Abstract
Bacillus coagulans has good potential as an industrial production organism for platform chemicals from renewable resources but has limited genetic tools available. Here, we present a targeted gene disruption system using the Cre-lox system, development of a LacZ reporter assay for monitoring gene transcription, and heterologous d-lactate dehydrogenase expression.
White biotechnology holds the promise to replace part of today's petrochemicals through the production of biofuels and green chemicals from renewable resources (19). Facultative anaerobic thermophilic Bacillus species, including the lactic acid-producing Bacillus coagulans (4, 10), appear to be attractive alternatives to mesophilic production hosts as potential next-generation microbial production organisms. Compared to the mesophilic and often aerobic strains, their fermentation requires less cooling and mixing and they can be used in a biorefinery concept (15, 18).
The development of genetic tools for B. coagulans has just emerged. Electroporation protocols that yield very small numbers of transformants have been described (22, 23), and an integration system, based on the thermosensitive lactococcal pSH71/pWV01 replicons, that works in combination with low transformation efficiencies is available (23). Still, metabolic engineering of B. coagulans is hampered by the availability of only a limited number of selective markers (23) and would greatly benefit from having a convenient markerless integration system. The Cre-lox system was previously applied for this purpose in B. subtilis (26). Here, we report the use of this system in the thermophilic B. coagulans for multiple genomic modifications and the reuse and removal of selectable markers.
Spore formation of genetically modified organisms is undesirable from both the biological safety and the industrial processing viewpoint. Therefore, we applied the Cre-lox system to construct a sporulation-deficient B. coagulans derivative by targeting the sigF (spoIIAC) gene, which has been successfully targeted for this purpose in Bacillus licheniformis and B. subtilis (7, 25, 27).
First an integration vector, pMH77 (Table 1), was constructed, carrying a lox66-cat-lox71 cassette encoding chloramphenicol resistance and flanked by restriction sites that can be used to clone the regions for homologous recombination (see the supplemental material). This vector is based on pNZ124 (21) that carries a thermosensitive replicon for B. coagulans (23) and has the cat gene under the control of a B. coagulans promoter. Mutant loxP sites, lox66 and lox71, that flank the cat gene can be recognized by the Cre recombinase, resulting in a double mutant and an inactive lox72 site (1). Subsequently, a plasmid for introduction of the Cre recombinase, pMH66, was constructed (see the supplemental material). The cre gene, encoding Cre recombinase, under the control of a Lactobacillus plantarum promoter (14) was cloned in a pNZ124 derivative with cat replaced by tetK, encoding tetracycline resistance.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant feature(s)a | Reference or source |
|---|---|---|
| B. coagulans strains | ||
| DSM 1 | Type strain | DSMZ collection |
| DSM 1 ΔsigF | DSM 1 ΔsigF | This study |
| DSM 1 ΔlacZ | DSM 1 ΔlacZ | This study |
| DSM 1 ΔlacZ ΔsigF | DSM 1 ΔlacZ ΔsigF | This study |
| Plasmids | ||
| pNZ124 | 2.8 kb, Cmr Ts, Gram-positive-E. coli shuttle vector | 21 |
| pNZ8048 | 3.3 kb, Cmr, pNZ124-based cloning vector for the use of the NICE system | 13 |
| pNW33n | 4.2 kb, Cmr, Geobacillus-E. coli shuttle vector | Bacillus Genetic Stock Centre |
| pMH66 | 5.1 kb, Tetr Ts, pNZ124-based Cre-encoding plasmid | This study |
| pMH77 | 3.0 kb, Cmr Ts, pNZ124-based cloning vector with lox66-cat-lox71 cassette | This study |
| pMH79 | 5.0 kb, Cmr Ts, pMH77-based integration vector for sigF deletion | This study |
| pJS27 | 5.5 kb, Cmr, pNW33n derivative containing Pamy-ldhA | This study |
| pLAC | 5.1 kb, Cmr Ts, pMH77-based integration vector for lacZ deletion | This study |
| pNZlac | 5.5 kb, Cmr Ts, pNZ124-based cloning vector with B. coagulans DSM 1 lacZ gene | This study |
| pSPOIIAA-LAC | 5.7 kb, Cmr Ts, pNZ8048 containing PspoIIAA-lacZ fusion | This study |
| pSPOIID-LAC | 5.9 kb, Cmr Ts, pNZ8048 containing PspoIID-lacZ fusion | This study |
| pDACF-LAC | 5.8 kb, Cmr Ts, pNZ8048 containing PdacF-lacZ fusion | This study |
| pCOTE-LAC | 5.7 kb, Cmr Ts, pNZ8048 containing PcotE-lacZ fusion | This study |
| pPTA-LAC | 5.8 kb, Cmr Ts, pNZ8048 containing Ppta-lacZ fusion | This study |
| pLDHL-LAC | 6.7 kb, Cmr Ts, pNZ8048 containing PldhL1-lacZ fusion | This study |
| pPGI-LAC | 6.2 kb, Cmr Ts, pNZ8048 containing Ppgi-lacZ fusion | This study |
Cmr, chloramphenicol resistant; Tetr, tetracycline resistant; Ts, thermosensitive replicon in B. coagulans.
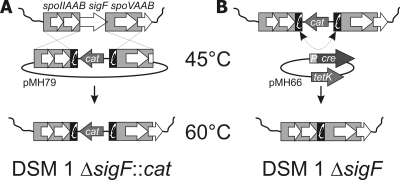
The sigF-flanking regions were cloned in integration vector pMH77, resulting in pMH79, which was introduced into B. coagulans DSM 1 by electroporation (see the supplemental material for details) (Fig. 1 A). One transformant having the presence of pMH79 confirmed by plasmid isolation was selected for integration (see the Fig. 1 legend for details). Single colonies were tested for integration, and both single and double crossovers were detected by PCR. The Cre resolvase-encoding plasmid pMH66 was transformed to a selected double crossover strain, DSM 1 ΔsigF::cat, while selecting for tetracycline resistance (Fig. 1B). By PCR, we detected the absence of cat in all tested transformants, indicating that Cre activity upon introduction was sufficient to cure cat from the transformant colonies and no extended incubation is required. Plasmid pMH66 was cured from the cat-free ΔsigF strain. Plasmid-free colonies were obtained, and one was selected as DSM 1 ΔsigF.
FIG. 1.
Schematic overview of Cre-lox-based chromosomal deletion system for construction of B. coagulans DSM 1 ΔsigF. (A) Exchange of sigF for cat by double crossover. The lox66-cat-lox71 cassette flanked by the homologous regions for crossover is introduced into B. coagulans on a thermosensitive plasmid, pMH79, at the permissive temperature of 45°C and with selection for chloramphenicol resistance (7 μg·ml−1). Double crossover recombination events are selected for by plating at 60°C, a nonpermissive temperature for the integration plasmid, while maintaining the antibiotic resistance pressure. (B) Removal of cat. The Cre-encoding plasmid is introduced on a thermosensitive plasmid (pMH66) carrying a tetracycline resistance gene for selection (1 μg·ml−1). Cre recombines the lox66 and lox71 sites to a lox72 site that is no longer recognized by Cre. The Cre-encoding plasmid is cured by cultivation at 60°C and subsequent plating of a dilution series.
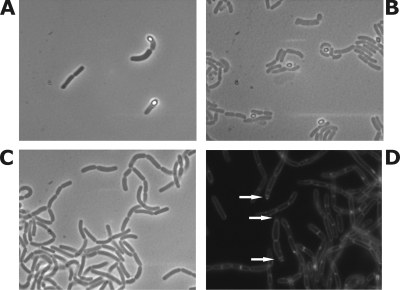
The B. coagulans DSM 1 cells were grown in a specially developed SM medium to quantify sporulation, a phenomenon which is rarely observed for this strain in Luria broth or BC broth (unpublished observations). Wild-type cells showed 3.1 × 10−4 sporulation efficiency, while the ΔsigF strain contained no visible spores when examined using confocal microscopy and gave no colonies in spore tests (described in the supplemental material). After staining of the cell membrane using FM 9-95 dye [N-(3-trimethylammonium propyl)-4-6-4-diethylamino phenol], the wild-type strain showed round spores, while the ΔsigF strain showed the presence of one or two asymmetric septa (Fig. 2), typical of sigF mutants of B. subtilis and B. licheniformis (3, 7).
FIG. 2.
Microscopic observation of wild-type B. coagulans (A and B) and the ΔsigF strain (C and D). Bright spores are observed in wild-type cells using confocal microscopy during early (A) and late (B) stationary phase. No spores are observed in the ΔsigF strain (C), while asymmetric septa can be visualized using FM 9-95 membrane stain during fluorescence microscopy (D).
We wanted to be able to monitor promoter activities of selected genes to assess the effect of the sigF deletion. After unsuccessfully testing several reporter systems (GFPmut1 and GFPuv [9], cyan fluorescent protein and yellow fluorescent protein [24], and anaerobic BsFbFP [B. subtilis FMN-based fluorescent protein] [5]), we decided to use the B. coagulans lacZ gene as a reporter gene. The promoterless B. coagulans lacZ gene identified in the 3.2-Mb draft genome sequence (unpublished results) was cloned, together with its ribosome-binding site (GGAGGAATGCGTGATG [start codon of lacZ is in bold]), in a pNZ124 derivative, resulting in the reporter vector pNZlac (for construction, see the supplemental material). To enable the use of the lacZ reporter system, we eliminated the lacZ gene from the genome by cloning the lacZ upstream and downstream fragments in the integration vector pMH77 (see the supplemental material). After electroporation of the resulting construct, single recombination integration was selected for by chloramphenicol resistance, followed by screening for white colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal)-containing plates, resulting in double-recombinant strains lacking the lacZ gene. The ΔlacZ mutation was also introduced into the DSM 1 ΔsigF strain, enabling a direct comparison between wild-type and ΔsigF mutant strains with the lacZ reporter system.
For evaluation of the effect of the sigF deletion on sporulation gene expression, we cloned promoters of the spoIIA operon, containing sigF (PspoIIA), spoIID (PspoIID), dacF (PdacF), and cotE (PcotE), whose homologues in B. subtilis code for proteins involved in sporulation (6, 20), in pNZlac. We determined the β-galactosidase activities as described before (12), using cultures of ΔlacZ and ΔlacZ ΔsigF strains grown overnight (see Table S2 in the supplemental material). While the promoter activity of PspoIIAA was not significantly changed in the ΔsigF strain, the β-galactosidase activities transcribed from PspoIID, PcotE, and PdacF in the ΔsigF strain were reduced 25, 10, and 50-fold, respectively, compared to those of the wild type. This is in line with the regulation reported for B. subtilis (6).
The effect of the sigF deletion on central metabolism was first determined by cloning the promoter regions of pgi, pta, and ldhL, encoding glucose-6-phosphate isomerase, phosphotransacetylase, and lactate dehydrogenase, respectively, in pNZlac. The reporter activities of the resulting reporter plasmids in the two strain backgrounds showed no significant differences (see Table S2 in the supplemental material), indicating that the sigF deletion has no major impact on central metabolism. This was further evaluated by analyzing the fermentation performance of wild-type and ΔsigF B. coagulans cultivated in parallel in serial-batch lactic acid fermentations. In 3 consecutive fermentations, there were no significant differences in the fermentation times and the organic acid profiles of the two strains (Table 2), demonstrating that the sigF deletion has no major effect on the fermentation performance.
TABLE 2.
Fermentation products of B. coagulans
| Strain | Amt of indicated organic acid produceda |
||
|---|---|---|---|
| Lactic acid | Acetic acid | Formic acid | |
| B. coagulans DSM 1 | 3.5 ± 0.3 | 0.06 ± 0.02 | 0.12 ± 0.05 |
| B. coagulans DSM 1 ΔsigF | 3.7 ± 0.2 | 0.03 ± 0.00 | 0.04 ± 0.00 |
Values are given as the percentage by weight and represent means ± standard deviations for 3 consecutive fermentations using an initial glucose concentration of 50 g·liter−1 and a 10% inoculum. Values for other organic acids were below the detection limit. The density of the fermentation broth was approximately 1.1 g·ml−1.
Lactic acid can serve as a building block for the production of poly(lactic acid) (PLA). Industrially produced lactic acid is mainly the l enantiomer, but stereocomplex PLA also requires the availability of low-cost d-lactic acid (8). Although several production strains for d-lactic acid are described in the literature (e.g., see references 2, 11, 16, 17, and 28), all are mesophilic. To demonstrate that B. coagulans may be used for thermophilic production of d-lactate, the Lactobacillus delbrueckii LMG 6901 ldhA gene was fused to the B. coagulans amylase gene promoter in the thermophilic cloning vector pNW33n, resulting in plasmid pJS27, which was transformed into strain DSM 1. Fermentation studies (Table 3) showed that the lactate produced by B. coagulans DSM 1 and B. coagulans DSM 1 harboring pNW33n was almost enantiopure in the l form, while a significant part of the lactate produced by B. coagulans DSM 1 harboring pJS27 was in the d form. No differences in lactic acid concentration or by-product formation were detected. These results demonstrate that d-lactate production by B. coagulans can be achieved by the introduction of a heterologous d-lactate dehydrogenase gene.
TABLE 3.
Chiral purity of lactic acid produced by B. coagulansa
| Strain | % of lactic acid that was: |
Total amt of lactic acid producedb | |
|---|---|---|---|
| d Form | l Form | ||
| DSM 1 | 0.2, 0.3 | 99.8, 99.7 | 2.5, 2.8 |
| DSM 1(pNW33n) | 0.4, 0.3 | 99.6, 99.7 | 2.3, 2.8 |
| DSM 1(pJS27) | 15.7, 16.9 | 84.3, 83.1 | 2.6, 2.7 |
Data are from two independent fermentations using initial glucose concentrations of 30 g·liter−1.
Values are given as the percentage by weight. The density of the fermentation broth was approximately 1.1 g·ml−1.
B. coagulans shows great potential as a next-generation production platform for building block chemicals or biofuels from renewable resources. We have extended the metabolic engineering toolkit, and we have demonstrated that B. coagulans can produce d-lactate. The construction of an industrial B. coagulans d-lactate production strain requires replacing the ldhL gene with the heterologous ldhA gene for d-lactate production and disruption of sigF for a sporulation-deficient phenotype.
Nucleotide sequence accession numbers.
Sequences used in this study have been deposited in GenBank under accession numbers GU323910 (PpMH71), GU323911 (PspoIIA), GU323912 (PspoIID), GU323913 (PcotE), GU323914 (PdacF), GU323915 (Ppta), GU323916 (PldhL), GU323917 (Ppgi), and GU323918 (PlacZ-lacZ).
Supplementary Material
Acknowledgments
We thank Jurgen Snelders and Esther van Mullekom for their contributions to cloning and fermentation activities.
Part of this work was financially supported by a SenterNovem subsidy, no. IS044081. A.T.K. was financially supported by grant 818.02.004 from the ALW-NWO Open program. This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print on 16 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albert, H., E. C. Dale, E. Lee, and D. W. Ow. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649-659. [DOI] [PubMed] [Google Scholar]
- 2.Chang, D. E., H. C. Jung, J. S. Rhee, and J. G. Pan. 1999. Homofermentative production of d- or l-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 65:1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppolecchia, R., H. DeGrazia, and C. P. Moran, Jr. 1991. Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J. Bacteriol. 173:6678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clerck, E., M. Rodriguez-Diaz, G. Forsyth, L. Lebbe, N. A. Logan, and P. DeVos. 2004. Polyphasic characterization of Bacillus coagulans strains, illustrating heterogeneity within this species, and emended description of the species. Syst. Appl. Microbiol. 27:50-60. [DOI] [PubMed] [Google Scholar]
- 5.Drepper, T., T. Eggert, F. Circolone, A. Heck, U. Krauss, J. K. Guterl, M. Wendorff, A. Losi, W. Gartner, and K. E. Jaeger. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443-445. [DOI] [PubMed] [Google Scholar]
- 6.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming, A. B., M. Tangney, P. L. Jorgensen, B. Diderichsen, and F. G. Priest. 1995. Extracellular enzyme synthesis in a sporulation-deficient strain of Bacillus licheniformis. Appl. Environ. Microbiol. 61:3775-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garlotta, D. 2001. A literature review of poly(lactic acid). J. Polym. Environ. 9:63-84. [Google Scholar]
- 9.Gat, O., I. Inbar, R. Aloni-Grinstein, E. Zahavy, C. Kronman, I. Mendelson, S. Cohen, B. Velan, and A. Shafferman. 2003. Use of a promoter trap system in Bacillus anthracis and Bacillus subtilis for the development of recombinant protective antigen-based vaccines. Infect. Immun. 71:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer, B. W. 1915. Bacteriological studies on the coagulation of evaporated milk. Iowa Agric. Exp. Stn. Res. Bull. 19:119-131. [Google Scholar]
- 11.Ishida, N., T. Suzuki, K. Tokuhiro, E. Nagamori, T. Onishi, S. Saitoh, K. Kitamoto, and H. Takahashi. 2006. d-Lactic acid production by metabolically engineered Saccharomyces cerevisiae. J. Biosci. Bioeng. 101:172-177. [DOI] [PubMed] [Google Scholar]
- 12.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 14.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maas, R. H., R. R. Bakker, M. L. Jansen, D. Visser, E. de Jong, G. Eggink, and R. A. Weusthuis. 2008. Lactic acid production from lime-treated wheat straw by Bacillus coagulans: neutralization of acid by fed-batch addition of alkaline substrate. Appl. Microbiol. Biotechnol. 78:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okano, K., Q. Zhang, S. Shinkawa, S. Yoshida, T. Tanaka, H. Fukuda, and A. Kondo. 2009. Efficient production of optically pure d-lactic acid from raw corn starch by using a genetically modified l-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum strain. Appl. Environ. Microbiol. 75:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okino, S., M. Suda, K. Fujikura, M. Inui, and H. Yukawa. 2008. Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 78:449-454. [DOI] [PubMed] [Google Scholar]
- 18.Ou, M. S., N. Mohammed, L. O. Ingram, and K. T. Shanmugam. 2009. Thermophilic Bacillus coagulans requires less cellulases for simultaneous saccharification and fermentation of cellulose to products than mesophilic microbial biocatalysts. Appl. Biochem. Biotechnol. 155:379-385. [DOI] [PubMed] [Google Scholar]
- 19.Patel, M., M. Crank, V. Dornburg, B. Hermann, L. Roes, B. Hüsing, L. Overbeek, F. Terragni, and E. Recchia. 2006. The BREW project: medium- and long-term opportunities and risks of the biotechnological production of bulk chemicals from renewable resources. http://www.chem.uu.nl/brew/.
- 20.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 21.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee, M. S., J. W. Kim, Y. Qian, L. O. Ingram, and K. T. Shanmugam. 2007. Development of plasmid vector and electroporation condition for gene transfer in sporogenic lactic acid bacterium, Bacillus coagulans. Plasmid 58:13-22. [DOI] [PubMed] [Google Scholar]
- 23.van Kranenburg, R., M. van Hartskamp, E. A. J. Heintz, E. J. G. van Mullekom, and J. Snelders. 2007. Genetic modification of homolactic thermophilic Bacilli. PCT WO/2007/085443.
- 24.Veening, J. W., W. K. Smits, L. W. Hamoen, J. D. Jongbloed, and O. P. Kuipers. 2004. Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl. Environ. Microbiol. 70:6809-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, J. J., W. B. Greenhut, and J. C. Shih. 2005. Development of an asporogenic Bacillus licheniformis for the production of keratinase. J. Appl. Microbiol. 98:761-767. [DOI] [PubMed] [Google Scholar]
- 26.Yan, X., H. J. Yu, Q. Hong, and S. P. Li. 2008. Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl. Environ. Microbiol. 74:5556-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yudkin, M. D. 1987. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J. Gen. Microbiol. 133:475-481. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, S., T. B. Causey, A. Hasona, K. T. Shanmugam, and L. O. Ingram. 2003. Production of optically pure d-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol. 69:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.