to the tangled intrageneric relationships underlying phylogeny in Euplotes (Ciliophora, Spirotrichea). Mol. Phylog. Evol. 22:118-130. [DOI] [PubMed] [Google Scholar]
- 36.Petroni, G., G. Rosati, C. Vannini, L. Modeo, F. Dini, and F. Verni. 2003. In situ identification by fluorescently labeled oligonucleotide probes of morphologically similar, closely related ciliate species. Microb. Ecol. 45:156-162. [DOI] [PubMed] [Google Scholar]
- 37.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. G. Ludwig, J. Peplies, and F. O. Glockner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard, S., P. Seng, P. Parola, D. Raoult, B. Davoust, and P. Brouqui. 2009. Detection of a new bacterium related to “Candidatus Midichloria mitochondrii” in bed bugs. Clin. Microbiol. Infect. 15:84-85. [DOI] [PubMed] [Google Scholar]
- 39.Rosati, G., L. Modeo, M. Melai, G. Petroni, and F. Verni. 2004. A multidisciplinary approach to describe protists: a morphological, ultrastructural, and molecular study on Peritromus kahli Villeneuve-Brachon, 1940 (Ciliophora, Heterotrichea). J. Eukaryot. Microbiol. 51:49-59. [DOI] [PubMed] [Google Scholar]
- 40.Sanogo, Y. O., P. Parola, S. Shpynov, J. L. Camicas, P. Brouqui, G. Caruso, and D. Raoult. 2003. Genetic diversity of bacterial agents detected in ticks removed from asymptomatic patients in Northeastern Italy. Ann. N. Y. Acad. Sci. 990:182-190. [DOI] [PubMed] [Google Scholar]
- 41.Sassera, D., T. Beninati, C. Bandi, E. A. P. Bouman, L. Sacchi, M. Fabbi, and N. Lo. 2006. “Candidatus Midichloria mitochondrii,” an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 56:2535-2540. [DOI] [PubMed] [Google Scholar]
- 42.Schabereiter-Gurtner, C., W. Lubitz, and S. Rolleke. 2003. Application of broad range 16S rRNA PCR amplification and DGGE fingerprinting for detection of tick-infecting bacteria. J. Microbiol. Methods 52:251-260. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 44.Silverman, D. J. 1991. Some contributions of electron microscopy to the study of rickettsiae. Eur. J. Epidemiol. 7:200-206. [DOI] [PubMed] [Google Scholar]
- 45.Spitalská, E., V. Boldis, Z. Kostanová, E. Kocianová, and K. Stefanidesová. 2008. Incidence of various tick-borne microorganisms in rodents and ticks of central Slovakia. Acta Virol. 52:175-179. [PubMed] [Google Scholar]
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons Ltd., Chichester, England.
- 47.Vannini, C., M. Pöckl, G. Petroni, Q. L. Wu, E. Lang, E. Stackebrandt, M. Schrallhammer, P. M. Richardson, and M. W. Hahn. 2007. Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria). Environ. Microbiol. 9:347-359. [DOI] [PubMed] [Google Scholar]
- 48.Vannini, C., F. Verni, G. Rosati, W. Ludwig, K. H. Schleifer, and G. Petroni. 2007. Characterization of Euplotes harpa secondary prokaryotic symbionts. Società Italiana di Protozoologia 26th Congresso Nazionale, 30 June to 1 July 2005, Macerata. J. Eukaryot. Microbiol. 54:30S. [Google Scholar]
- 49.Vannini, C., S. Lucchesi, and G. Rosati. 2007. Polynucleobacter: symbiotic bacteria in ciliates compensate for a genetic disorder in glycogenolysis. Symbiosis 44:85-91. [Google Scholar]
- 50.Vannini, C., G. Petroni, F. Verni, and G. Rosati. 2005. Polynucleobacter bacteria in the brackish-water species Euplotes harpa (Ciliata, Hypotrichia). J. Eukaryot. Microbiol. 52:116-122. [DOI] [PubMed] [Google Scholar]
- 51.Vannini, C., G. Petroni, F. Verni, and G. Rosati. 2005. A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49:434-442. [DOI] [PubMed] [Google Scholar]
- 52.Vannini, C., G. Rosati, F. Verni, and G. Petroni. 2004. Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of “Candidatus Devosia euplotis.” Int. J. Syst. Evol. Microbiol. 54:1151-1156. [DOI] [PubMed] [Google Scholar]
- 53.van Overbeek, L., F. Gassner, C. L. van der Plas, P. Kastelein, U. Nunes-da Rocha, and W. Takken. 2008. Diversity of Ixodes ricinus tick-associated bacterial communities from different forests. FEMS Microbiol. Ecol. 66:72-84. [DOI] [PubMed] [Google Scholar]
- 54.Venzal, J. M., A. Estrada-Peña, A. Portillo, A. J. Mangold, O. Castro, C. G. de Souza, M. L. Félix, L. Pérez-Martínez, S. Santibánez, and J. A. Oteo. 2008. Detection of alpha- and gamma-proteobacteria in Amblyomma triste (Acari: Ixodidae) from Uruguay. Exp. Appl. Acarol. 44:49-56. [DOI] [PubMed] [Google Scholar]
- 55.Vitorino, L., I. M. Chelo, F. Bacellar, and L. Ze-Ze. 2007. Rickettsiae phylogeny: a multigenic approach. Microbiology 153:160-168. [DOI] [PubMed] [Google Scholar]
- 56.Weinert, L. A., J. H. Werren, A. Aebi, G. N. Stone, and F. M. Jiggins. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wisotzkey, J. D., P. Jurtshuk, and G. E. Fox. 1990. PCR amplification of 16S rDNA from lyophilized cell cultures facilitates studies in molecular systematics. Curr. Microbiol. 21:325-327. [DOI] [PubMed] [Google Scholar]