Abstract
Caprazamycins are antimycobacterials produced by Streptomyces sp. MK730-62F2. Previously, cosmid cpzLK09 was shown to direct the biosynthesis of caprazamycin aglycones, but not of intact caprazamycins. Sequence analysis of cpzLK09 identified 23 genes involved in the formation of the caprazamycin aglycones and the transfer and methylation of the sugar moiety, together with genes for resistance, transport, and regulation. In this study, coexpression of cpzLK09 in Streptomyces coelicolor M512 with pRHAM, containing all the required genes for dTDP-l-rhamnose biosynthesis, led to the production of intact caprazamycins. In vitro studies showed that Cpz31 is responsible for the attachment of the l-rhamnose to the caprazamycin aglycones, generating a rare acylated deoxyhexose. An l-rhamnose gene cluster was identified elsewhere on the Streptomyces sp. MK730-62F2 genome, and its involvement in caprazamycin formation was demonstrated by insertional inactivation of cpzDIII. The l-rhamnose subcluster was assembled with cpzLK09 using Red/ET-mediated recombination. Heterologous expression of the resulting cosmid, cpzEW07, led to the production of caprazamycins, demonstrating that both sets of genes are required for caprazamycin biosynthesis. Knockouts of cpzDI and cpzDV in the l-rhamnose subcluster confirmed that four genes, cpzDII, cpzDIII, cpzDIV, and cpzDVI, are sufficient for the biosynthesis of the deoxysugar moiety. The presented recombineering strategy may provide a useful tool for the assembly of biosynthetic building blocks for heterologous production of microbial compounds.
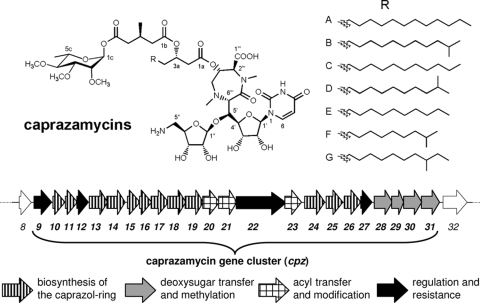
Caprazamycins are potent antimycobacterials isolated from Streptomyces sp. MK730F-62F2 (23). In a pulmonary tuberculosis mouse model, they showed a therapeutic effect but no significant toxicity (24). The caprazamycins are assigned to the translocase I inhibitors (27) due to their structural similarity to the liposidomycins (34, 43), which have been studied in more detail. Translocase I catalyzes the first step in the membrane-linked reaction cycle of bacterial cell wall formation (52): the transfer of phospho-N-acetylmuramic acid-l-Ala-γ-d-Glu-m-diaminopimelic acid-d-Ala-d-Ala from UMP to the lipid carrier undecaprenyl phosphate. Structurally unique in nature, the caprazamycins and liposidomycins share a 5′-β-O-aminoribosyl-glycyluridine and a rare N-methylated diazepanone as their characteristic feature (Fig. 1) (25, 53). Attached at the 3″ position are β-hydroxylated fatty acid groups of different chain lengths, carrying a 3-methylglutarate. While the liposidomycins are sulfated, the caprazamycins lack this group. Instead, they are glycosylated with a 2,3,4-O-methyl-l-rhamnose and therefore belong to the large number of bioactive compounds containing 6-deoxyhexoses. Usually, these moieties contribute significantly to the compounds' properties, influencing, e.g., molecule-target interactions, cell import and export, pharmacokinetics, and solubility (56). The biosynthesis of deoxysugars has been studied in detail and generally starts from NDP-activated hexoses via 4-keto-6-deoxy intermediates (36). The formation of l-rhamnose involves four enzymes, a dTDP-d-glucose synthase, a dTDP-d-glucose 4,6-dehydratase, a dTDP-3,5-deoxyglucose epimerase, and a 4-ketoreductase. Recent advances in combinatorial biosynthesis have led to a variety of novel natural products with an engineered glycosylation pattern and altered bioactivity (37, 44). The key step in this approach is the attachment of different deoxysugars to the aglycones, which demands substrate-flexible glycosyltransferases.
FIG. 1.
Structure of caprazamycins and organization of the caprazamycin gene cluster (cpz9 to cpz31) lacking the genes for deoxysugar formation. Assignments of genes to different steps in the biosynthesis are indicated.
We recently reported cloning and heterologous expression of cosmid cpzLK09 containing the first identified gene cluster of a translocase I inhibitor, the caprazamycins. However, genes for the formation of the dTDP-l-rhamnose could not be identified on this cosmid (Fig. 1) and therefore only caprazamycin aglycones were produced by the heterologous host, Streptomyces coelicolor M512/cpzLK09 (31). The absence of genes for the formation of the l-rhamnose moiety within the corresponding biosynthetic gene cluster has previously been reported for aranciamycin (38), steffimycin (16), spinosyn (55), and elloramycin (9). Since potential genes for O methylation (cpz28 to cpz30) and a glycosyltransferase (cpz31) are encoded in the caprazamycin gene cluster, we speculated that only four genes are missing for successful heterologous production of intact caprazamycins.
Here we report the identification of the genes required for the biosynthesis of the caprazamycin deoxysugar moiety elsewhere on the genome of Streptomyces sp. MK730-62F2. A new strategy was developed, based on Red/ET-mediated recombination, to assemble the identified subcluster into cosmid cpzLK09. Expression of the assembled cluster readily resulted in the production of intact caprazamycins in the heterologous producer strain. Moreover, in vitro studies demonstrated that Cpz31 is the glycosyltransferase in caprazamycin biosynthesis.
MATERIALS AND METHODS
Bacterial strains and general methods.
Chemicals and microbiological and molecular biological agents were purchased from standard commercial sources. Streptomyces sp. MK730-62F2 and S. coelicolor M512 (SCP1− SCP2−, ΔactIIorf4 ΔredD) and their respective derivatives were maintained and grown on either MS agar (2% soy flour, 2% mannitol, 2% agar [components purchased from Carl Roth, Karlsruhe, Germany]) or trypic soy broth (TSB) medium (Becton Dickinson, Heidelberg, Germany). Escherichia coli strains were cultivated in LB medium (components purchased from Carl Roth, Karlsruhe, Germany) supplemented with appropriate antibiotics. DNA isolation and manipulations were carried out according to standard methods for E. coli (51) and Streptomyces (33).
Production, extraction, and detection of caprazamycin derivatives.
Fifty milliliters of TSB medium was inoculated with a spore suspension of Streptomyces sp. MK730-62F2, S. coelicolor M512, or a derivative thereof. The cultures were incubated for 2 days at 30°C and 200 rpm. For the production of caprazamycins, 1 ml of the precultures was inoculated into 100 ml of a medium containing 1% soytone, 1% soluble starch, and 2% d-maltose adjusted to pH 6.7 (components purchased from Becton Dickinson, Heidelberg, Germany). The cultures were incubated for 7 days at 30°C and 200 rpm. Partial purification of caprazamycins was achieved by extraction of the culture supernatant (adjusted to pH 4) with an equal volume of n-butanol. The organic phase was evaporated, and extracts were resolved in 500 μl methanol. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed on a Surveyor high-performance LC (HPLC) system equipped with a Reprosil-Pur Basic C18 column (5 μm; 250 by 2 mm; Maisch, Ammerbuch, Germany) coupled to a Thermo Finnigan TSQ Quantum triple-quadrupole mass spectrometer (heated capillary temperature, 320°C; sheath gas, nitrogen; collision gas, argon). Analysis of extracts from the coexpression of pRHAM experiments was performed with a linear gradient of 2% to 40% acetonitrile in aqueous formic acid (0.1%) over 5 min followed by a linear gradient of 40% to 100% acetonitrile in aqueous formic acid (0.1%) over 20 min. Otherwise, a linear gradient of 2% to 40% acetonitrile in aqueous formic acid (0.1%) over 4 min was used followed by a linear gradient of 40% to 100% acetonitrile in aqueous formic acid (0.1%) over 31 min. The flow rate was 0.2 liter min−1, and detection was at 262 nm. Positive electrospray ionization [(+)-ESI] was performed with an electrospray voltage of 3.8 kV, and collision-induced dissociation (CID) spectra were recorded with a collision energy of 35 eV.
Coexpression of plasmid pRHAM.
Plasmid pRHAM (49), containing all required genes for the biosynthesis of dTDP-l-rhamnose oleL, oleS, oleE, and oleU from Streptomyces antibioticus ATCC 1891, was introduced into the nonmethylating strain E. coli ET12567 (40) and reisolated. The isolated plasmid DNA was used for polyethylene glycol (PEG)-mediated protoplast transformation as described by Kieser et al. (33) with either Streptomyces sp. MK730-62F2 ΔcpzDIII for complementation of the cpzDIII knockout or with S. coelicolor/cpzLK09 for the production of intact caprazamycins. For the preparation of protoplasts, the corresponding Streptomyces strain was cultivated in 100 ml S-YEME broth (34% sucrose, 1% glucose, 0.5% peptone, 0.3% yeast extract, 0.3% malt extract, 5 mM MgCl2; components purchased from Carl Roth, Karlsruhe, Germany) for 48 h at 30°C. Cells were harvested and washed with an aqueous solution of 10.3% sucrose. The degradation of the cell wall was performed at 30°C in P buffer [10.3% sucrose, 0.025% K2SO4, 0.202% MgCl2·6H2O, 0.005% KH2PO4, 0.368% CaCl2·2H2O, and 0.573% N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] supplemented with 4 mg/ml lysozyme (Serva, Heidelberg, Germany) and monitored by microscopy. Subsequently, PEG-mediated protoplast transformation was performed. Thiostrepton resistance mutants were selected and termed either Streptomyces sp. MK730-62F2/ΔcpzDIII/pRHAM-(1-3) or S. coelicolor/cpzLK09/pRHAM-(1-3).
Overexpression and purification of Cpz31.
cpz31 was amplified from cosmid DNA using the primers cpzGTEco_fw, GAATTCATGCGCGTGCTCTTCACG, and cpzGTHind_rv, AAGCTTCTAGGCCCTCCTCGCCAG (restriction sites for EcoRI and HindIII are underlined). A 1.2-kb PCR product was cloned into the pGEM-T cloning vector (Promega, Mannheim, Germany). The resulting plasmid, pLK01, was verified by sequencing. cpz31 was subsequently cloned into the expression vector pHis8 (29), taking advantage of the EcoRI and HindIII restriction sites, and confirmed by sequencing. The resulting plasmid was named pLK02.
E. coli Rosetta2 (DE3)pLys (Novagen, Darmstadt, Germany) cells containing plasmid pLK02 were cultivated in 1 liter of TB broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 0.23% KH2PO4, 1.25% K2HPO4) supplemented with 50 μg ml−1 kanamycin and 50 μg ml−1 chloramphenicol at 37°C. At an optical density at 600 nm of 0.7, the temperature was adjusted to 20°C, and isopropyl thiogalactoside (IPTG) was added to a 0.5 mM final concentration. After an additional 10-h cultivation at 20°C, the culture was harvested and 10 ml of buffer A (50 mM Tris-HCl [pH 8], 1 M NaCl, 10% glycerol, 20 mM imidazole, 10 mM β-mercaptoethanol) supplemented with 0.5 mg ml−1 lysozyme and 0.5 mM phenylmethylsulfonyl fluoride was added to the pellet (12 g). Cells were disrupted by sonication (Branson, Danbury, CT) at 4°C. The lysate was centrifuged (55,000 × g, 45 min), and the supernatant was applied to an affinity chromatography column with 4 ml of Ni-nitrilotriacetic acid (NTA)-agarose resin (Qiagen, Hilden, Germany) according to the manufacturer's instructions. His8-Cpz31 was eluted from the column with 250 mM imidazole in buffer A. Buffer exchange was carried out with PD10 columns (Amersham Biosciences, Freiburg, Germany) and buffer A without β-mercaptoethanol and imidazole. The low solubility of His8-Cpz31 resulted in a yield of 0.46 mg of partially purified protein per liter of culture. The protein was stored at −80°C in aliquots of 50 μl.
Assay for glycosyltransferase activity.
Partially purified caprazamycin aglycones were obtained from a culture extract of S. coelicolor M512/cpzLK09. Five microliters of caprazamycin aglycones in methanolic solution was applied to a reaction tube, air dried, and dissolved in a 50-μl reaction mixture containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 5 mM dTDP-l-rhamnose, and 9 μM protein. The assay was performed at 30°C for 2 h and stopped by the addition of 150 μl methanol. The activity of His8-Cpz31 was measured by LC-ESI-MS.
Screening of the cosmid library.
The construction of the genomic cosmid library of Streptomyces sp. MK730-62F2 has been described previously (31). A 0.5-kb fragment of cpzDIII was amplified from genomic DNA by using a degenerated primer as described by Decker et al. (8). Perfect matching primers GDHspf_fw, CGTAGTTGTTGGAGCAGCGCG, and GDHspf_rv, GGTTCATCGGCTCCCGCTACG, amplifying a 0.5-kb fragment of cpzDIII, were applied in a PCR screening of the cosmid library.
DNA sequencing and computer-assisted sequence analysis.
Double-stranded sequencing of the entire cosmid clone 4H11 (39,442-bp insert) was performed by GenoTech (Baejeon, South Korea) by using a shotgun library with DNA fragments of approximately 0.5 to 1.0 kb in length. The DNASIS software package (Hitachi Software Engineering, Tokyo, Japan) and Artemis (Wellcome Trust Genome Campus, Cambridge, United Kingdom) were used for sequence analysis and annotation. Database comparisons were carried out in the GenBank database by using the BLAST program (1). Alignment and comparison of sequences were performed using the ClustalX algorithm (28) and GeneDoc alignment editor (http://www.psc.edu/biomed/genedoc).
Inactivation of cpzDIII in Streptomyces sp. MK730-62F2.
An apramycin resistance cassette [aac(3)IV] was amplified from plasmid pIJ773 (18) using the primer pair gdhKO_fw, GTGCTGGACAAGCTCACCTACGCCGGCACCCTCGACGACATTCCGGGGATCCGTCGACC, and gdhKO_rv, GAGCTGGCACGGGCCGTAGTTGTTGGAGCAGCGCGTGACTGTAGGCTGGAGCTGC TTC. Italic letters represent 39-nucleotide (nt) homologous extensions for Red/ET-mediated recombination. The gene was replaced in E. coli BW25113/pIJ790/4H11 by using Red/ET-mediated recombination (18), and the resulting cosmid, cpzLK01, was confirmed by restriction analysis. cpzLK01 was transferred into the nonmethylating strain E. coli ET12567/pUZ8002 (46) and introduced into Streptomyces sp. MK730-62F2 by conjugation. Exconjugants, resistant to apramycin, were isolated and tested for the loss of their kanamycin resistance, indicating a successful double crossover. The mutants were further analyzed by PCR with chromosomal DNA as the template. Mutant strains were designated Streptomyces sp. MK730-62F2ΔcpzDIII-(1-3). Complementation of the mutants was performed by coexpression of plasmid pRHAM as described above.
Assembly of the caprazamycin biosynthetic gene cluster and the l-rhamnose subcluster.
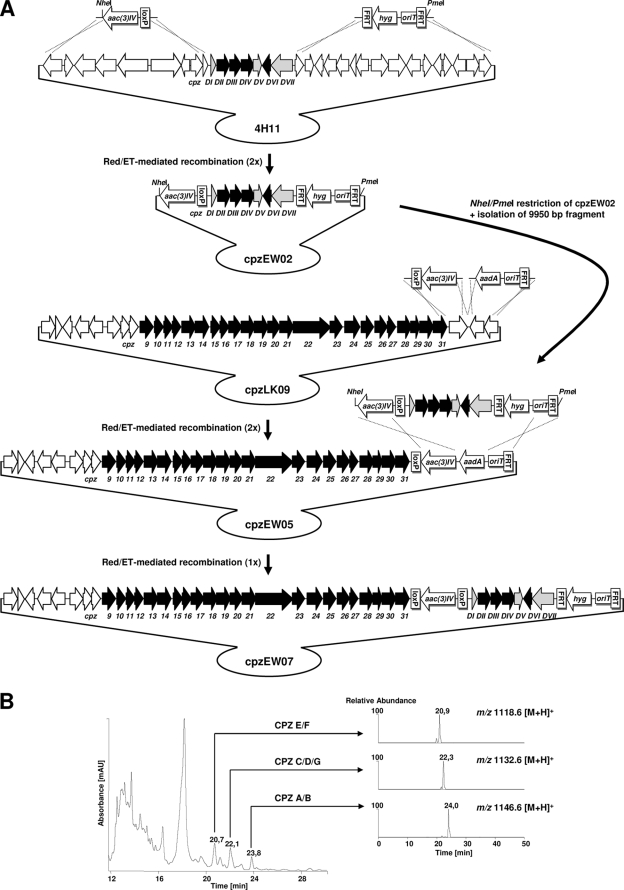
An apramycin resistance cassette [aac(3)IV] was amplified by PCR from plasmid pIJ774 (19) with primers 4H11_774_fw, GCCCGAGCACCCGCAGCCGGTTCGCCCGCGAGCCCAGCAGCTAGCTTATGAGCAGCCAATCGAC, including a NheI restriction site (bold), and 4H11_774_rv, CAGAGGGTGCGCTCGGAAGACGGGCGCGGAAGCCGCCCGACTAGTATTCCGGGGATCCGTCGACC, including a SpeI restriction site (bold). Italic letters represent 39-nt homologous extensions to an internal region of orf1 and to a region downstream of orf9, respectively. The resulting 1,310-bp PCR product contained the aac(3)IV gene and a single loxP site. The PCR product was inserted into cosmid 4H11 by Red/ET-mediated recombination (18), thereby replacing a 13,531-bp region (orf1 to orf9) upstream of cpzDI, resulting in the apramycin- and kanamycin-resistant cosmid cpzEW01. A hygromycin resistance cassette (hyg) was amplified from pIJ10700 by PCR with primers 4H11_10700_fw, TGACGGGCGGGCGGGGCTCGCAGCCCCTAGGCTGAGGAGATTTAAATTGTAGGCTGGAGCTGCTTC, including a SwaI restriction site (bold), and 4H11_10700_rv, GGGGGCCGTCGCCGAAGTGCGGGCCGCCGGGCGGCACGGGTTTAAACATTCCGGGGATCCGTCGACC, including a PmeI restriction site (bold). Italic letters represent 39-nt homologous extensions to sequences upstream of orf10 and to an internal region of orf25, respectively. The resulting 1,678-bp PCR product contained the hygromycin resistance gene hyg, an origin of transfer, oriT, and was flanked by two FLP recombinase recognition sites (FRT). The PCR product was inserted into cosmid cpzEW01 by Red/ET-mediated recombination, thereby replacing a 16,144-bp region (orf10 to orf25) upstream of cpzDVII and resulting in the hygromycin-, apramycin-, and kanamycin-resistant cosmid cpzEW02. Restriction of cpzEW02 with NheI and PmeI generated the 9,950-bp l-rhamnose subcluster fragment (cpzDI to cpzDVII) flanked by the apramycin and the hygromycin resistance cassettes.
Cosmid cpzLK09, which already contained the ΦC31 integration cassette from pIJ787 (10), was prepared as the l-rhamnose subcluster acceptor cosmid. An apramycin resistance cassette [aac(3)IV] was amplified from pIJ774 (18) by PCR with primers 31C2_774_fw2, TGCCGTACCCGCGGTCACCGGTTCCGCCTCGGCGGGTGCACTAGTTGTAGGCTGGAGCTGCTTC, including a SpeI restriction site (bold), and 31C2_774_rv, CGTGCTGGTGCTCGCCAACCACCCCATGCGGCTGGGCATGCTAGCAAATGCCGGCCTTTGAATGG, including a NheI restriction site (bold). Italic letters represent 39-nt homologous extensions to sequences downstream of cpz31 and to an internal region of cpz33, respectively. The resulting 1,079-bp PCR product contained the aac(3)IV gene and a single loxP site. The PCR product was inserted into cosmid cpzLK09 by Red/ET-mediated recombination, thereby replacing a 1,000-bp region downstream of cpz31 and resulting in the apramycin- and kanamycin-resistant cosmid cpzEW03. A spectinomycin/streptomycin resistance cassette (aadA) was amplified from pIJ778 (18) by PCR with primers 31C2_778_fw, TCGTCAGATGCAGATGGACGTCGTGCCCGGCCGCGTTCTATTTAAATTTATTTGCCGACTACCTTGG, including a SwaI restriction site (bold), and 31C2_778_rv, CCGCGGTGGCGCACCACGACGAACTGGTGCCGCGGGCCGGTTTAAACATTCCGGGGATCCGTCGACC, including a PmeI restriction site (bold). Italic letters represent 39-nt homologous extensions to an internal sequence of cpz34 and to a region 351 bp upstream of cpz34, respectively. The resulting 1,406-bp PCR product contained the aadA gene, an oriT, and a single FRT site. The PCR product was inserted into cosmid cpzEW03, thereby replacing the 613-bp region between aac(3)IV and the beginning of the SuperCos1 backbone and resulting in the spectinomycin-streptomycin-, apramycin-, and kanamycin-resistant cosmid cpzEW05.
For the generation of cpzEW07, the cosmid containing the caprazamycin gene cluster, and the l-rhamnose subcluster, Red/ET-mediated recombination was used. A Red/ET-proficient E. coli strain harboring cpzEW05 was transformed with the 9,950-bp NheI/PmeI fragment from cpzEW02. Homologous recombination took place between a 924-bp region, represented by the apramycin resistance gene aac(3)IV, and a 523-bp region of identical sequence between the hygromycin and the spectinomycin-streptomycin resistance cassettes. Positive selection with hygromycin was possible for this recombination event due to the exchange of the resistance gene aadA with hyg. Note that after Red/ET-mediated recombination, the apramycin resistance cassette is now flanked by loxP sites and the hygromycin resistance cassette by FRT sites, allowing removal of the resistance cassettes by the use of Cre recombinase or FLP recombinase, respectively. The isolated cosmid DNA of cpzEW07 was verified by restriction analysis. cpzEW07 was introduced into S. coelicolor M512 by triparental intergeneric conjugation with the help of E. coli ET12567/pUB307 (14). Apramycin-, hygromycin-, and kanamycin-resistant clones were selected and termed S. coelicolor M512/cpzEW07-(1-3).
Inactivation of cpzDI and cpzDV in S. coelicolor M512/cpzEW07.
The apramycin resistance cassette in cosmid cpzEW07 was eliminated by SpeI restriction and religation as described elsewhere (17). The resulting cosmid cpzEW08 was verified by restriction analysis. An apramycin resistance cassette [aac(3)IV] was amplified from plasmid pIJ774 (18) by PCR with primers containing an AflII site (bold), KOcpzDI_fw, GTGGAAGCAGAAGGAAGATGATGAGTCGTGGAGTCCATGCTTAAGATTCCGGGGATCCGTCGACC, and KOcpzDI_rv, GCTCGGATCGTCACCGCGCTGTCCACGAGGGAGGCGTCACTTAAGTGTAGGC TGGAGCTGCTTC, for the inactivation of cpzDI, or KOcpzDV_fw, GCATCCGCAAGGAAGGTCTTCCTCGTGAAACGCCATGAGCTTAAGATTCCGGG GATCCGTCGACc, and KOcpzDV_rv, CGCCGGCCGCCCCCGGGCGGTGGCCGGCGCCGTCGTTCACTTAAGTGTAGGCTGGAGCTGCttc, for the inactivation of cpzDV. Italic letters represent 39-nt homologous extensions to sequences up- and downstream of the target genes for Red/ET-mediated recombination. Resulting cosmids cpzEW09 [ΔcpzDI::aac(3)IV] and cpzEW11 [ΔcpzDV::aac(3)IV] were confirmed by restriction analysis. Excision of the cassette was performed by in vitro application of Cre recombinase (New England Biolabs, Frankfurt am Main, Germany) according to the manufacturer's instructions, taking advantage of the loxP recognition sites flanking the apramycin resistance cassette. The obtained cosmids cpzEW10 (ΔcpzDI) and cpzEW12 (ΔcpzDV) were screened for apramycin sensitivity and verified by restriction analysis. Cosmids cpzEW10 and cpzEW12 were transferred into E. coli ET12567 (40) and introduced into S. coelicolor M512 by triparental intergeneric conjugation with the help of E. coli ET12567/pUB307 (14). Kanamycin resistance exconjugants were selected and designated S. coelicolor M512/cpzEW10-(1-3) (ΔcpzDI) and S. coelicolor M512/cpzEW12-(1-3) (ΔcpzDV).
Nucleotide sequence accession number.
The nucleotide sequence of the gene cluster for dTDP-l-rhamnose biosynthesis, cpzDI through cpzDVII, has been deposited in GenBank under the accession number HM051054.
RESULTS
Coexpression of pRHAM in S. coelicolor containing the caprazamycin gene cluster.
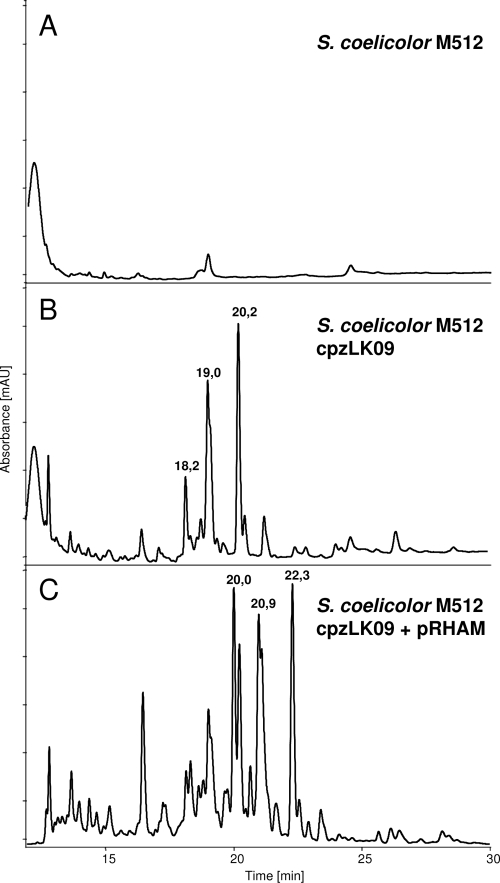
Previously, we reported that heterologous expression of cosmid cpzLK09, containing the caprazamycin gene cluster, resulted in the accumulation of caprazamycin aglycones in S. coelicolor M512 (31). The production of the nonglycosylated caprazamycin derivatives correlates with the absence of putative genes for the deoxysugar formation within the gene cluster. In order to investigate if all other genes required for the biosynthesis of the intact caprazamycins are functionally expressed on cpzLK09, we introduced plasmid pRHAM (49) into the heterologous host S. coelicolor M512/cpzLK09. pRHAM contains all genes required for dTDP-l-rhamnose biosynthesis from S. antibioticus: oleS, encoding a dTDP-glucose synthase, oleE, a dTDP-glucose 4,6-dehydratase, oleL, a dTDP-deoxyglucose 3,5-epimerase, and oleU, a 4-ketoreductase. Three individual clones termed S. coelicolor M512/cpzLK09/pRHAM-(1-3) were selected by their thiostrepton resistance and cultivated in production medium. Analysis of the culture extracts by LC-ESI-MS revealed that in comparison to S. coelicolor M512/cpzLK09, which only produced the caprazamycin aglycones (Fig. 2 B; CPZ aglycones E/F with a retention time [Rt] of 18.2 min, CPZ aglycones C/D/G with Rt of 19.0 min, and CPZ aglycones A/B with Rt of 20.2 min), all mutant strains now accumulated additional metabolites (Fig. 2C). These new compounds correspond to the caprazamycins E/F (Rt, 20.0 min), caprazamycins C/D/G (Rt, 20.9 min), and caprazamycins A/B (Rt, 22.3 min), as found by ESI-MS product ion scans (see Fig. S1 in the supplemental material). MS/MS spectra from CID experiments (see Fig. S1) matched exactly the characteristic fragmentation profiles of the intact caprazamycins (31). This verified that the enzymes for methylation and attachment of the l-rhamnose moiety have to be encoded on the gene cluster. A set of genes, cpz28 to -31, encoding three hypothetical sugar O-methyltransferases and a putative rhamnosyltransferase, are the most likely candidates for these reactions.
FIG. 2.
HPLC chromatograms of n-butanolic extracts from S. coelicolor M512 (A), S. coelicolor M512 containing the caprazamycin gene cluster cpzLK09 (B) and S. coelicolor M512/cpzLK09 containing pRHAM coexpressing the biosynthetic genes for dTDP-l-rhamnose (C).
In vitro investigation of Cpz31 with dTDP-l-rhamnose as substrate.
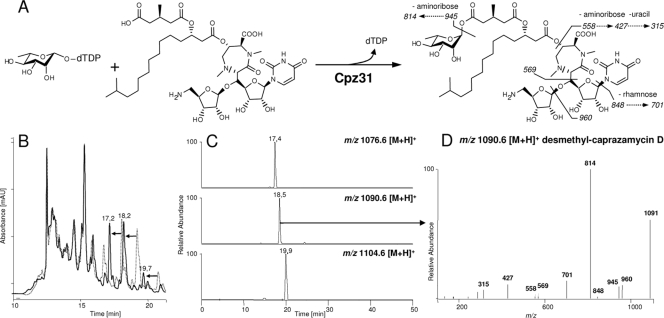
Cpz31 shows high sequence homology to known rhamnosyltransferases from other antibiotic gene clusters, e.g., ElmGT from S. olivaceus (38% identity, 50% similarity) (4). pLK02 containing cpz31 was overexpressed in E. coli, and the resulting protein was purified for in vitro studies. His8-Cpz31 consists of 420 amino acids with a calculated molecular mass of 44.7 kDa. Expression of a corresponding protein could be observed after induction with IPTG (see Fig. S2 in the supplemental material). Solubility of the enzyme was low, and the yield could not be improved using different culture temperatures or IPTG concentrations for expression. However, His8-Cpz31 was partially purified by Ni-NTA chromatography, resulting in 0.85 mg of protein from a 1-liter culture (see Fig. S2). A butanolic extract from an S. coelicolor M512/cpzLK09 culture containing caprazamycin aglycones was incubated with His8-Cpz31 in the presence of dTDP-l-rhamnose. After 2 h, caprazamycin aglycones could not be observed in the assay mixture anymore, and instead three new peaks appeared in the LC chromatograms (Fig. 3 B; at Rts of 17.2 min, 18.2 min, and 19.7 min), indicating product formation. Precursor ions for the rhamnosylated caprazamycins E/F with m/z 1,076.6 [M+H]+, C/D/G with m/z 1,090.6 [M+H]+, and A/B with m/z 1,104.6 [M+H]+ were readily detected by ESI-MS product ion scans at Rts of 17.4 min, 18.5 min, and 19.9 min, respectively (Fig. 3C). The MS/MS spectrum obtained from the parent ion with m/z 1,090.6 [M+H]+ (Fig. 3D) was identical to the predicted fragmentation of the rhamnosylated caprazamycins C/D/G (Fig. 3A). Characteristic ions with m/z 1,091, m/z 960, and m/z 848 displayed the presence of the rhamnosyl moiety. Similar spectra were produced by the parent ions with m/z 1,076.6 [M+H]+ and m/z 1,104.6 [M+H]+, representing the rhamnosylated caprazamycins with different fatty acid chain lengths (data not shown). These compounds were not detectable in assay mixtures without Cpz31 or dTDP-l-rhamnose. Therefore, we conclude that Cpz31 glycosylates the caprazamycin aglycones by using dTDP-l-rhamnose as a substrate (Fig. 3A).
FIG. 3.
Glycosylation of caprazamycins by Cpz31. (A) Reaction scheme of Cpz31 with dTDP-l-rhamnose and caprazamycin D aglycone as substrates. The suggested fragmentation of the reaction product 2c,3c,4c-desmethyl-caprazamycin D is depicted. (B) Overlay of HPLC chromatograms (262 nm) from activity assays with Cpz31 (black line) and without protein (dashed line). The observed shift in retention time by the complete conversion of caprazamycin aglycones to the rhamnosylated product is indicated by bold arrows. (C) LC-ESI-MS product ion scans in positive mode for desmethyl-caprazamycins E/F with m/z 1,076.6 [M+H]+, for desmethyl-caprazamycins C/D/G with m/z 1,090.6 [M+H]+, and for desmethyl-caprazamycins A/B with m/z 1,104.6 [M+H]+ in the activity assay with Cpz31. (D) Mass spectrometric fragmentation pattern in the CID experiment for 2c,3c,4c-desmethyl-caprazamycin C/D/G.
Identification of genes for biosynthesis of dTDP-l-rhamnose.
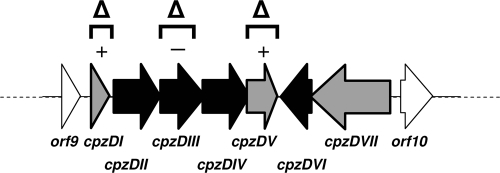
In bacteria, genes directing the biosynthesis of a certain secondary metabolite are generally clustered together. However, cosmid 31C2, containing the caprazamycin gene cluster, missed the genes for the formation of the l-rhamnose moiety (31). Because these genes are required for caprazamycin biosynthesis, they have to be located elsewhere on the chromosome of the original producer strain Streptomyces sp. MK730-62F2. Using degenerated primers for the amplification of NDP-glucose 4,6-dehydratases (8), we obtained a 500-bp partial sequence of cpzDIII from genomic DNA of the caprazamycin producer. This fragment was cloned and sequenced. Annotation revealed high homology to dTDP-glucose 4,6-dehydratases on the protein level. Perfect matching primers were constructed to screen a cosmid library of Streptomyces sp. MK730-62F2 (31). Sequencing and annotation of a positive cosmid, 4H11, revealed a stretch of 6.8 kb of DNA putatively involved in the biosynthesis of dTDP-l-rhamnose (Fig. 4). According to the gene function predictions (Table 1), cpzDII encodes a putative dTDP-glucose synthase, cpzDIII encodes a dTDP-glucose 4,6-dehydratase, cpzDIV encodes a 4-ketoreductase, and cpzDVI encodes a dTDP-deoxyglucose 3,5-epimerase. The identified gene cluster consists of two putative operons (Fig. 4). cpzDII, cpzDIII, and cpzDIV are translationally coupled to cpzDV, as indicated by overlap of the stop and start codons. The deduced gene product of cpzDV shows homology to hypothetical proteins from various Streptomyces strains which are annotated as methyltransferases. In the opposite direction, cpzDVI is translationally coupled with cpzDVII, which encodes a putative glycosyltransferase. Homologues of CpzDVII have been assigned to bacterial cell wall biosynthesis. The putative gene cpzDI, 105 bp upstream of cpzDII, encodes a small protein of 57 amino acids with several homologues from Streptomyces genomes. Many of them, e.g., SvirD4_37404 from Streptomyces viridochromogenes DSM40736 and SAMR0798 from Streptomyces ambofaciens ATCC 23877, are located next to hypothetical NDP-glucose dehydratase genes. Since orf9, a putative oxidoreductase gene, and orf10, a possible phosphodiesterase gene, have no apparent role in deoxysugar metabolism, we proposed that the l-rhamnose biosynthetic gene cluster spans from cpzDI to cpzDVII.
FIG. 4.
Organization of the l-rhamnose gene cluster cpzDI to cpzDVII on cosmid 4H11. Black arrows indicate genes proposed to be essential in dTDP-l-rhamnose biosynthesis. Gray arrows indicate genes with unknown function. Squared brackets above the cluster mark the gene deletions performed in this study. The minus sign indicates that the deletion of the respective region led to an abolishment of caprazamycin production. Plus signs indicate that caprazamycin production was not influenced.
TABLE 1.
Deduced functions of genes within the l-rhamnose subcluster
| Genea | No. of amino acids | Protein homologue (accession no.) | Overall homologyb | Proposed function |
|---|---|---|---|---|
| orf9 | 140 | SvirD4_010100037399 of S. viridochromogenes DSM 40736 (ZP_05536129) | 87/93 | Oxidoreductase |
| cpzDI | 58 | SAMR0798 of S. ambofaciens ATCC 23877 (CAJ88507) | 64/75 | Unknown |
| cpzDII | 356 | MtmD of S. argillaceus (CAK50774) | 76/86 | dTDP-glucose synthase |
| cpzDIII | 327 | NanG2 of S. nanchangensis (AAP42865) | 77/83 | dTDP-glucose dehydratase |
| cpzDIV | 296 | OleU of S. antibioticus (AAD55455) | 63/71 | 4-Ketoreductase |
| cpzDV | 268 | SvirD4_010100037424 of S. viridochromogenes DSM 40736 (ZP_05536134) | 96/99 | Unknown |
| cpzDVI | 202 | StrM of S. glaucescens (CAA07389) | 65/77 | Sugar 3,5-epimerase |
| cpzDVII | 647 | SghaA1_010100002568 of S. ghanaensis ATCC 14672 (ZP_04684037) | 76/82 | Cell wall biosynthesis glycosyltransferase |
| orf10 | 170 | SAMR0794 of S. ambofaciens ATCC 23877 (CAJ88503) | 83/89 | Phosphoesterase |
Genes shown in bold represent the proposed l-rhamnose subcluster.
Homology is reported as percent identity/percent similarity.
Inactivation of cpzDIII and complementation with pRHAM.
In order to confirm the involvement of the postulated l-rhamnose biosynthetic gene cluster in caprazamycin biosynthesis, the hypothetical dTDP-glucose 4,6-dehydratase gene cpzDIII was inactivated. The resulting cosmid, cpzLK01, was introduced into Streptomyces sp. MK730-62F2 by intergeneric conjugation. Four double-crossover mutants of Streptomyces sp. MK730-62F2 ΔcpzDIII were finally selected and confirmed by PCR. After cultivation, extracts of the mutant strains were applied to LC-ESI-MS analysis. In contrast to the wild-type strain, production of intact caprazamycins was completely abolished in the mutants (see Fig. S3 in the supplemental material). However, caprazamycin aglycones could clearly be detected, and extracts of the mutants still showed bioactivity in an agar diffusion assay against Mycobacterium phlei (data not shown). In order to validate that the disruption of cpzDIII is responsible for the abolishment of caprazamycin production, we complemented the ΔcpzDIII mutant by introducing pRHAM. After cultivation, LC-ESI-MS analysis showed that caprazamycin production was restored (see Fig. S3). Analysis of the ΔcpzDIII mutant and the complementation confirmed that the l-rhamnose biosynthetic gene cluster on cosmid 4H11 is required for the formation of the l-rhamnose moiety in caprazamycin biosynthesis.
Assembly of the entire gene cluster for heterologous production of intact caprazamycins.
In order to prove that the identified genes, cpzDI to -VII, are sufficient for l-rhamnose biosynthesis, we assembled the subcluster to cosmid cpzLK09 containing the caprazamycin gene cluster (Fig. 5 A). It was shown previously that the genes cpz32 to cpz34 are not involved in caprazamycin biosynthesis, based on inactivation experiments (31). Therefore, an apramycin resistance cassette and a spectinomycin-streptomycin resistance cassette were used to replace cpz32 to cpz34 by Red/ET-mediated recombination, generating cosmid cpzEW05. Within cosmid 4H11, containing the l-rhamnose subcluster, a 14-kb stretch of DNA upstream of cpzDI was replaced with an apramycin resistance cassette, introducing a unique NheI restriction site. A 17-kb fragment upstream of cpzDVII was replaced by a hygromycin resistance cassette, introducing a unique PmeI restriction site. From the generated cosmid cpzEW02, a 10-kb PmeI/NheI restriction fragment, containing the cpzDI-DVII subcluster, was transferred into Red/ET-proficient E. coli cells harboring cpzEW05. Homologous recombination of the restriction fragment with cosmid cpzEW05 was possible between the apramycin resistance gene aac(3)IV (924 bp) and 523-bp identical sequences of the hygromycin and the spectinomycin-streptomycin resistance cassettes. These sequences include an oriT and an FRT site. Red/ET-mediated recombination would lead to an apramycin-, hygromycin-, and kanamycin-resistant transformant only if homologous recombination occurred within these regions.
FIG. 5.
(A) Strategy for assembly of the caprazamycin gene cluster and the l-rhamnose subcluster. aac(3)IV, apramycin resistance gene; aad4, spectinomycin-streptomycin resistance gene; hyg, hygromycin resistance gene; loxP, Cre recognition site; oriT, origin of transfer. The size of the resistance cassettes and their components are not drawn to scale. (B) LC-ESI-MS UV chromatogram and product ion chromatograms from mass scans for caprazamycins E/F with m/z 1,118.6 [M+H]+, caprazamycins C/D/G with m/z 1,132.6 [M+H]+, and caprazamycins A/B with m/z 1,146.6 [M+H]+ in butanolic extracts of S. coelicolor M512/cpzEW07.
The resulting cosmid, cpzEW07, was introduced into S. coelicolor M512 by intergeneric triparental conjugation. Positive clones were selected by their apramycin, hygromycin, and kanamycin resistance and termed S. coelicolor M512/cpzEW07. The mutant strain was cultivated, and extracts were analyzed by LC-ESI-MS. Three new peaks appeared in the UV chromatograms at retention times of 20.7 min, 22.1 min, and 23.8 min (Fig. 5B). These peaks were missing in extracts of S. coelicolor M512 and S. coelicolor M512/cpzLK09 (data not shown). Corresponding mass peaks were detected by product ion scanning for the caprazamycins E/F with m/z 1,118.8 [M+H]+ (Rt 20.9 min), caprazamycins C/D/G with m/z 1,132.8 [M+H]+ (Rt, 22.3 min), and the caprazamycins A/B with m/z 1,146.8 [M+H]+ (Rt, 24.0 min) (Fig. 5B). MS/MS fragmentation patterns matched exactly the intact caprazamycins (data not shown). The heterologous production of caprazamycins in S. coelicolor M512 containing the assembled gene cluster proves that the cpzDI to -VII subcluster is sufficient for the biosynthesis of dTDP-l-rhamnose.
Deletion of nonessential genes in the l-rhamnose subcluster.
The production of intact caprazamycins by expression of the assembled gene cluster now enabled us to investigate other genes in caprazamycin biosynthesis in a heterologous background. However, to allow further gene deletion experiments, the existing apramycin resistance cassette needed to be eliminated from cosmid cpzEW07. This was achieved by a SpeI digestion and religation procedure, generating cosmid cpzEW08. Corresponding SpeI sites were added to the primer sequence during the generation of cosmids cpzEW02 and cpzEW05 (see Materials and Methods).
Three of the hypothetical genes in the l-rhamnose subcluster have no obvious role in dTDP-l-rhamnose biosynthesis. The potential glycosyltransferase CpzDVII contains several conserved domains typical for glycan synthases and is most likely involved in cell wall biogenesis. cpzDI is similar to small hypothetical genes often located next to genes from deoxysugar metabolism, and cpzDV encodes a putative protein with unknown function. The genes cpzDI and cpzDV were individually deleted by replacement with an apramycin resistance cassette flanked by loxP sites. Subsequently, the resistance cassettes were eliminated in vitro using Cre recombinase. The resulting cosmids were termed cpzEW10 (ΔcpzDI) and cpzEW12 (ΔcpzDV). After introduction of the cosmids into S. coelicolor M512, hygromycin- and kanamycin-resistant exconjugants were selected and designated S. coelicolor M512/cpzEW10 and S. coelicolor M512/cpzEW12, respectively. After cultivation, culture extracts of the mutants showed no differences in LC-ESI-MS analysis in comparison to S. coelicolor M512/cpzEW07 containing the complete l-rhamnose cluster (see Fig. S4 in the supplemental material). Caprazamycins were accumulated by all of the strains. Therefore, we conclude that cpzDI and cpzDV have no role in caprazamycin biosynthesis and are probably involved in primary metabolism.
DISCUSSION
Based on the identification of the l-rhamnose biosynthetic genes in this study, we propose two gene clusters, the caprazamycin and the l-rhamnose subcluster, to participate in caprazamycin biosynthesis. Coexpression of pRHAM, containing all genes required for the biosynthesis of dTDP-l-rhamnose, led to the accumulation of intact caprazamycins in a heterologous producer strain harboring cosmid cpzLK09. This clearly demonstrates that only the genes for the formation of dTDP-l-rhamnose are missing in the heterologous caprazamycin producer. Thus, the genes directing the attachment and methylation of the l-rhamnosyl moiety have to be carried on cosmid cpzLK09. Three putative methyltransferases, Cpz28, Cpz29, and Cpz30, show the highest sequence homology to known rhamnose O-methyltransferases from elloramycin (47) and spinosyn (54) biosynthesis. Most likely, these proteins have similar functions in caprazamycin formation. Notably, the recently identified gene cluster of liposidomycins, nonglycosylated caprazamycin derivatives, did not contain any cpz28, cpz29, or cpz30 orthologues (32).
Translationally coupled to the genes cpz28 to cpz30 is the putative glycosyltransferase gene cpz31. An in vitro assay with His8-Cpz31 using dTDP-l-rhamnose and caprazamycin aglycones as substrates showed a complete conversion of the aglycones after a 2-h incubation. The products were unambiguously identified as 2c,3c,4c-desmethyl caprazamycins by LC-ESI-MS/MS analysis. This result implies that attachment of the deoxysugar occurs prior to its sequential methylation. A similar order of sugar transfer and subsequent tailoring reactions has been found in the biosynthesis of other glycosylated compounds, such as tylosin (2), oleandomycin (50), mycinamycin (26), and chromomycin (42). Cpz31, together with most glycosyltransferases involved in natural product biosynthesis, belongs to the GT-1 family of glycosyltransferases as classified by the CAZy system (6). Cpz31 exhibits significantly higher homology to, for example, the 4,6-dideoxy-4-hydroxylamino-α-d-glucosyltransferase CalG3 (36% identity, 50% similarity) from Micromonospora echinospora (59) than to the rhamnosyltransferases AraGT (29% identiy, 38% similarity) from S. echinatus (38) and SpnG (29% identity, 39% similarity) from Saccharopolyspora spinosa (5). Luzhetskyy et al. (39) proposed that similarity-based relationships of glycosyltransferases are better understood with consideration of the aglycone substrate than of the sugar donor. In this aspect, Cpz31 is distinctive from other known glycosyltransferases involved in natural product biosynthesis. The rhamnosyl moiety of caprazamycins is linked to the methylglutaryl moiety by an ester bond and not by a glycosidic bond. While acylated deoxyhexoses are common in plants, they are rare in bacterial secondary metabolism. So far, only the phenazine derivatives aestivophoenins (35) and phenazoviridins (30) and the rhamnopyranosides (21) have been reported. Thus, Cpz31 may serve as a new enzymatic tool for the modification of bioactive compounds in the future.
Rhamnosyltransferases have been frequently exploited for the structural diversification of glycosylated antibiotics (4, 15, 38, 44), as they often exhibit high flexibility towards the donor substrate. However, heterologous expression of cosmid cpzLK09 in S. fradiae A0, which produces the deoxysugars d-olivose and l-rhodinose, and Streptomyces sp. TÜ6071ΔplaA6, producing l-amicetose, did not lead to the identification of new caprazamycin derivatives (data not shown). Since the caprazamycin aglycones were readily detected in the culture extracts, it can be speculated that Cpz31 is more specific for dTDP-l-rhamnose than related glycosyltransferases, e.g., ElmGT from S. olivaceus (4) or StfG from S. steffisburgensis (44). Further investigations will have to prove this assumption.
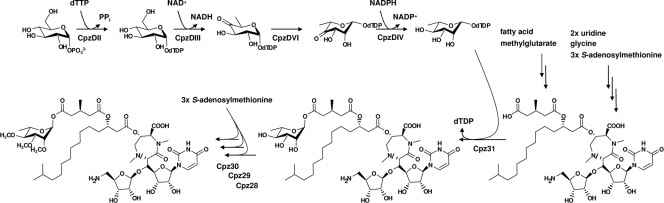
Screening of a cosmid library of the caprazamycin producer Streptomyces sp. MK730F-62F2 led to the identification of cpzDII, cpzDIII, cpzDIV, and cpzDVI elsewhere on the genome. The deduced gene products match the required enzymes for the biosynthesis of dTDP-l-rhamnose. Involvement in caprazamycin biosynthesis was demonstrated by inactivation of cpzDIII, resulting in the accumulation of caprazamycin aglycones. In contrast, inactivation of the colocated genes cpzDI and cpzDV had no influence on caprazamycin formation. Based on the presented results, we propose the biosynthetic pathway for the latter steps in caprazamycin biosynthesis as follows (Fig. 6). Generation of the deoxysugar would start with the activation of glucose-1-phosphate by the addition of deoxythymidyldiphosphate (dTDP), catalyzed by the putative NDP-glucose synthase CpzDII. Subsequently, the glucose dehydratase CpzDIII would dehydrate dTDP-glucose to dTDP-4-keto-6-deoxyglucose. This product would serve as a substrate for the sugar 3,5-epimerase CpzDVI, resulting in dTDP-4-keto-l-rhamnose, which would be further reduced to dTDP-l-rhamnose by the 4-ketoreductase CpzDIV. Cpz31 would than attach the l-rhamnose to the caprazamycin aglycones, generating 2c,3c,4c,-demethyl caprazamycins. Sequential methylation of the deoxysugar moiety by the putative O-methyltransferases Cpz28, Cpz29, and Cpz30 would finally lead to the caprazamycins.
FIG. 6.
Model for formation, attachment, and methylation of the l-rhamnosyl moiety in caprazamycin biosynthesis.
Usually, genes involved in the formation of a bacterial secondary metabolite are clustered together. However, biosynthetic gene clusters of rhamnosylated compounds often lack the genes for l-rhamnose formation (9, 16, 38, 54). In the case of elloramycin (48) and spinosyn (41), these genes were identified elsewhere in the genome. Inactivation experiments in Saccharopolyspora spinosa suggested that the gtt, gdh, epi, and kre genes provide the rhamnosyl moiety for both spinosyn and cell wall biosynthesis. The presence of cpzDVII, a putative cell wall biosynthesis glycosyltransferase gene, may indicate a similar role for the l-rhamnose cluster in Streptomyces sp. MK730F-62F2. However, l-rhamnose seems not to be essential for the strain, as inactivation of the glucose dehydratase gene cpzDIII did result in a viable corresponding mutant.
In order to allow investigation of the entire biosynthetic pathway of caprazamycins, we assembled the l-rhamnose and caprazamycin gene cluster on cosmid cpzLK09, using a new strategy based on Red/ET-mediated recombination. This technology, designated recombineering, was initially established for DNA manipulations in E. coli (7, 60, 61) and has been adapted to Streptomyces (18). It has been successfully applied for functional studies and engineering of antibiotic biosynthetic machineries (17), e.g., heterologous expression (10) and combinatorial biosynthesis (11). More recently, recombineering was used for the reconstitution of large biosynthetic gene clusters from overlapping inserts (3, 22, 57). This “stitching” overcomes the problem that genomic library clones rarely contain an entire gene cluster due to limitations of the average insert sizes. While “stitching” relies on the presence of overlapping, identical DNA sequences in both clones, we now show for the first time that assembly is possible without the need of overlapping DNA regions. For the presented strategy, Red/ET-mediated recombination is restricted to sequences in the form of resistance cassettes. These cassettes were introduced into the l-rhamnose donor cosmid and the recipient cosmid harboring the caprazamycin gene cluster. Final assembly was achieved by Red/ET-mediated recombination between a restriction fragment from the donor cosmid with the recipient cosmid. FRT and loxP recognition sites and SpeI restriction sites were introduced for the latter excision of the resistance cassettes. The in vivo usage of Cre (13) and the Flp recombinase (12) has now been successfully established in Streptomyces and allows the rapid generation of unmarked deletions. However, if Cre or Flp recombinase is used, a “scar” sequence comprising a loxP or an FRT site is maintained, which can cause undesirable recombination events in further recombineering experiments. Therefore, elimination of the apramycin resistance cassette was performed with SpeI digestion and religation of the cosmid DNA. In this study, cpzDI and cpzDV were successfully deleted on the assembled cosmid and the corresponding mutants were analyzed.
Since manipulation of natural producer strains can be difficult and time-consuming, heterologous expression of entire biosynthetic gene clusters in more-amenable and fully sequenced host strains is desirable. In addition, formation of a natural compound often relies on the supply of specific precursors provided by genes encoded outside the gene cluster, e.g., deoxysugars (48), isoprenoid derived moieties (20), or other small subunits (45, 58). Therefore, the new assembly approach described in this study may help to establish heterologous expression and genetic engineering of scattered gene clusters.
Supplementary Material
Acknowledgments
We thank Björn Boll (Tübingen University, Germany) for help with the purification of Cpz31 and are grateful to Lutz Heide (Tübingen University, Germany) for comments on the manuscript. We thank the John Innes Center (Norwich, United Kingdom) for the kind provision of plasmid pIJ10700 and Andriy Luzhetskyy (Freiburg University, Germany) for the strains S. fradiae A0 and Streptomyces sp. TÜ6071ΔplaA6.
This work was supported by a grant from the European Commission (IP005224, ActinoGen).
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bate, N., and E. Cundliffe. 1999. The mycinose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. J. Ind. Microbiol. Biotechnol. 23:118-122. [DOI] [PubMed] [Google Scholar]
- 3.Binz, T. M., S. C. Wenzel, H. J. Schnell, A. Bechthold, and R. Muller. 2008. Heterologous expression and genetic engineering of the phenalinolactone biosynthetic gene cluster by using red/ET recombineering. Chembiochem 9:447-454. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, G., E. P. Patallo, A. F. Brana, A. Trefzer, A. Bechthold, J. Rohr, C. Mendez, and J. A. Salas. 2001. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem. Biol. 8:253-263. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. L., Y. H. Chen, Y. C. Lin, K. C. Tsai, and H. T. Chiu. 2009. Functional characterization and substrate specificity of spinosyn rhamnosyltransferase by in vitro reconstitution of spinosyn biosynthetic enzymes. J. Biol. Chem. 284:7352-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker, H., S. Gaisser, S. Pelzer, P. Schneider, L. Westrich, W. Wohlleben, and A. Bechthold. 1996. A general approach for cloning and characterizing dNDP-glucose dehydratase genes from actinomycetes. FEMS Microbiol. Lett. 141:195-201. [DOI] [PubMed] [Google Scholar]
- 9.Decker, H., J. Rohr, H. Motamedi, H. Zahner, and C. R. Hutchinson. 1995. Identification of Streptomyces olivaceus Tu 2353 genes involved in the production of the polyketide elloramycin. Gene 166:121-126. [DOI] [PubMed] [Google Scholar]
- 10.Eustaquio, A. S., B. Gust, U. Galm, S. M. Li, K. F. Chater, and L. Heide. 2005. Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters. Appl. Environ. Microbiol. 71:2452-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eustaquio, A. S., B. Gust, T. Luft, S. M. Li, K. F. Chater, and L. Heide. 2003. Clorobiocin biosynthesis in Streptomyces: identification of the halogenase and generation of structural analogs. Chem. Biol. 10:279-288. [DOI] [PubMed] [Google Scholar]
- 12.Fedoryshyn, M., L. Petzke, E. Welle, A. Bechthold, and A. Luzhetskyy. 2008. Marker removal from actinomycetes genome using Flp recombinase. Gene 419:43-47. [DOI] [PubMed] [Google Scholar]
- 13.Fedoryshyn, M., E. Welle, A. Bechthold, and A. Luzhetskyy. 2008. Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 78:1065-1070. [DOI] [PubMed] [Google Scholar]
- 14.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 15.Gaisser, S., I. Carletti, U. Schell, P. R. Graupner, T. C. Sparks, C. J. Martin, and B. Wilkinson. 2009. Glycosylation engineering of spinosyn analogues containing an L-olivose moiety. Org. Biomol. Chem. 7:1705-1708. [DOI] [PubMed] [Google Scholar]
- 16.Gullon, S., C. Olano, M. S. Abdelfattah, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2006. Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl. Environ. Microbiol. 72:4172-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gust, B. 2009. Cloning and analysis of natural product pathways. Methods Enzymol. 458:159-180. [DOI] [PubMed] [Google Scholar]
- 18.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107-128. [DOI] [PubMed] [Google Scholar]
- 20.Haagen, Y., K. Gluck, K. Fay, B. Kammerer, B. Gust, and L. Heide. 2006. A gene cluster for prenylated naphthoquinone and prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042. Chembiochem 7:2016-2027. [DOI] [PubMed] [Google Scholar]
- 21.Hu, J. F., D. Wunderlich, I. Sattler, A. Hartl, I. Papastavrou, S. Grond, S. Grabley, X. Z. Feng, and R. Thiericke. 2000. New 1-O-acyl alpha-L-rhamnopyranosides and rhamnosylated lactones from Streptomyces sp., inhibitors of 3 alpha-hydroxysteroid-dehydrogenase (3α-HSD). J. Antibiot. (Tokyo) 53:944-953. [DOI] [PubMed] [Google Scholar]
- 22.Hu, Y., V. V. Phelan, C. M. Farnet, E. Zazopoulos, and B. O. Bachmann. 2008. Reassembly of anthramycin biosynthetic gene cluster by using recombinogenic cassettes. Chembiochem 9:1603-1608. [DOI] [PubMed] [Google Scholar]
- 23.Igarashi, M., N. Nakagawa, N. Doi, S. Hattori, H. Naganawa, and M. Hamada. 2003. Caprazamycin B, a novel anti-tuberculosis antibiotic, from Streptomyces sp. J. Antibiot. (Tokyo) 56:580-583. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi, M., S. Nakagawa, S. Hattori, T. Doi, T. Nasuda, M. Mijake, H. Ishizuka, H. Naganawa, T. Shomura, and M. Hamada. 2002. Caprazamycins A-F, novel anti-TB antibiotics, from Streptomyces sp., p. 232, abstr. F-2031. 42nd Intersci. Conf. Antimicrob. Agents Chemother., San Diego, CA. American Society for Microbiology, Washington, DC.
- 25.Igarashi, M., Y. Takahashi, T. Shitara, H. Nakamura, H. Naganawa, T. Miyake, and Y. Akamatsu. 2005. Caprazamycins, novel lipo-nucleoside antibiotics, from Streptomyces sp. II. Structure elucidation of caprazamycins. J. Antibiot. (Tokyo) 58:327-337. [DOI] [PubMed] [Google Scholar]
- 26.Inouye, M., H. Suzuki, Y. Takada, N. Muto, S. Horinouchi, and T. Beppu. 1994. A gene encoding mycinamicin III O-methyltransferase from Micromonospora griseorubida. Gene 141:121-124. [DOI] [PubMed] [Google Scholar]
- 27.Isono, K., M. Uramoto, H. Kusakabe, K. Kimura, K. Isaki, C. C. Nelson, and J. A. McCloskey. 1985. Liposidomycins: novel nucleoside antibiotics which inhibit bacterial peptidoglycan synthesis. J. Antibiot. (Tokyo) 38:1617-1621. [DOI] [PubMed] [Google Scholar]
- 28.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 29.Jez, J. M., J. L. Ferrer, M. E. Bowman, R. A. Dixon, and J. P. Noel. 2000. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39:890-902. [DOI] [PubMed] [Google Scholar]
- 30.Kato, S., K. Shindo, Y. Yamagishi, M. Matsuoka, H. Kawai, and J. Mochizuki. 1993. Phenazoviridin, a novel free radical scavenger from Streptomyces sp. Taxonomy, fermentation, isolation, structure elucidation and biological properties. J. Antibiot. (Tokyo) 46:1485-1493. [DOI] [PubMed] [Google Scholar]
- 31.Kaysser, L., L. Lutsch, S. Siebenberg, E. Wemakor, B. Kammerer, and B. Gust. 2009. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J. Biol. Chem. 284:14987-14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaysser, L., S. Siebenberg, B. Kammerer, and B. Gust. 2010. Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. Chembiochem 11:191-196. [DOI] [PubMed] [Google Scholar]
- 33.Kieser, T., M. Bibb, M. Buttner, K. Chater, and D. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 34.Kimura, K., N. Miyata, G. Kawanishi, Y. Kamio, K. Izaki, and K. Isono. 1989. Liposidomycin-C inhibits phospho-N-acetylmuramyl-pentapeptide transferase in peptidoglycan synthesis of Escherichia coli Y-10. Agric. Biol. Chem. 53:1811-1815. [Google Scholar]
- 35.Kunigami, T., K. Shin-Ya, K. Furihata, Y. Hayakawa, and H. Seto. 1998. A novel neuronal cell protecting substance, aestivophoenin C, produced by Streptomyces purpeofuscus. J. Antibiot. (Tokyo) 51:880-882. [DOI] [PubMed] [Google Scholar]
- 36.Liu, H. W., and J. S. Thorson. 1994. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 48:223-256. [DOI] [PubMed] [Google Scholar]
- 37.Luzhetskyy, A., J. Hoffmann, S. Pelzer, S. E. Wohlert, A. Vente, and A. Bechthold. 2008. Aranciamycin analogs generated by combinatorial biosynthesis show improved antitumor activity. Appl. Microbiol. Biotechnol. 80:15-19. [DOI] [PubMed] [Google Scholar]
- 38.Luzhetskyy, A., A. Mayer, J. Hoffmann, S. Pelzer, M. Holzenkamper, B. Schmitt, S. E. Wohlert, A. Vente, and A. Bechthold. 2007. Cloning and heterologous expression of the aranciamycin biosynthetic gene cluster revealed a new flexible glycosyltransferase. Chembiochem 8:599-602. [DOI] [PubMed] [Google Scholar]
- 39.Luzhetskyy, A., A. Vente, and A. Bechthold. 2005. Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides. Mol. Biosyst. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 40.MacNeil, D. J. 1988. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J. Bacteriol. 170:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madduri, K., C. Waldron, and D. J. Merlo. 2001. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J. Bacteriol. 183:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menendez, N., E. A. M. Nur, A. F. Brana, J. Rohr, J. A. Salas, and C. Mendez. 2004. Tailoring modification of deoxysugars during biosynthesis of the antitumour drug chromomycin A by Streptomyces griseus ssp. griseus. Mol. Microbiol. 53:903-915. [DOI] [PubMed] [Google Scholar]
- 43.Muroi, M., K. Kimura, H. Osada, M. Inukai, and A. Takatsuki. 1997. Liposidomycin B inhibits in vitro formation of polyprenyl (pyro)phosphate N-acetylglucosamine, an intermediate in glycoconjugate biosynthesis. J. Antibiot. (Tokyo) 50:103-104. [DOI] [PubMed] [Google Scholar]
- 44.Olano, C., M. S. Abdelfattah, S. Gullon, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2008. Glycosylated derivatives of steffimycin: insights into the role of the sugar moieties for the biological activity. Chembiochem 9:624-633. [DOI] [PubMed] [Google Scholar]
- 45.Ostash, B., A. Saghatelian, and S. Walker. 2007. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem. Biol. 14:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patallo, E. P., G. Blanco, C. Fischer, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2001. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 276:18765-18774. [DOI] [PubMed] [Google Scholar]
- 48.Ramos, A., F. Lombo, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2008. Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. Microbiology 154:781-788. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez, L., C. Oelkers, I. Aguirrezabalaga, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2000. Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J. Mol. Microbiol. Biotechnol. 2:271-276. [PubMed] [Google Scholar]
- 50.Rodriguez, L., D. Rodriguez, C. Olano, A. F. Brana, C. Mendez, and J. A. Salas. 2001. Functional analysis of OleY L-oleandrosyl 3-O-methyltransferase of the oleandomycin biosynthetic pathway in Streptomyces antibioticus. J. Bacteriol. 183:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Struve, W. G., R. K. Sinha, and F. C. Neuhaus. 1966. On the initial stage in peptidoglycan synthesis. Phospho-N-acetylmuramyl-pentapeptide translocase (uridine monophosphate). Biochemistry 5:82-93. [DOI] [PubMed] [Google Scholar]
- 53.Ubukata, M., K. Kimura, K. Isono, C. C. Nelson, J. M. Gregson, and J. A. McCloskey. 1992. Structure elucidation of liposidomycins, a class of complex lipid nucleoside antibiotics. J. Org. Chem. 57:6392-6403. [Google Scholar]
- 54.Waldron, C., K. Madduri, K. Crawford, D. J. Merlo, P. Treadway, M. C. Broughton, and R. H. Baltz. 2000. A cluster of genes for the biosynthesis of spinosyns, novel macrolide insect control agents produced by Saccharopolyspora spinosa. Antonie Van Leeuwenhoek 78:385-390. [DOI] [PubMed] [Google Scholar]
- 55.Waldron, C., P. Matsushima, P. R. Rosteck, Jr., M. C. Broughton, J. Turner, K. Madduri, K. P. Crawford, D. J. Merlo, and R. H. Baltz. 2001. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8:487-499. [DOI] [PubMed] [Google Scholar]
- 56.Williams, G. J., R. W. Gantt, and J. S. Thorson. 2008. The impact of enzyme engineering upon natural product glycodiversification. Curr. Opin. Chem. Biol. 12:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolpert, M., L. Heide, B. Kammerer, and B. Gust. 2008. Assembly and heterologous expression of the coumermycin A1 gene cluster and production of new derivatives by genetic engineering. Chembiochem 9:603-612. [DOI] [PubMed] [Google Scholar]
- 58.Yu, T. W., L. Bai, D. Clade, D. Hoffmann, S. Toelzer, K. Q. Trinh, J. Xu, S. J. Moss, E. Leistner, and H. G. Floss. 2002. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. U. S. A. 99:7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, C., E. Bitto, R. D. Goff, S. Singh, C. A. Bingman, B. R. Griffith, C. Albermann, G. N. Phillips, Jr., and J. S. Thorson. 2008. Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis. Chem. Biol. 15:842-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y., J. P. Muyrers, G. Testa, and A. F. Stewart. 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18:1314-1317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.