Abstract
The research described here was aimed at the selection of oral bacteria that displayed properties compatible with their potential use as probiotics for the pharyngeal mucosa. We included in the study 56 bacteria newly isolated from the pharynges of healthy donors, which were identified at the intraspecies level and characterized in vitro for their probiotic potential. The experiments led us to select two potential probiotic bacterial strains (Streptococcus salivarius RS1 and ST3) and to compare them with the prototype oral probiotic S. salivarius strain K12. All three strains efficiently bound to FaDu human epithelial pharyngeal cells and thereby antagonized Streptococcus pyogenes adhesion and growth. All were sensitive to a variety of antibiotics routinely used for the control of upper respiratory tract infections. Immunological in vitro testing on a FaDu layer revealed different responses to RS1, ST3, and K12. RS1 and ST3 modulated NF-κB activation and biased proinflammatory cytokines at baseline and after interleukin-1β (IL-1β) induction. In conclusion, we suggest that the selected commensal streptococci represent potential pharyngeal probiotic candidates. They could display a good degree of adaptation to the host and possess potential immunomodulatory and anti-inflammatory properties.
Metagenomics and functional molecular immunology substantiate the interpretation of humans as holobionts, in the sense of functional superorganisms, combining the self and microbes acting in concert to produce phenomena governed by the collective (25, 42). The association between host and symbionts affects the fitness of the holobiont within its environment, and it often governs the physiological homeostasis of the narrow balance between host well being and dysfunction (13, 35).
The mechanisms underlying the cross talk between a human host and microbes are only marginally understood. Their elucidation at a molecular level could supply the theoretical bases to develop strategies for preventing or treating several human dysfunctions, such as autoimmune diseases, through the reconstitution of a proper human-microbe mutualism.
The probiotic approach, in its widest sense, falls into this context, since it consists of the modification of a human microbiota by exogenous administration of microbial cells (or cell components), aimed at benefiting the host's health. A most commonly accepted definition comes from FAO/WHO, which states that probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (17).
So far, probiotics have been most predominantly investigated for and applied to the intestinal tract. Nevertheless, a few applications beyond the gut have proposed the potential beneficial role of probiotics for the stomach (23), vaginal mucosa (36), urinary tract (6), skin (27), and oral cavity (39). With respect to oral probiotics, particularly noticeable are the studies done by J. R. Tagg and coworkers of Streptococcus salivarius strain K12. Tagg and others, in fact, showed that, following oral administration, the bacterial strain K12 can colonize the oral mucosae of infants and adults (20, 34), downregulate the innate immune responses of human epithelial cells (11), and reduce oral volatile sulfur compound levels (8). Strain K12 was also revealed to produce two plasmid-encoded lantibiotic peptides (22, 38) that are active against Streptococcus pyogenes, the main etiological agent of bacterial pharyngitis. These investigations demonstrated the potential effectiveness of the probiotic intervention in the oropharyngeal tract.
Encouraged by the promising results obtained in J. R. Tagg's experiments, in the present study, we screened oral bacteria for their potential use as probiotics in the pharyngeal mucosa. We tested the ability of bacteria, which were newly isolated from the pharynges of healthy volunteers, to adhere to a human pharyngeal cell layer and to antagonize S. pyogenes on two different epithelial cell lines. The study allowed the selection of two bacterial strains, which were further investigated from an immunological point of view for their ability to cross talk with human epithelial cells in vitro.
MATERIALS AND METHODS
Isolation of bacteria from pharyngeal mucosa, bacterial strains, and culture conditions.
To isolate bacteria from the pharynx, specimens were collected using polyester fiber-tipped applicator swabs (VWR, Milan, Italy) taken from 4 healthy donors (3 females at 58, 32, and 29 years old, and a 32-year-old male). After serial dilutions in 0.1% peptonized saline, specimens were plated on MRS agar (Fluka Feinchemikalien GmbH, Neu-Ulm, Germany) supplemented with 0.05% cysteine-HCl (cMRS), M17 (Fluka Feinchemikalien GmbH) containing 2% lactose (LM17), and 2% glucose-tryptic soy agar (Difco, Detroit, MI). All 56 colonies grown at the highest dilutions were picked and spread on a plate with a loop. This procedure was repeated at least four times in order to obtain pure cultures. Table 1 lists the oral bacterial isolates used in this study. If not differently specified, oral streptococci and Streptococcus thermophilus DSM20617T were routinely grown overnight at 37°C in LM17, while Streptococcus pyogenes was grown overnight at 37°C in brain heart infusion (BHI) medium (Difco, Detroit, MI) supplemented with 0.3% yeast extract (yeBHI). S. pyogenes C11 has been clinically isolated from a pharyngitis patient and has been ascribed to emm type 77 through emm gene sequence analysis (data not shown).
TABLE 1.
Bacteria isolated from pharyngeal mucosa
| Human source designation | BOX-PCR genotypea | Isolate(s)b | Isolation medium usede | Taxonomic identificationc | Additional identification methodd |
|---|---|---|---|---|---|
| IS | IS-A1 | IS1 | cMRS | S. salivarius | |
| IS8, IS10, IS12 | LM17 | ||||
| IS-A2 | IS5, IS6 | LM17 | S. salivarius | ||
| IS7 | gTSA | ||||
| IS-B | IS3, IS4 | cMRS | S. sanguinis | ||
| IS-C | IS9 | LM17 | Staphylococcus aureus | ||
| IS15 | gTSA | ||||
| IS-D | IS11, IS13 | LM17 | Rothia mucilaginosa | ||
| IS14 | gTSA | ||||
| RS | RS-A1 | RS1, RS8, RS10 | LM17 | S. salivarius | |
| RS3, RS6 | cMRS | ||||
| RS13, RS14 | gTSA | ||||
| RS-A2 | RS9 | LM17 | S. salivarius | ||
| RS-B | RS4 | cMRS | Alloscardovia omnicolens | ||
| RS-C | RS5 | cMRS | Lactococcus lactis subsp. lactis | his | |
| RS-D | RS11 | LM17 | Micrococcus sp. (M. luteus) | ||
| RS-E | RS12 | gTSA | Rothia mucilaginosa | ||
| RS-F | RS15 | gTSA | Bacillus subtilis | ||
| SM | SM-A1 | SM1, SM2, SM4, SM5 | cMRS | S. salivarius | |
| SM6, SM8, SM9 | LM17 | S. salivarius | |||
| SM11, SM14, SM15 | gTSA | S. salivarius | |||
| SM-A2 | SM12 | gTSA | S. salivarius | ||
| SM-B | SM3 | cMRS | S. oralis | gdh | |
| SM-C | SM7 | LM17 | Rothia mucilaginosa | ||
| SM-D | SM13 | gTSA | Neisseria sp. (N. subflava) | ||
| ST | ST-A1 | ST3 | cMRS | S. salivarius | |
| ST-A2 | ST12 | LM17 | S. salivarius | ||
| ST-B | ST2, ST5p | cMRS | S. oralis | gdh | |
| ST-C | ST5g | cMRS | S. sanguinis | ||
| ST-D | ST4 | cMRS | S. oralis | gdh | |
| ST15 | gTSA | ||||
| ST-E | ST7, ST11 | LM17 | S. infantis | gdh | |
| ST-F | ST1, ST8 | LM17 | Bacillus sp. (B. cereus) | ||
| ST-G | ST9 | LM17 | S. oralis | gdh | |
| ST-H | ST6 | cMRS | Staphylococcus epidermidis | ||
| ST-I | ST13, ST14 | gTSA | Rothia mucilaginosa |
Similar (but not identical) BOX-PCR electrophoretic patterns are indicated with the same letter but a different number.
Representative strains chosen for 16S rRNA gene sequence analysis are in boldface. Representative isolates were deposited in the culture collection of the Department of Food Science and Microbiology, Industrial Microbiology Section, University of Milan, Italy (MIM culture collection).
Species designations in parentheses refer to those with 16S rRNA gene sequence similarities below 95%.
gdh, taxonomic identification based on the sequence of an approximately 500-bp internal fragment of the glucose-6-phosphate dehydrogenase gene (gdh) (5); his, taxonomic identification based on the length polymorphism of a PCR-amplified fragment from a histidine biosynthesis operon (4).
gTSA, 2% glucose-tryptic soy agar.
Identification and molecular characterization of bacterial isolates.
The isolates from each subject have been clustered by means of a BOX-PCR assay (Table 1), which was performed with the primer BoxA1 (5′-CTACGGCAAGGCGACGCTGACG-3′).
The 16S rRNA gene was amplified from at least one representative isolate from each BOX genotypic group (Table 1) by PCR, using primers P0 (5′-GAAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′). The resulting amplicons (each about 1.5 kb long) were then completely sequenced. Streptococcus salivarius isolates were further characterized by random amplified polymorphic DNA (RAPD) analysis, performed with primers OPI17 (5′-CGAGGGTGGTGATC-3′), OPI02-mod (5′-GCTCGGAGGAGAGG-3′), M13 (5′-GTAAAACGACGGCCAGT-3′), and PedAF (5′-ATACTACGGTAATGGGGT-3′).
A similarity dendrogram was built using NTSYSpc version 2.01 (Applied Biostatistics Inc., NY).
In vitro cultivation of epithelial cell lines.
FaDu (human pharynx carcinoma cell line; ATCC HTB-43) and HaCaT (human keratinocytes from a spontaneous immortalized, nontumorigenic cell line) cells were routinely grown in 24-well tissue culture plates in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (30 min at 56°C), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 0.1 mM nonessential amino acids, 2 mM l-glutamine and incubated at 37°C in a water-jacketed incubator in an atmosphere of 95% air and 5% carbon dioxide, until a confluent monolayer was formed.
Bacterial adhesion to the FaDu cell layer.
FaDu cells were grown in 3-cm petri plates on microscope cover glasses as described above. Cell monolayers were carefully washed with phosphate-buffered saline (PBS; pH 7.3) before bacterial cells were added. The bacterial cell concentration of an overnight culture was determined microscopically with a Neubauer-improved counting chamber (Marienfeld GmbH, Lauda-Königshofen, Germany). Approximately 2 × 108 cells of each strain resuspended in PBS (pH 7.3) were incubated with a monolayer of FaDu cells. After 1 h at 37°C, all monolayers were washed three times with PBS to release unbound bacteria. Cells were then fixed with 3 ml of methanol and incubated for 8 min at room temperature. After methanol was removed, cells were stained with 3 ml of Giemsa stain solution (1:20; Carlo Erba, Milan, Italy) and left for 30 min at room temperature. Wells were then washed until no color was observed in the washing solution and dried in an incubator at 30°C for 1 h. Microscope cover glasses were then removed from the petri plate and examined microscopically (magnification of ×100) immersed in oil. Adherent bacteria in 20 randomly selected microscopic fields were counted and averaged.
Preparation of bioluminescent Streptococcus pyogenes.
Reporter vector pCSS945, carrying a phage T5 promoter-lac operator upstream of the insect luciferase gene lucGR (29), was used to obtain the luminescent phenotype in Streptococcus pyogenes C11, according to conventional electrotransformation methods. Transformants were selected on yeBHI agar plates with 5 μg ml−1 of chloramphenicol. The selected luminescent S. pyogenes clone was named C11LucFF.
Antagonistic activity against Streptococcus pyogenes.
The antagonism against S. pyogenes was studied through exclusion and competition assays. Exclusion consisted of an hour of preincubation of the FaDu layer with 1 ml of a tester strain suspension (5 × 108 cells ml−1), followed by a washing step with PBS and incubation with 1 ml of the indicator strain (S. pyogenes C11LucFF) suspension (2 × 108 cells ml−1) for 1 h. The concentration of 5 × 108 tester cells ml−1 was chosen because it corresponded to the plateau of dose-response curves which were prepared during the setup of the experiment by measuring the antagonistic activity as a function of tester cell concentration (data not shown). Competition consisted of an hour of coincubation of the same number of tester and indicator strains (2 × 108 cells ml−1).
The bacterial cell concentration was determined from an overnight culture microscopically by means of a Neubauer-improved counting chamber (Marienfeld GmbH). After incubation, FaDu layers were quickly washed twice with 1 ml PBS (pH 7.3), and d-luciferin (Sigma-Aldrich, Steinheim, Germany) was added at the concentration of 12.5 μM in citrate buffer, pH 5. Immediately, the luminescence signal was measured with a Victor 3 luminometer (PerkinElmer, Monza, Italy). Each tester strain was analyzed in triplicate with at least two independent experiments. An unpaired Student's t test was performed to find statistically significant differences.
Antibacterial activity against Streptococcus pyogenes and PCR detection of bacteriocin-encoding genes.
In the first set of experiments, tester bacterial strains were spread with a loop on an agar plate containing LM17 medium and incubated overnight at 37°C. Then, 15 ml of soft yeBHI agar containing about 106 cells of the indicator strain (S. pyogenes C11) was poured over the plates. The plates were checked for inhibition zones after incubation at 37°C for 24 and 48 h. The production of antimicrobial substances was also tested through disk diffusion. Briefly, tester strains were grown until stationary growth phase in LM17 medium. Culture supernatants were neutralized to pH 7, filter sterilized, and spotted (0.1 ml) on a filter paper disk, which was previously placed on yeBHI soft agar plates inoculated with about 106 S. pyogenes cells. The presence of an inhibition halo was checked after 24 and 48 h.
The PCRs used to detect previously characterized bacteriocin structural genes (salivaricin A, salivaricin B, streptin, and peptide SA-FF22) were performed as described by Wescombe et al. (41).
Stimulation of FaDu monolayers and enzyme-linked immunosorbent assay (ELISA) measurement of cytokine production.
Human pharyngeal carcinoma cells (FaDu) were seeded into 24-well plates and grown, as previously described. Bacterial cells were added to monolayers of FaDu cells in 0.5 ml of fresh antibiotic-free Eagle's minimum essential medium (EMEM) containing 100 mM HEPES (pH 7.4) and incubated overnight at 37°C. Each bacterial strain was used at a multiplicity of infection (MOI) of about 1,000, while EMEM/HEPES medium without bacterial cells was used as a control. After overnight incubation, the supernatants were collected by pipetting, centrifuged to remove cells, and kept at −80°C. The same experiment was also performed by incubating bacteria and FaDu cells in the presence of 2 ng ml−1 of interleukin-1β (IL-1β). Finally, different cytokines in the supernatants were determined with a Bio-Plex array reader (Luminex 100; Bio-Rad Laboratories, Hercules, CA) using the Bio-Plex human cytokine 17-plex panel (Bio-Rad), according to the human cytokine Bio-Plex panel assay protocol (Bio-Rad). The list of tested cytokines and the corresponding detection limits were as follows: IL-1β, 0.3 pg ml−1; IL-2, 0.2 pg ml−1; IL-4, 0.1 pg ml−1; IL-5, 0.3 pg ml−1; IL-6, 0.2 pg ml−1; IL-7, 0.3 pg ml−1; IL-8, 0.3 pg ml−1; IL-10, 0.2 pg ml−1; IL-12 (p70), 0.4 pg ml−1; IL-13, 0.3 pg ml−1; IL-17, 0.5 pg ml−1; granulocyte colony-stimulating factor (G-CSF), 0.2 pg ml−1; granulocyte-macrophage colony-stimulating factor (GM-CSF), 1.1 pg ml−1; gamma interferon (IFN-γ), 2.6 pg ml−1; monocyte chemotactic protein 1 (MCP-1), 0.8 pg ml−1; macrophage inflammatory protein 1β (MIP-1β), 0.6 pg ml−1; and tumor necrosis factor alpha (TNF-α), 0.6 pg ml−1.
Construction of stable NF-κB reporting FaDu cells.
Stable transfectants of the FaDu cell line were obtained after transfection with the plasmid pNiFty2-Luc (InvivoGen, Rho, Italy). This plasmid combines five NF-κB sites with the insect luciferase reporter gene luc. The presence of active NF-κB molecules in the cell activates the promoter, resulting in the expression of the luciferase gene. Transfection was performed by means of the StoS transfection kit (GeneSpin, Milan, Italy), in accordance with the manufacturer's protocol. Afterward, cells were resuspended in fresh DMEM, seeded in 24-well plates, and incubated for 48 h, in order to obtain the expression of the antibiotic resistance. Finally, stable recombinant clones were selected by adding into the culture medium 50 μg ml−1 of zeocin (InvivoGen, Rho, Italy).
Study of NF-κB activation.
Recombinant FaDu cells were cultured using the same protocol as that used for nontransfected FaDu cells, in the presence of 50 μg ml−1 of zeocin. After growth, the FaDu layer was detached by trypsinization, and cells were resuspended in DMEM at a concentration of 250,000 cells ml−1 in the presence of 100 mM HEPES (pH 7.4). Subsequently, 50 μl of tester bacterial suspension, containing 2 × 109, 2 × 108, or 2 × 107 cells ml−1, was added to 450 μl of a FaDu suspension, resulting in MOIs of about 1,000, 100, or 10, respectively. After incubation at 37°C for 4 h, samples were kept in ice and sonicated at maximum frequency for 5 s (Bandelin sonicator; Bandelin electronic GmbH & Co., Berlin, Germany). Bacterial cells and insoluble particles were removed by centrifugation, and the supernatants were transferred into a new tube. At this point, 100 μl of supernatants was aliquoted in duplicate into the wells of a 96-well white microtiter plate (PerkinElmer, Monza, Italy); then, 12.5 μl of a 10 mM ATP solution and 12.5 μl of 0.1 mM d-luciferin were added, and the emitted bioluminescence was immediately recorded every 90 s with a Victor 3 luminometer (PerkinElmer). The maximum of the light production curve was considered for comparison of the results. In a different set of experiments, recombinant FaDu cells were simultaneously stimulated with IL-1β (2 ng ml−1). In the setup of the experiment, we confirmed that IL-1β was not modified or digested by bacteria during the incubation step (data not shown). All strains were analyzed in duplicate in at least three independent experiments per each single MOI. An unpaired Student's t test was performed to find statistically significant differences.
Antibiotic susceptibility of selected bacteria.
The inhibitory concentrations of several antimicrobial agents were determined, according to conventional broth microdilution protocols, in commercial 96-well microtiter plates for the following concentration ranges: ampicillin, chloramphenicol, erythromycin, oxytetracycline, and vancomycin, 1 to 16 μg ml−1; gentamicin, 8 to 64 μg ml−1; and kanamycin and streptomycin, 16 to 128 μg ml−1. The following three different liquid media were used in this experiment: LM17, MRS, and BHI.
Determination of urease activity and PCR detection of the ureC gene.
Urease activity was tested by evaluating the release of ammonia by means of the phenol red assay, as described in the literature (28).
The amplification of the gene coding for the main subunit of the urease complex (ureC) was carried out as previously described (31), using primers ureIAf (5′-GGAATTGTAACAGCTTGGAT-3′) and ureCr (5′-GTCGTATGGATTGGTTCACA-3′).
RESULTS
Isolation and characterization of bacteria from pharyngeal mucosa.
A total of 56 isolates were obtained from pharyngeal swab samples taken from four healthy donors by using three different culture media (Table 1). After a preliminary grouping of all isolated bacteria through BOX-PCR fingerprinting (data not shown), 16S rRNA genes sequence analysis revealed that 39 of the isolates belonged to the Streptococcus genus. Streptococcus salivarius was the most represented species (28 isolates). We also isolated Streptococcus oralis (6 isolates) and Streptococcus infantis (2 isolates), which were distinguished by glucose-6-phosphate dehydrogenase gene (gdh) sequence analysis (5). The only species isolated from all four pharyngeal samples were S. salivarius and Rothia mucilaginosa (7 isolates). All the other bacteria are listed in Table 1.
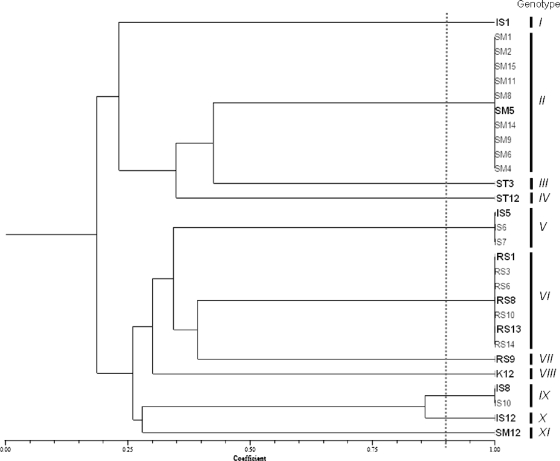
Streptococcus salivarius isolates were further characterized at the intraspecies level by means of BOX-PCR and RAPD-PCR analyses (data not shown). A computer evaluation of similarities and clustering resulted in a total of 11 unique S. salivarius genotypes out of 28 pharyngeal isolates and the K12 commercial probiotic strain (Fig. 1). The isolates that were included in a single genotypic group originated from the same pharyngeal sample, suggesting that they were probably multiple isolates.
FIG. 1.
Unweighted-pair group method using average linkages (UPGMA) dendrogram derived from similarity coefficients calculated by the Jaccard method (simple Jaccard [Sj] coefficients; shown on the scale at the bottom), showing the relationship among Streptococcus salivarius pharyngeal isolates, analyzed by BOX-PCR and RAPD analysis using primers M13, OPI02mod, OPI17mod, and PedAF. Samples with a similarity coefficient higher than 0.9 have been included in the same genotype. Selected bacterial isolates included in antagonism experiments are indicated in boldface.
Several oral isolates adhere efficiently to the FaDu epithelial layer.
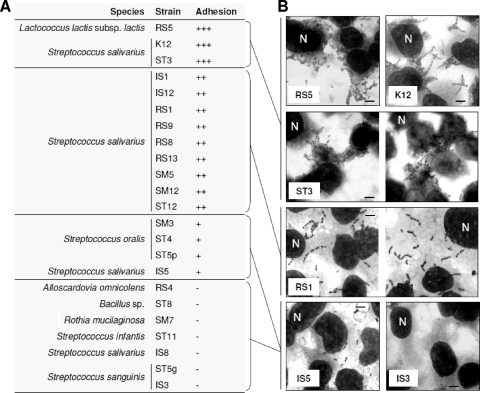
Twenty-three bacterial strains were tested for their ability to adhere to the FaDu epithelial cell layer. We studied 13 S. salivarius strains, including at least one representative isolate from each genotypic cluster and the commercial probiotic strain K12. We also included 10 other pharyngeal isolates that were arbitrarily selected. After being extensively washed with PBS, a significant proportion of cells from many bacterial strains remained attached to the FaDu monolayer, providing evidence that the adhesion was not only nonspecific physical entrapment. In particular, the following three strains displayed strong adhesive phenotypes, coinciding with an adhesion index (ADI; bacterial cells per 100 FaDu cells) of more than 2,500 (Fig. 2): Lactococcus lactis subsp. lactis RS5, S. salivarius K12, and S. salivarius ST3. Good adhesive ability was also displayed by nine other strains (ADI between 2,500 and 500), all belonging to the species S. salivarius.
FIG. 2.
Adhesion of bacterial strains to the FaDu epithelial cell layer according to their adhesion indexes (ADI; number of bacteria/100 FaDu cells). (A) +++, strong adhesion (ADI of >2,500); ++, good adhesion (ADI of between 2,500 and 500); +, weak adhesion (ADI of between 500 and 100); −, no adhesion (ADI of <100). (B) Adhesion to FaDu cell monolayers, as observed with Giemsa staining under a light microscope. Bars, 8 μm. One FaDu nucleus for each layer is indicated with the letter N.
The antagonistic activity against Streptococcus pyogenes on human epithelial cell lines is strain dependent.
All the oral S. salivarius strains studied in the adhesion assay, together with the other bacteria displaying a significant adhesion on the FaDu cell layer (Fig. 2), were investigated with a three-component system consisting of the epithelial cell layer, the S. pyogenes C11LucFF indicator bioluminescent strain, and the tester bacterium. In this experiment, we measured the reduction of bioluminescence produced by S. pyogenes C11LucFF as an indication of the antagonistic activity exerted by the tester strains.
Antagonism through exclusion was tested on the layers of two different human epithelial cell lines, FaDu and HaCaT. The results showed that antagonistic exclusion against S. pyogenes C11LucFF was generally stronger on FaDu hypopharyngeal carcinoma cells than HaCaT keratinocytes. The results also demonstrated that the exclusion activity is strain specific, since significant differences were also observed among strains of the same species.
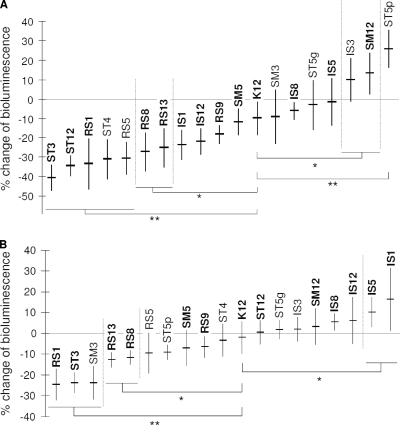
We found a correlation between the results obtained with the two cell lines. In fact, with only a few exceptions, those strains that significantly reduced the light production in HaCaT cells also did so in the FaDu cell layer. Particularly, the following two strains were the most active in both cell lines: S. salivarius ST3 (with average reductions of bioluminescence of 40% in FaDu and 24% in HaCaT) and S. salivarius RS1 (−33% in FaDu and −25% in HaCaT) (Fig. 3). Their activity was significantly stronger than that of the S. salivarius K12 reference oral probiotic strain. Therefore, strains RS1 and ST3 were selected and included in all of the following experiments.
FIG. 3.
Antagonistic exclusion activity of bacterial pharyngeal isolates against bioluminescent Streptococcus pyogenes C11LucFF on FaDu hypopharyngeal carcinoma cells (A) and HaCaT keratinocytes (B). Data reported as percent variation of light emission, which referred to the cell layer treated with only PBS buffer before incubation with S. pyogenes. Numerical results are given as arithmetic means ± standard deviations. Each sample was processed in triplicate in at least two independent experiments. Strains belonging to species S. salivarius are indicated in boldface. Statistically significant differences compared to strain K12 were calculated according to an unpaired Student's t test (**, P < 0.001; *, P < 0.05).
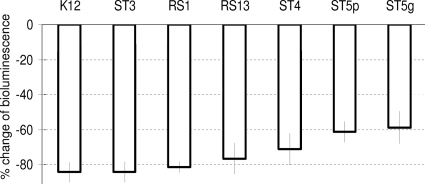
The antagonism by competition was tested only on the FaDu cell layer for the two previously selected strains (S. salivarius ST3 and RS1), the reference oral probiotic K12, and four other strains (S. salivarius RS13, S. oralis ST4 and ST5p, and S. sanguinis ST5g), which were selected in order to have at least one representative strain for each of the four typologies of bacterial adhesion on the FaDu layer (Fig. 2). In this experiment, all tested strains markedly inhibited S. pyogenes bioluminescence, with particular evidence for S. salivarius strains (∼80% reduction) (Fig. 4). We also noticed a partial dependence of competition on adhesion ability. The antagonism efficacy was in fact slightly weaker for poorly adhesive strains (e.g., ST5p and ST5g) than strongly adhesive bacteria (e.g., K12 and ST3; P values of <0.001, according to an unpaired Student's t test) (Fig. 4).
FIG. 4.
Antagonistic competition activity of bacterial pharyngeal isolates against bioluminescent Streptococcus pyogenes C11LucFF on FaDu hypopharyngeal carcinoma cells. Data reported as percent variation of light emission, which referred to the cell layer treated with only S. pyogenes cells. Numerical results are given as arithmetic means ± standard deviations. All samples resulted as being significantly different compared to the control (P < 0.001, according to an unpaired Student's t test).
Inhibition of S. pyogenes and PCR detection of bacteriocin-encoding genes.
After 24 h of incubation at 37°C, we observed clear inhibition zones corresponding to the colonies of the tester strains K12, RS1, and ST3 (data not shown). On the contrary, when cell-free neutralized broths were used in disk diffusion tests, inhibition zones were observed only for the reference strain S. salivarius K12. Accordingly, PCR experiments suggested that strains RS1 and ST3 could not possess bacteriocin genes, while strain K12 gave a positive signal for two lantibiotics, salivaricins A and B (data not shown), which are known to be carried by strain K12 on a 190-kb transmissible plasmid (22). In contrast, strains RS1 and ST3 are plasmid-free strains (G. Ricci, personal communication). Therefore, it appears plausible that the pharyngeal isolates ST3 and RS1 inhibited the growth of S. pyogenes in the plate probably simply due to their acid production.
S. salivarius K12, RS1, and ST3 drive different immune responses in vitro.
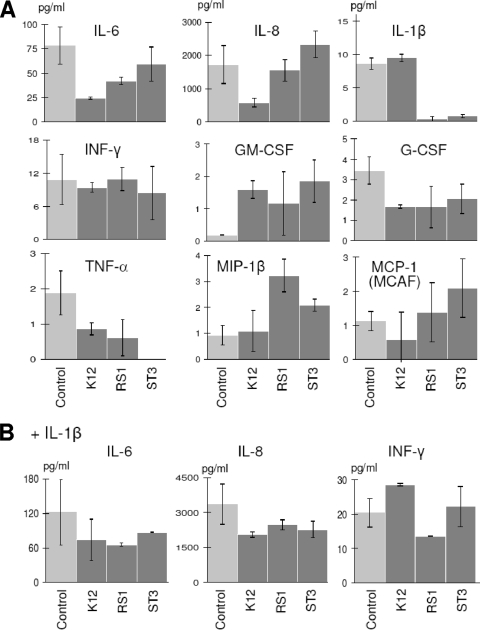
Overnight incubation of a FaDu monolayer with selected bacteria resulted in the modulation of cytokine production profiles, as determined by the Bio-Plex human cytokine 17-plex array system (Bio-Rad) (Table 2 and Fig. 5). Under baseline conditions, the oral isolate S. salivarius RS1 nearly eliminated IL-1β secretion and reduced IL-6 and TNF-α in the FaDu supernatant from 78.1 to 42.0 pg ml−1 and from 1.9 to 0.6 pg ml−1, respectively. Similarly, S. salivarius ST3 eliminated the secretion of IL-1β and TNF-α but, in addition, enhanced the production of IL-8 (from 1,721 to 2,331 pg ml−1), GM-CSF (from 0.2 to 1.9 pg ml−1), and MIP-1β (from 0.9 to 3.2 pg ml−1). The commercial oral probiotic Streptococcus salivarius K12 decreased IL-6 and IL-8 at levels greater than those of the other two streptococci (from 78.1 to 24.2 pg ml−1 and from 1,721 to 579 pg ml−1, respectively). Furthermore, strain K12 induced a reduction of the secretion of TNF-α (from 1.9 to 0.9 pg ml−1) and G-CSF (from 3.5 to 1.7 pg ml−1) and increased GM-CSF (from 0.2 to 1.6 pg ml−1) (Table 2 and Fig. 5A).
TABLE 2.
Cytokine levels
| Cytokine | Cytokine level (pg ml−1) in EMEM culture brotha |
|||||||
|---|---|---|---|---|---|---|---|---|
| Without IL-1β |
With IL-1β |
|||||||
| Control | K12 | RS1 | ST3 | Control | K12 | RS1 | ST3 | |
| IL-1β | 8.60 ± 0.85 | 9.50 ± 0.54 | 0.34 ± 0.37 | 0.77 ± 0.25 | 254.2 ± 3.3 | 298.1 ± 64.1 | 204.7 ± 20.0 | 229.8 ± 18.7 |
| IL-2 | 0.35 ± 0.49 | 1.18 ± 0.00 | 0.35 ± 0.49 | 1.16 ± 0.37 | 0.86 ± 0.49 | 2.75 ± 0.45 | 0.15 ± 0.05 | 0.98 ± 0.04 |
| IL-4 | 0.22 ± 0.01 | 0.15 ± 0.21 | 0.19 ± 0.19 | 0.21 ± 0.09 | 0.27 ± 0.04 | 0.76 ± 0.06 | 0.57 ± 0.16 | 0.46 ± 0.03 |
| IL-5 | BDL | BDL | BDL | BDL | BDL | BDL | BDL | BDL |
| IL-6 | 78.10 ± 19.18 | 24.23 ± 1.03 | 41.95 ± 3.74 | 59.01 ± 17.47 | 121.5 ± 56.6 | 73.66 ± 36.03 | 65.23 ± 2.69 | 86.72 ± 0.29 |
| IL-7 | 0.28 ± 0.03 | 0.20 ± 0.28 | 0.45 ± 0.26 | 0.86 ± 0.27 | 0.61 ± 0.23 | 0.37 ± 0.06 | 0.83 ± 0.12 | 0.46 ± 0.22 |
| IL-8 | 1,721 ± 565 | 579,0 ± 134.7 | 1,547 ± 327 | 2,331 ± 402 | 3,353 ± 872 | 2,043 ± 124 | 2,464 ± 213 | 2,261 ± 357 |
| IL-10 | 0.07 ± 0.10 | 0.26 ± 0.04 | 0.02 ± 0.03 | 0.16 ± 0.22 | BDL | 0.33 ± 0.22 | BDL | 0.15 ± 0.05 |
| IL-12 (p70) | 0.40 ± 0.57 | 0.80 ± 0.28 | 0.34 ± 0.07 | BDL | BDL | 1.95 ± 1.06 | 0.60 ± 0.85 | 0.70 ± 0.14 |
| IL-13 | 0.61 ± 0.09 | 0.36 ± 0.01 | 0.09 ± 0.03 | 0.18 ± 0.25 | 0.21 ± 0.05 | 1.16 ± 0.37 | 0.48 ± 0.04 | 0.67 ± 0.11 |
| IL-17 | 0.29 ± 0.17 | 0.94 ± 0.36 | 0.47 ± 0.66 | 0.91 ± 0.21 | 0.32 ± 0.028 | 3.08 ± 0.77 | 1.66 ± 0.57 | 2.06 ± 0.57 |
| G-CSF | 3.45 ± 0.67 | 1.67 ± 0.08 | 1.65 ± 1.73 | 2.06 ± 0.72 | 9.55 ± 2.52 | 7.62 ± 0.41 | 5.02 ± 1.08 | 13.14 ± 5.17 |
| GM-CSF | 0.19 ± 0.01 | 1.59 ± 0.28 | 1.17 ± 0.98 | 1.85 ± 0.65 | 2.84 ± 0.75 | 5.55 ± 2.12 | 2.88 ± 0.95 | 2.37 ± 0.51 |
| IFN-γ | 10.83 ± 4.60 | 9.42 ± 0.86 | 10.90 ± 2.09 | 8.38 ± 4.82 | 20.31 ± 4.12 | 28.54 ± 0.35 | 13.52 ± 0.21 | 22.11 ± 5.90 |
| MCP-1 | 1.13 ± 0.28 | 0.58 ± 0.81 | 1.39 ± 0.87 | 2.09 ± 0.85 | 3.08 ± 0.66 | 3.36 ± 0.40 | 2.57 ± 0.02 | 2.73 ± 0.45 |
| MIP-1β | 0.92 ± 0.38 | 0.25 ± 0.18 | 1.08 ± 0.79 | 3.23 ± 0.63 | 1.79 ± 0.24 | 2.46 ± 0.76 | 2.37 ± 1.59 | 3.54 ± 0.97 |
| TNF-α | 1.88 ± 0.62 | 0.86 ± 0.17 | 0.62 ± 0.52 | BDL | 3.51 ± 0.15 | 5.99 ± 1.87 | 3.18 ± 1.51 | 7.26 ± 1.49 |
Determined by using Bio-Plex assays after overnight incubation of FaDu layers in the presence of bacterial cells (2 × 108 cells ml−1) with or without IL-1β (2 ng ml−1). Control samples were obtained by incubating FaDu layers without bacterial cells. BDL, below detection limit.
FIG. 5.
Cytokine secretions that changed significantly after treatment of the FaDu layer with bacterial cells, as determined by using the Bio-Plex assay. The same results are also included in Table 2. FaDu layers were incubated overnight with bacterial cells (2 × 108 cells ml−1) without (A) and in the presence of (B) 2 ng ml−1 of IL-1β. The values are the means from two experiments conducted in duplicate. The vertical bars indicate standard deviations.
When the FaDu layer was stimulated with 2 ng ml−1 of the proinflammatory cytokine IL-1β, all strains tested noticeably reduced the concentrations of IL-6 and IL-8 in the FaDu culture supernatant. To be specific, strains K12, RS1, and ST3 reduced IL-6 from 121.5 to 73.7, 65.2, and 86.7 pg ml−1, respectively, and IL-8 from 3,353 to 2,043, 2,464, and 2,261 pg ml−1, respectively. Furthermore, S. salivarius RS1 induced a reduction of IFN-γ secretion from 20.3 to 13.5 pg ml−1 (Table 2 and Fig. 5B).
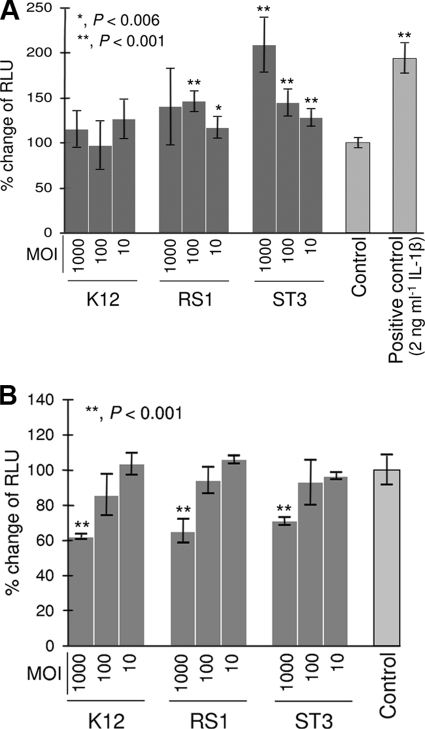
To further explore the immunomodulatory properties of selected oral bacteria, we tested the effect of the microorganisms on NF-κB activation using a recombinant cell line, obtained by transfecting FaDu cells with a luciferase reporter vector inducible by NF-κB. In FaDu cells, strains RS1 and ST3 showed at baseline a stimulatory effect on NF-κB-dependent production of bioluminescence. NF-κB activation was particularly evident for strain ST3, which doubled the bioluminescence produced by FaDu cells when we employed an MOI of about 1,000. In contrast, S. salivarius K12 did not significantly affect NF-κB at baseline (Fig. 6A).
FIG. 6.
Effects of selected bacterial strains on FaDu cells stably transfected with an NF-κB/luciferase reporter vector, without (A) or with (B) stimulation with IL-1β (2 ng ml−1). Luciferase activity is expressed as percent change of relative luminescence units (RLU), assuming the control as 100%. Control, FaDu layers incubated without bacterial cells. The values are the means (± standard deviations) from at least three independent experiments conducted in duplicate. Asterisks indicate statistically significant differences compared to the control. MOI, multiplicity of infection (bacterial cells per FaDu cell).
In the series of experiments which followed, the effect of bacterial strains on NF-κB activation was assessed during stimulation of FaDu cells with the proinflammatory cytokine IL-1β. The addition of 2 ng ml−1 of IL-1β to FaDu culture medium caused an almost 2-fold increase in NF-κB activity after 4 h of incubation. The IL-1β-dependent increase in bioluminescence was partially inhibited only at the highest MOI tested (1,000) by S. salivarius strains K12 (−38%), RS1 (−34.5%), and ST3 (−29.2%) (Fig. 6B).
Safety assessment of selected bacteria by antibiotic susceptibility testing.
We studied the antibiotic resistance of S. salivarius K12, RS1, and ST3 with reference to the European Food Safety Authority (EFSA) breakpoints for Streptococcus thermophilus (16), because S. salivarius is phylogenetically close to S. thermophilus. Bacterial strains were found to be sensitive to ampicillin, chloramphenicol, erythromycin, oxytetracycline, and vancomycin. A variable resistance was observed with the aminoglycoside antibiotics (gentamicin, kanamycin, and streptomycin), depending on the strain and the growth medium. All strains were assessed to be resistant to kanamycin, with the only exception being S. salivarius ST3 grown in LM17 medium (Table 3). Strain K12 was resistant to gentamicin in LM17 medium and to streptomycin in BHI medium. Strain RS1 displayed resistance to gentamicin in LM17 and MRS media and to streptomycin in BHI and LM17 media. Finally, S. salivarius ST3 was resistant to gentamicin in MRS medium and to streptomycin in BHI and MRS media (Table 3).
TABLE 3.
Antibiotic sensitivities of selected bacterial strains
| Antibiotic | MIC (μg ml−1)c |
EFSA MIC (μg ml−1)b | |||||||
|---|---|---|---|---|---|---|---|---|---|
| K12 |
RS1 |
ST3 |
|||||||
| BHI | LM17 | BHI | LM17 | MRS | BHI | LM17 | MRS | ||
| Ampicillin | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 2 |
| Chloramphenicol | <1 | <4, >1 | <1 | <4, >1 | <4, >1 | <1 | <4, >1 | <4, >1 | 4 |
| Erythromycin | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 2 |
| Gentamicina | <32 | >64 | <16 | <64 | >64 | <16 | <8 | >64 | 32 |
| >16 | >8 | >32 | >8 | ||||||
| Kanamycina | >128 | >128 | >128 | >128 | >128 | >128 | <16 | >128 | 64 |
| Oxytetracycline | <1 | <1 | <1 | <4, >1 | <1 | <1 | <1 | <1 | 4 |
| Streptomycin1 | <128, >64 | <16 | <128, >64 | <128, >64 | <64, >32 | <128, >64 | <32, >16 | <128, >64 | 64 |
| Vancomycin | <1 | <4, >1 | <1 | <4, >1 | <4, >1 | <4, >1 | <4, >1 | <4, >1 | 4 |
ST3 is a natural urease-negative S. salivarius strain.
The potential safety of selected S. salivarius isolates was also tested, considering their ability to hydrolyze urea. S. salivarius K12 and RS1 were markedly urealytic when grown in a medium containing nickel cations. In contrast, strain ST3 did not hydrolyze urea. According to the phenotypic assay, we failed to amplify from the strain ST3 ureC gene, which encodes the alpha subunit protein containing the active site and conserved nickel binding ligands of the urease complex. In contrast, a PCR fragment of the expected length was obtained from the other S. salivarius strains using primers targeting ureC (data not shown).
DISCUSSION
Upper respiratory tract infections (URTIs) are the most frequent reason for a visit to a pediatrician, and Streptococcus pyogenes is a major cause of acute pharyngeal infections, especially in children 5 to 12 years of age (12). At present, the treatment of acute bacterial pharyngitis consists of the administration of broad-spectrum antibiotics. These have been estimated to be prescribed in as high as 90% of the pediatric visits for URTIs. A probiotic strategy effective in the prophylaxis of pharyngitis, therefore, could provide a significant social benefit.
In accordance with this objective, in this study we aimed to select oral bacteria with potential probiotic features for the pharyngeal mucosa. We included bacteria newly isolated from the pharynges of healthy donors. Potentially, pharyngeal isolates could, in fact, display better performances in the colonization of the oral ecosystem than traditional dairy or intestinal probiotic bacteria.
In this research, half of the pharyngeal isolates were ascribed to the species Streptococcus salivarius (28 out of 56 isolates), in accordance with previous studies showing these bacteria to be the dominant cultivable species in the oropharyngeal tract (24). Nonpathogenic streptococci are the bacteria most largely present at the oropharyngeal level, and they have been proposed to exert a key role in the protection against pathogenic agents, which cause inflammation and infections (39). In particular, Streptococcus salivarius already becomes a stable colonizer of the oral microbiota a few days after birth and represents, in adults, the major species at the levels of the pharyngeal mucosa and dorsal tongue.
The main criterion we adopted for the selection of potential pharyngeal probiotics was the ability to antagonize Streptococcus pyogenes on human epithelial cell layers. The experimental system set up during this study consisted of the use of a recombinant S. pyogenes strain, expressing a firefly luciferase. Since luciferase catalyzes a bioluminescent reaction that depends stoichiometrically on ATP, the measurement of light production, after addition of the substrate d-luciferin, is dependent on both the number and the metabolic state of bacterial cells.
We also included in this part of the study immortalized HaCaT cells that have been reported to retain many characteristics of the human keratinocytes from which they were originally derived (7). Keratinocytes are, in fact, a constitutive part of the stratified oral epithelium and represent a primary target of adhesion for invading S. pyogenes cells (1, 14). In exclusion experiments, the ability of the tested bacteria to antagonize S. pyogenes was found to be strain specific. In fact, the most active strains on both of the epithelial layers were S. salivarius RS1 and ST3, while strain SM12, belonging to the same species, was unable to antagonize S. pyogenes.
Adhesion experiments performed on the FaDu cell layer can give a partial explanation of the strong antagonizing activity displayed by S. salivarius ST3 and RS1 (Fig. 3). Strains ST3 and RS1, in fact, adhered efficiently to the epithelial layer (Fig. 2). It can be hypothesized, therefore, that the competition for adhesion sites is a major mechanism through which these bacteria antagonize S. pyogenes on FaDu cells.
One mechanism of action of probiotics is suggested to be their modulation of host immune responses. In a recent study, Cosseau and collaborators showed that the oral probiotic S. salivarius K12 can induce in vitro anti-inflammatory responses in epithelial cells, indicating a potential promotion of cellular health and homeostasis (11). In that study, after coculture of human bronchial epithelial cells (16HBE14O- cells) with strain K12, they observed an inhibition of the baseline secretion of the chemokine IL-8, in coincidence with the inhibition of the activation of the NF-κB pathway (11).
In our study, the immunomodulatory properties of S. salivarius K12, RS1, and ST3 were tested on a FaDu layer by means of ELISA quantification of 17 secreted cytokines.
Subsequently, in order to elucidate the possible mechanisms involved in the effects on cytokine production, we studied the modulation of NF-κB activation.
In these experiments, none of the tested strains exhibited potential proinflammatory effects, suggesting that they could be well tolerated by human epithelial cells in vivo. This statement is consistent with the induced reduction of baseline TNF-α secretion, which was observed with all tested strains. In details, we found two different behaviors among the bacteria under study. While strain K12 reduced baseline IL-8 and IL-6 secretion, in contrast, RS1 and ST3 inhibited drastically IL-1β and stimulated the MIP-1β and MCP-1 cytokines. These results have been partially explained by the experiments on NF-κB activation. The reduced secretion of IL-8 and IL-6 provoked by strain K12 can be attributed to the inhibition of NF-κB activation, as already proposed by Cosseau et al. (11). In contrast, S. salivarius RS1 and ST3 promoted the baseline activation of NF-κB. Greten and collaborators have recently demonstrated that NF-κB activates the secretion of a selective inhibitor of caspase-1, a peptidase required for pro-IL-1β maturation (18). Therefore, we can reasonably speculate that the inhibition of IL-1β secretion by strains RS1 and ST3 could also be mediated by a mechanism involving inhibition of the enzyme caspase-1. The oral S. salivarius isolates RS1 and ST3, additionally, stimulated the secretion of MIP-1β and MCP-1 by FaDu cells. Similar behavior has been previously described for the well-known intestinal probiotic Escherichia coli Nissle 1917 (40). Even if MIP-1β and MCP-1 are proinflammatory cytokines, it has been proposed that, upon contact with commensal microbes, local induction of proinflammatory immune responses by way of the upregulation of the MCP-1 and MIP cytokines might reflect part of the host defense process against pathogenic bacteria by establishing a protective immunological barrier (40).
Cytokine secretion and modulation of NF-κB activity by selected bacteria were also tested on FaDu cells stimulated with IL-1β, a prototypical proinflammatory cytokine that plays a central role in the inflammation amplification cascade. After stimulation by IL-1β, we observed that, at an MOI of 1,000, strains K12, RS1, and ST3 reduced the NF-κB activation in a statistically significant manner, while the other conditions tested had no significant effect. The stimulatory activity of RS1 and ST3 bacterial cells on NF-κB activation was, therefore, eliminated in the presence of the inflammatory stimulus due to IL-1β. Similarly, it is noteworthy that in IL-1β-treated FaDu cells, S. salivarius strains can considerably reduce IL-6 and IL-8 secretion, suggesting their potential anti-inflammatory activity.
Recently, the European Food Safety Authority (EFSA) assigned a “Qualified Presumption of Safety” (QPS) status (15) to several lactic acid bacterial species, including Streptococcus thermophilus but not Streptococcus salivarius. In Europe, S. salivarius belongs to risk group 2 (like S. pyogenes or S. pneumoniae), while the very closely related Streptococcus species S. thermophilus, S. uberis, and S. vestibularis (32) belong to risk group 1. Presumably, S. salivarius is considered an opportunistic pathogen because, as with many food-grade lactobacilli, there have been sporadic reports of infections, generally in subjects under adverse medical conditions (2, 3, 10). Quite the opposite, in other parts of the world, S. salivarius has already acquired the status of safe microorganism and has been commercialized for several years as a probiotic without any reported adverse consequences (9). In the light of the above-mentioned facts, the optimal strategy to assess the safety of S. salivarius would be considering every specific strain independently, in accordance with FAO/WHO guidelines on probiotics (17), as has been done for S. salivarius strain K12 (9, 11). From this perspective, the absence of transmissible antibiotic resistances is considered a key safety prerequisite for the selection of a probiotic microorganism (15, 17). In this study, S. salivarius strains, according to the EFSA breakpoints suggested for S. thermophilus, resulted in sensitivity to a variety of antibiotics that are routinely used for the control of URTIs. Differently, they showed resistance exclusively to the aminoglycosidic antibiotics gentamicin, kanamycin, and streptomycin, for which an intrinsic resistance is known for several lactic acid bacteria (10, 16, 21, 30).
Another bacterial feature that exerts an important role in the interaction with the human host is urease activity. Ammonia production from ureolysis in saliva and crevicular fluids is, in fact, a primary source of amino nitrogen and is thought to inhibit the initiation and progression of dental caries by reducing acidity (26, 33). At the same time, a high concentration of ammonia can have deleterious effects on host cells (19), such as fibroblasts and polymorphonuclear leukocytes, and may therefore contribute to tissue damage (19). Among the species of oral bacteria that have been identified as ureolytic, S. salivarius is known to produce high levels of urease (37). Unexpectedly, S. salivarius strain ST3, selected for this work for its potential probiotic properties, was unable to hydrolyze urea. Accordingly, the gene coding for the main subunit of the urease complex (ureC) was missing. In the consideration of its use as the pharynx's probiotic, this bacterium would interact directly with oropharyngeal mucosae and tonsil crypts. Therefore, the inability to hydrolyze urea could be of potential benefit in preserving the host's mucosal cells from ammonia injury.
In summary, during this research, an in vitro rational process led us to select two potential probiotic bacterial strains for the pharynx mucosa. These bacteria efficiently adhered to human epithelial pharyngeal cell lines, antagonizing the adhesion of Streptococcus pyogenes. Furthermore, immunological tests suggested a good degree of adaptation to the host and potential immunomodulatory and anti-inflammatory abilities by selected commensal streptococci. Future work will aim to characterize the probiotic potential of these bacteria in the oral tract by means of in vivo murine models of upper respiratory tract infections.
Acknowledgments
We thank Elisa Bertolasi and Giovanni Ricci for technical support. Special thanks go to Marco Oggioni for providing Streptococcus pyogenes C11.
Part of this study was financed by PUR 2008.
Footnotes
Published ahead of print on 23 April 2010.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9:1822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Afek, S., A. D. Sperber, and Y. Almog. 2004. Carcinoma of the colon presenting as Streptococcus salivarius sepsis. J. Clin. Gastroenterol. 38:86-87. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, R., T. Hassall, B. Morland, and J. Gray. 2003. Viridans streptococcus bacteremia in children on chemotherapy for cancer: an underestimated problem. Pediatr. Hematol. Oncol. 20:439-444. [PubMed] [Google Scholar]
- 4.Beimfohr, C., W. Ludwig, and K. H. Schleifer. 1997. Rapid genotypic differentiation of Lactococcus lactis subspecies and biovar. Syst. Appl. Microbiol. 20:216-221. [Google Scholar]
- 5.Bek-Thomsen, M., H. Tettelin, I. Hance, K. E. Nelson, and M. Kilian. 2008. Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy. Infect. Immun. 76:1889-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borchert, D., L. Sheridan, A. Papatsoris, Z. Faruquz, J. M. Barua, I. Junaid, Y. Pati, F. Chinegwundoh, and N. Buchholz. 2008. Prevention and treatment of urinary tract infection with probiotics: review and research perspective. Indian J. Urol. 24:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, J. P., C. N. Chilcott, C. J. Moore, G. Speiser, and J. R. Tagg. 2006. A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 100:754-764. [DOI] [PubMed] [Google Scholar]
- 9.Burton, J. P., P. A. Wescombe, C. J. Moore, C. N. Chilcott, and J. R. Tagg. 2006. Safety assessment of the oral cavity probiotic Streptococcus salivarius K12. Appl. Environ. Microbiol. 72:3050-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corredoira, J. C., M. P. Alonso, J. F. Garcia, E. Casariego, A. Coira, A. Rodriguez, J. Pita, C. Louzao, B. Pombo, M. J. Lopez, and J. Varela. 2005. Clinical characteristics and significance of Streptococcus salivarius bacteremia and Streptococcus bovis bacteremia: a prospective 16-year study. Eur. J. Clin. Microbiol. Infect. Dis. 24:250-255. [DOI] [PubMed] [Google Scholar]
- 11.Cosseau, C., D. A. Devine, E. Dullaghan, J. L. Gardy, A. Chikatamarla, S. Gellatly, L. L. Yu, J. Pistolic, R. Falsafi, J. R. Tagg, and R. E. Hancock. 2008. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 76:4163-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberl, G., and M. Lochner. 2009. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2:478-485. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, M. L., P. K. Fagan, R. J. Towers, B. J. Currie, and K. S. Sriprakash. 2004. Inhibition of Streptococcus pyogenes adherence to HaCaT cells by a peptide corresponding to the streptococcal fibronectin-binding protein, SfbI, is strain dependent. Microbes Infect. 6:926-928. [DOI] [PubMed] [Google Scholar]
- 15.EFSA. 13-14 December 2004. Scientific Colloquium Summary Report. QPS: qualified presumption of safety of microorganisms in food and feed. EFSA, Brussels, Belgium.
- 16.EFSA. 2008. Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. Prepared by the Panel on Additives and Products or Substances Used in Animal Feed. EFSA J. 732:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FAO/WHO. 2002. Report of a joint FAO/WHO expert consultation on guidelines for the evaluation of probiotics in food. World Health Organization and Food and Agriculture Organization of the United Nations, London Ontario, Canada.
- 18.Greten, F. R., M. C. Arkan, J. Bollrath, L. C. Hsu, J. Goode, C. Miething, S. I. Goktuna, M. Neuenhahn, J. Fierer, S. Paxian, N. Van Rooijen, Y. Xu, T. O'Cain, B. B. Jaffee, D. H. Busch, J. Duyster, R. M. Schmid, L. Eckmann, and M. Karin. 2007. NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell 130:918-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helgeland, K. 1985. pH and the effect of NH4Cl on human gingival fibroblasts. Scand. J. Dent. Res. 93:39-45. [DOI] [PubMed] [Google Scholar]
- 20.Horz, H. P., A. Meinelt, B. Houben, and G. Conrads. 2007. Distribution and persistence of probiotic Streptococcus salivarius K12 in the human oral cavity as determined by real-time quantitative polymerase chain reaction. Oral Microbiol. Immunol. 22:126-130. [DOI] [PubMed] [Google Scholar]
- 21.Hummel, A. S., C. Hertel, W. H. Holzapfel, and C. M. Franz. 2007. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 73:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyink, O., P. A. Wescombe, M. Upton, N. Ragland, J. P. Burton, and J. R. Tagg. 2007. Salivaricin A2 and the novel lantibiotic salivaricin B are encoded at adjacent loci on a 190-kilobase transmissible megaplasmid in the oral probiotic strain Streptococcus salivarius K12. Appl. Environ. Microbiol. 73:1107-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson-Henry, K. C., D. J. Mitchell, Y. Avitzur, E. Galindo-Mata, N. L. Jones, and P. M. Sherman. 2004. Probiotics reduce bacterial colonization and gastric inflammation in H. pylori-infected mice. Dig. Dis. Sci. 49:1095-1102. [DOI] [PubMed] [Google Scholar]
- 24.Kang, J. G., S. H. Kim, and T. Y. Ahn. 2006. Bacterial diversity in the human saliva from different ages. J. Microbiol. 44:572-576. [PubMed] [Google Scholar]
- 25.Kelly, K. 1994. Out of control: the new biology of machines, social systems and the economic world. Addison-Wesley, Boston, MA.
- 26.Kleinberg, I. 1967. Effect of urea concentration on human plaque pH levels in situ. Arch. Oral Biol. 12:1475-1484. [DOI] [PubMed] [Google Scholar]
- 27.Krutmann, J. 2009. Pre- and probiotics for human skin. J. Dermatol. Sci. 54:1-5. [DOI] [PubMed] [Google Scholar]
- 28.Lanyi, B. 1987. Classical and rapid identification: methods for medically important bacteria, p. 1-67. In R. R. Colwell and R. Grigorova (ed.), Methods in microbiology, vol. 19. Academic Press, New York, NY. [Google Scholar]
- 29.Loimaranta, V., J. Tenovuo, L. Koivisto, and M. Karp. 1998. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob. Agents Chemother. 42:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maragkoudakis, P. A., M. Papadelli, M. Georgalaki, E. G. Panayotopoulou, B. Martinez-Gonzalez, A. F. Mentis, K. Petraki, D. N. Sgouras, and E. Tsakalidou. 2009. In vitro and in vivo safety evaluation of the bacteriocin producer Streptococcus macedonicus ACA-DC 198. Int. J. Food Microbiol. 133:141-147. [DOI] [PubMed] [Google Scholar]
- 31.Mora, D., E. Maguin, M. Masiero, C. Parini, G. Ricci, P. L. Manachini, and D. Daffonchio. 2004. Characterization of urease genes cluster of Streptococcus thermophilus. J. Appl. Microbiol. 96:209-219. [DOI] [PubMed] [Google Scholar]
- 32.Mora, D., G. Ricci, S. Guglielmetti, D. Daffonchio, and M. G. Fortina. 2003. 16S-23S rRNA intergenic spacer region sequence variation in Streptococcus thermophilus and related dairy streptococci and development of a multiplex ITS-SSCP analysis for their identification. Microbiology 149:807-813. [DOI] [PubMed] [Google Scholar]
- 33.Peterson, S., J. Woodhead, and J. Crall. 1985. Caries resistance in children with chronic renal failure: plaque pH, salivary pH, and salivary composition. Pediatr. Res. 19:796-799. [DOI] [PubMed] [Google Scholar]
- 34.Power, D. A., J. P. Burton, C. N. Chilcott, P. J. Dawes, and J. R. Tagg. 2008. Preliminary investigations of the colonisation of upper respiratory tract tissues of infants using a paediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur. J. Clin. Microbiol. Infect. Dis. 27:1261-1263. [DOI] [PubMed] [Google Scholar]
- 35.Proal, A. D., P. J. Albert, and T. Marshall. 2009. Autoimmune disease in the era of the metagenome. Autoimmun. Rev. 8:677-681. [DOI] [PubMed] [Google Scholar]
- 36.Reid, G., J. Dols, and W. Miller. 2009. Targeting the vaginal microbiota with probiotics as a means to counteract infections. Curr. Opin. Clin. Nutr. Metab. Care 12:583-587. [DOI] [PubMed] [Google Scholar]
- 37.Sissons, C. H., and T. W. Cutress. 1987. In-vitro urea-dependent pH-changes by human salivary bacteria and dispersed, artificial-mouth, bacterial plaques. Arch. Oral Biol. 32:181-189. [DOI] [PubMed] [Google Scholar]
- 38.Tagg, J. R. 2004. Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J. Med. Res. 119(Suppl.):13-16. [PubMed] [Google Scholar]
- 39.Tagg, J. R., and K. P. Dierksen. 2003. Bacterial replacement therapy: adapting ‘germ warfare’ to infection prevention. Trends Biotechnol. 21:217-223. [DOI] [PubMed] [Google Scholar]
- 40.Ukena, S. N., A. M. Westendorf, W. Hansen, M. Rohde, R. Geffers, S. Coldewey, S. Suerbaum, J. Buer, and F. Gunzer. 2005. The host response to the probiotic Escherichia coli strain Nissle 1917: specific up-regulation of the proinflammatory chemokine MCP-1. BMC Med. Genet. 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wescombe, P. A., J. P. Burton, P. A. Cadieux, N. A. Klesse, O. Hyink, N. C. Heng, C. N. Chilcott, G. Reid, and J. R. Tagg. 2006. Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie Van Leeuwenhoek 90:269-280. [DOI] [PubMed] [Google Scholar]
- 42.Zilber-Rosenberg, I., and E. Rosenberg. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32:723-735. [DOI] [PubMed] [Google Scholar]