Abstract
We have adapted molecular inversion probe technology to identify microbes in a highly multiplexed procedure. This procedure does not require growth of the microbes. Rather, the technology employs DNA homology twice: once for the molecular probe to hybridize to its homologous DNA and again for the 20-mer oligonucleotide barcode on the molecular probe to hybridize to a commercially available molecular barcode array. As proof of concept, we have designed, tested, and employed 192 molecular probes for 40 microbes. While these particular molecular probes are aimed at our interest in the microbes in the human vagina, this molecular probe method could be employed to identify the microbes in any ecological niche.
The substantial majority of the earth's bacteria have not been grown in culture and do not form colonies on agar plates (for examples, see references 19 and 21). These statements are particularly true of the bacteria living in or on human beings (for examples, see references 2, 6, and 7). The Human Microbiome Project (for examples, see references 2, 5, 24, and 27) is employing DNA sequencing and other genome-based technologies to reveal the plethora of microbes living in or on humans. Our goal was to develop a massively multiplex method employing currently commercially available reagents to identify those microbes at the species level.
Several useful approaches to the identification of microbes that do not form colonies on agar plates have been published. Many scientists have employed dideoxy sequencing of the PCR-amplified 16S rRNA gene to identify microbes (for an example, see reference 22). Dideoxy sequencing is expensive, cumbersome, and slow. “Next-generation” sequencing reduces the cost of sequencing but produces much shorter read lengths. As examples, Tarnberg et al. (26), Jonasson et al. (11), and Sundquist et al. (25) employed pyrosequencing of small portions of the 16S rRNA gene to identify microbes. These scientists could not always identify the microbes down to the species level. “Checkerboard DNA-DNA hybridization” (CDH) is a technology that is more than a decade old (23). Nikolaitchouk et al. (14) have applied CDH to identify the microbes in the human female genital tract and achieved a 13-plex reaction. Dumonceaux et al. (4) coupled microbe-specific oligonucleotides to fluorescently labeled microspheres. Subsequently, the microbes were detected and counted by flow cytometry. Dumonceaux et al. (4) achieved a 9-plex reaction. DeSantis et al. (3) designed and employed a microarray containing 297,851 oligonucleotide probes derived from the 16S rRNA gene from 842 subfamilies of prokaryotes. DeSantis et al. (3) concluded that their microarray revealed greater prokaryotic diversity than dideoxy sequencing of a “typically sized clone library.”
None of these ingenious methods meets the requirements for a robust, commercially available, highly multiplex technology. Therefore, we have adapted an independent method to identify microbes: molecular inversion probes (8) coupled with a commercial molecular barcode array. This method does not require growth of the microbes. Rather, molecular probe technology is a nucleic acid-based technology employing DNA homology twice: once for the molecular probe to hybridize to its homologous DNA and again for the 20-mer oligonucleotide barcode on the molecular probe to hybridize to a commercially available oligonucleotide barcode array. We present here data demonstrating proof of concept in which molecular probes were designed, tested, and employed to detect microbes in simulated clinical samples. Because of our ongoing interest in the bacteria that inhabit the adult human vagina (10), we focus on that ecological niche. However, this method is sufficiently general that it can be applied to detect the microbes in any ecological niche, e.g., soil and the ocean.
MATERIALS AND METHODS
Molecular probe design.
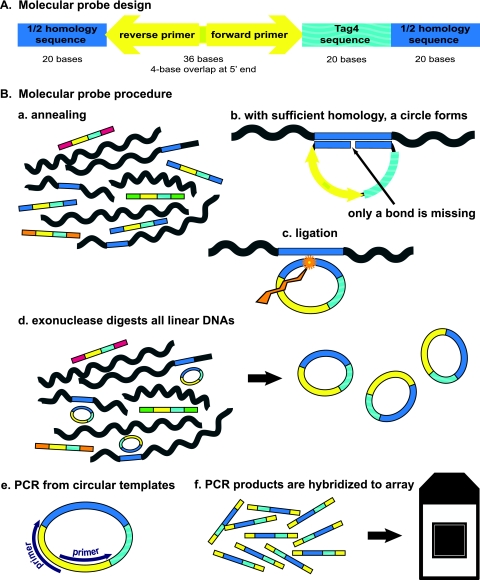
The design of our molecular probes was based upon the previously published designs of padlock probes (15) and molecular inversion probes (8). There are three domains within our molecular probes (Fig. 1 A). The first domain is a contiguous 40-base sequence (a “homer”) unique to the genome of the microbe of choice, divided into two 20-mers. The second domain is a 20-base barcode from the Affymetrix GenFlex Tag16K array v2 (hereinafter the “Tag4 array”; Affymetrix, San Carlos, CA). The Tag4 array contains 8-μm features (also known as 20-mers or barcodes), each replicated and dispersed five times on the array (16). The third domain is a 36-base universal PCR amplification sequence (1). Thus, these molecular probes are 96 bases in length. Six examples of molecular probe design are given in Fig. 2. The designs for all 192 molecular probes are given in Table S1 in the supplemental material. Since the oligonucleotide probes contain fewer than 100 bases, they can be purchased commercially. We purchased the 5′-phosphorylated, PAGE-purified molecular probes from Integrated DNA Technologies (Coralville, IA). We arbitrarily chose to give the molecular probe designs sequential “ED” numbers. The list of microbial DNAs employed in these experiments is given in Table S2 in the supplemental material.
FIG. 1.
(A) Molecular probe design. The deep blue color represents the 40-base homology domain (the “homer”), which is divided into two 20-base segments. The aquamarine color represents the 20-base oligonucleotide barcode from the Tag4 array. The yellow color represents the 36-base domain for the 4-base-overlapped PCR primers. The total length is 96 bases. The 5′ end is phosphorylated. (B) Outline of the molecular probe procedure. (a) In the first step, the molecular probe mixture is incubated with the denatured target DNA under annealing conditions. The molecular probes have different-colored ends to indicate the different homers. (b) Where sufficient homology exists between the molecular probe and the target single-stranded DNA (indicated by the blue color), 40 bp of duplex DNA are formed. The 5′-phosphorylated end of the molecular probe is adjacent to the 3′ end of the probe with no bases missing. (c) A ligation reaction creates a phosphodiester bond between the 5′ and 3′ ends of the molecular probe, as indicated by the jagged gold arrow. The molecular probe is now circular. (d) All linear DNA is digested by added exonucleases. The circular probe remains. (e) 5′-Biotinylated universal amplification primers (purple) are added, and PCR is performed. (f) The PCR products are hybridized to a Tag4 array. This step makes use of the Tag4 bar codes (aquamarine). Washing, staining, and washing of the Tag4 array follow.
FIG. 2.
Design of six molecular probes for L. gasseri DNA. The lowercase letters designate the 40-base (2 × 20 base) homer. The uppercase letters in boldface designate the 36-base combined amplification sequence. The uppercase letters not in boldface designate the 20-mer bar code from the Tag4 array.
We have written custom software, Blaster.rb, to identify 40-base homers unique to the genome of the microbe of interest. Blaster.rb is freely available at http://med.stanford.edu/sgtc/research/blaster.html. Blaster.rb divides an input sequence into 40-base oligomers, offset by 20 bases. Then, Blaster.rb applies the following screens. (i) The 40-mer must be between 45% and 55% G+C, such that multiplexed molecular probes will hybridize at the maximum rate at the same temperature. (ii) The 40-mer cannot contain more than three of any base in a row, because purines, especially guanine, tend to stack, and stacking interferes with hybridization. (iii) The 20th and 21st bases of the homer must be a G or a C. The later ligation event will take place between these two bases. We want three, rather than two, hydrogen bonds holding each base in place. (iv) The 40-mer cannot contain an inverted repeat. Such a repeat could form a stem-and-loop structure and reduce or abolish the homer's ability to form hydrogen bonds with its homologous sequence in the target DNA. (v) Since human DNA was the most likely contaminating DNA in a vaginal swab, the 40-mers were BLASTed against the GenBank Human Genome database. Any homer with a good match to human DNA was excluded. A “good match” was defined as more than 17 perfectly matched bases in a row or a single mismatch within 20 bases. (vi) The 40-mers that passed all of the previous screens were BLASTed against the GenBank Other Genomes database. Blaster.rb reported the 40-mers unique to the species in one list. A second list reported 40-mers unique to the species plus other members of the genus. Blaster.rb is not restricted to the rRNA genes or even to genes. As examples, for the six Lactobacillus gasseri molecular probes (Fig. 2), the homers were derived from the nucleotidyltransferase gene, the lactate dehydrogenase gene, a transcriptional regulator, and two different hypothetical protein genes, respectively. (vii) Blaster.rb does not require a complete genome sequence. For Atopobium vaginae (GenBank accession number ADNA01000000) and Gardnerella vaginalis (GenBank accession number ADNB01000000), we had robust contigs but not complete genome sequences. For each, we linked together about a dozen contigs, placing 20 As between each contig. (viii) Since sequences are added to GenBank daily, identifying unique 40-mers was a moving target. BLAST was repeated over time.
The 96-base molecular probe design was entered into the Rensselaer bioinformatics web server (http://frontend.bioinfo.rpi.edu/applications/mfold/) and folded into all possible secondary structures (28). Any design for which any form had 8 or more contiguous base pairs anywhere among the 96 bases was rejected. Such a structure could compete with the annealing process. Any design that could form six or more contiguous base pairs within 20 bases of the 5′ end or of the 3′ end was rejected. (GT was counted as a base pair.) Approximately half of our designs were rejected by these criteria.
Molecular probe procedure.
Fig. 1B presents an outline of the molecular probe procedure. In the first step (step a), a pool of molecular probes was annealed to the denatured target DNAs in Ampligase buffer (Epicentre Technologies, Madison, WI). We titrated most molecular probes and concluded that the optimal concentration for each molecular probe in the stock solution was 100 attomol/μl (60 × 106 molecules/μl; 10 times higher for Lactobacillus crispatus and Lactobacillus jensenii probes). One microliter of the mixed-probe stock solution was diluted into the final volume of the annealing reaction mixture (7.8 μl). The annealing reaction mixture was heated to 95°C for 5 min. Then, the temperature was dropped one degree at a time for 1 min at each temperature, until 65°C was reached and held at 65°C overnight. For step b, where there was sufficient homology between the homer of the molecular probe and the target DNA, the probe hybridized to the target and yielded 40 bp of duplex DNA. That hybridization brought the 5′-phosphorylated end of the probe adjacent to the 3′ end of the probe with no bases missing. For step c, ligation formed a phosphodiester bond between the 5′ and 3′ bases of the molecular probe. Ampligase (1 unit; Epicentre Technologies) was added, and the reaction mixture was heated to 58°C for 2 min. The volume was 14.2 μl. For step d, all linear DNA was digested by exonuclease treatment: 0.65 units of exonuclease I and 3.3 units of exonuclease III (Epicentre Technologies) were added to each reaction mixture, with a negligible change in reaction mixture volume. The reaction conditions were 37°C for 15 min, followed by 80°C for 15 min to abolish the enzymatic activities. The circular molecular probes remained. The fifth step (step e) was a conventional PCR. We employed AmpliTaq Gold kits purchased from ABI (Foster City, CA) and followed the manufacturer's instructions. We purchased the 5′-biotinylated and high-pressure liquid chromatography-purified universal forward and reverse amplification primers (1) 5′-biotin-TACTGAGGTCGGTACACTCT and 5′-biotin-AGTAGCCGTGACTATCGACT from Integrated DNA Technologies. Two units of AmpliTaq DNA polymerase were added per final 50-μl amplification reaction mixture. The cycling conditions were as follows: 95°C for 10 min; 25 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. For step f, we used 10-μl amounts to electrophorese through a 3% composite gel. The remaining 40-μl amounts (without purification) of the 5′-biotinylated PCR products were hybridized to a Tag4 array (17; also see Materials and Methods in the supplemental material).
Additional information is in the Materials and Methods section in the supplemental material.
Genome accession numbers.
The Atopobium vaginae Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number ADNA00000000. The version described in this paper is the first version, ADNA01000000. The Gardnerella vaginalis Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number ADNB00000000. The version described in this paper is the first version, ADNB01000000.
RESULTS
Specificity.
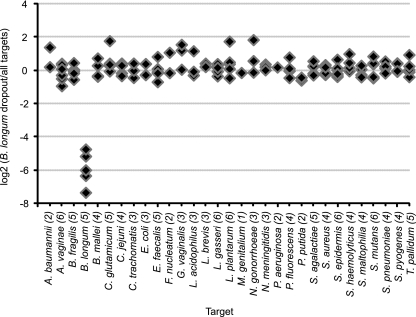
To test the specificity of our molecular probes, we reacted the probe set with different “dropout” pools of genomic DNAs (see Materials and Methods in the supplemental material). Each of the 36 pools was lacking one of the 36 target DNAs recognized by our probe set. The positive control contained all DNAs. For example, the data for the Bifidobacterium longum DNA dropout pool is shown in Fig. 3. The only negative probes were the five probes directed against B. longum DNA. These molecular probes could not hybridize to the absent B. longum DNA and did not hybridize to any of the other 35 prokaryotic genomic DNAs in the dropout pool. Included in these tests for specificity were molecular probes for different species of the same genus. As one example, Neisseria gonorrhoeae probes did not hybridize to Neisseria meningitidis DNA and, conversely, N. meningitidis probes did not hybridize to N. gonorrhoeae DNA (data not shown). As a second example, there were three species of the Pseudomonas genus in the probe mix and the dropout pools: P. aeruginosa, P. fluorescens, and P. putida. The molecular probes for each species hybridized only to the species for which they were designed and not to the DNA of the other two species in the genus (data not shown).
FIG. 3.
Results of a representative dropout experiment demonstrating the specificity of the B. longum molecular probes. One-hundred-twenty-two molecular probes were reacted with a pool of DNA representing 31 microbial targets and an identical pool that lacked only B. longum DNA. The ordinate is the logarithm base 2 of the fluorescence ratio of each molecular probe for these two reactions: [log2(B. longum dropout/all targets)]. The abscissa is each molecular probe arranged alphabetically by microbe, with the number of probes targeting each genome indicated in parentheses. For all but the probes targeting B. longum DNA, the logarithm base 2 of the normalized ratio is within the experimental error of zero, meaning that there was equivalent fluorescence of the probes reacting with the DNAs in the dropout pool relative to that in the complete pool. The five B. longum molecular probes are negative, as they should be, because B. longum DNA was absent from this dropout pool. Organisms not mentioned in the text are Acinetobacter baumannii, Bacteroides fragilis, Campylobacter jejuni, Chlamydia trachomatis, Enterococcus faecalis, Fusobacterium nucleatum, Mycoplasma genitalium, Streptococcus agalactiae, Staphylococcus aureus, Staphylococcus epidermis, Staphylococcus haemolyticus, Streptococcus mutans, Streptococcus pneumoniae, and Streptococcus pyogenes.
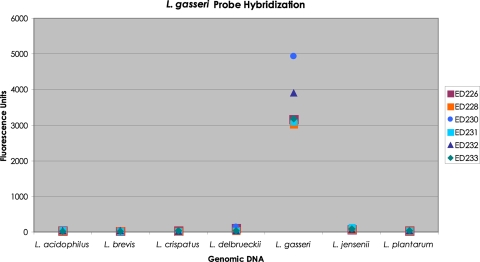
Because of the importance of Lactobacillus bacteria in the adult vagina, additional experiments were performed to test the specificity of the Lactobacillus species probes. As an example, a mixture of six L. gasseri molecular probes was hybridized separately to each of seven Lactobacillus DNAs: L. acidophilus, L. brevis, L. crispatus, L. delbrueckii, L. gasseri, L. jensenii, and L. plantarum. The results are shown in Fig. 4. The only strong fluorescence resulted from each molecular probe reacted with L. gasseri genomic DNA.
FIG. 4.
Histogram of the amount of fluorescence produced by each of six L. gasseri probes against seven Lactobacillus DNAs. A mixture of the six L. gasseri molecular probes was separately hybridized to 10 ng of each of seven genomic Lactobacillus DNAs, L. acidophilus, L. brevis, L. crispatus, L. delbrueckii, L. gasseri, L. jensenii, and L. plantarum. Each hybridization reaction was carried through our complete standard procedure. The fluorescence units corresponding to each molecular probe for each Lactobacillus DNA were recorded. The data are presented as a histogram.
The possible complication of the presence of human DNA in the target.
When collecting vaginal swabs, human cells are sometimes inadvertently also collected, leading to the presence of contaminating human DNA. During our annealing reaction, we did not want any of our molecular probes to hybridize to human DNA, since such hybridization would compromise the interpretation of the data. Therefore, three tests were undertaken. (i) All potential 40-base homers were BLASTed against the human DNA sequence in GenBank. Any potential homer with more than negligible homology was discarded (Materials and Methods). (ii) The sequences of the two universal amplification primers were BLASTed against the human DNA sequence in GenBank. “No significant similarity” was found. (iii) An experiment was undertaken to determine if human DNA would act as a target for any of the molecular probes (see Materials and Methods in the supplemental material). The resulting fluorescence signals are depicted in Fig. S1 in the supplemental material. No probe produced significant signal consistently with all three amounts of human DNA. Only one probe was found to react significantly with the highest concentration of human DNA. However, even this probe yielded a very low signal. Therefore, we concluded that the reactivity of our molecular probes with the assayed amounts of human DNA was negligible.
Minimum detection limit.
Fig. S2 in the supplemental material presents a histogram of the minimum detection limit for the molecular probes (see Materials and Methods in the supplemental material). Notably, molecular probes targeting the same whole-genome amplified (WGA) microbial DNA did not necessarily have the same minimum detection limit. Among the molecular probes with a minimum detection limit of, at most, 0.5 pg were probes for Burkholderia mallei, Corynebacterium glutamicum, L. brevis, L. gasseri, Stenotrophomonas maltophilia, and Treponema pallidum.
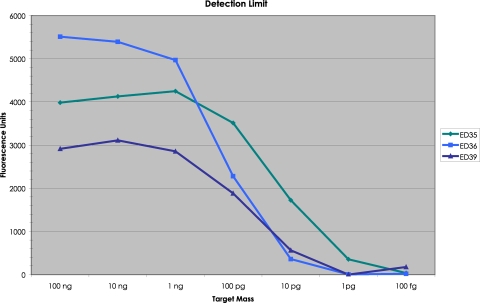
ATCC genomic Escherichia coli DNA was employed to measure the minimum detection limit without the complication of WGA. A mixture of the three E. coli molecular probes (ED35, ED36, and ED39) was hybridized to decreasing concentrations of E. coli DNA and taken through our procedure. The Tag4 array results are shown in Fig. 5. ED35 had a minimum detection limit of 1 pg (326 yoctomol; 2,000 molecules). ED36 and ED39 had a minimum detection limit of 10 pg (3.26 zeptomol; 20,000 molecules). For the same amount of E. coli genomic DNA, the three molecular probes produced a different amount of fluorescence. For example, at apparent saturation (100 ng), ED35 produced 3,984 fluorescence units (FU), ED36 produced 5,513 FU, and ED39 produced 2,914 FU (Fig. 5).
FIG. 5.
Minimum detection limit for three molecular probes for E. coli DNA. A mixture of the three E. coli molecular probes was annealed to decreasing amounts of ATCC E. coli genomic DNA, including a negative control containing no DNA. In the annealing reaction, there were 100 attomol (60 × 106 molecules) of each probe. The amount of E. coli DNA ranged from a high of 100 ng (32.6 attomol; 1.96 × 107 molecules) to a low of 100 fg (32.6 yoctomol; 196 molecules). For the background on the Tag4 array, the mean signal intensity from the unused features was employed. The amount of fluorescence present with no DNA in the annealing reaction was subtracted from each value when DNA was present.
Use of molecular probes with simulated clinical samples.
To simulate the total DNA derived from vaginal swabs, we constructed five mixtures of DNAs wherein each mixture had very different mass amounts of 3 to 5 WGA genomic DNAs relevant to the human vagina, called simulated clinical samples A through E (see Table S3 in the supplemental material). As an example, the data for simulated clinical sample E is shown in Fig. 6. The data for the other four simulated clinical samples are shown in Fig. S3 to S6 in the supplemental material. In all cases, the molecular probes detected only the DNAs known to be in the simulated clinical sample. There were no false positives, and there were no false negatives.
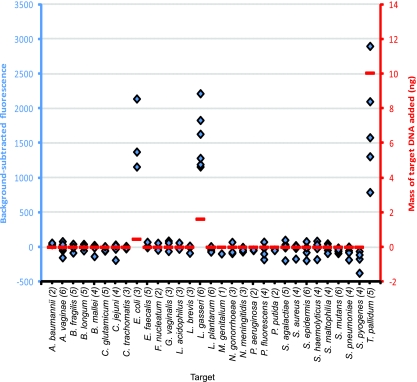
FIG. 6.
Characterization of simulated clinical sample E. The background-subtracted fluorescence is on the left ordinate and shown in blue. The abscissa lists the target 31 microbes in alphabetical order. Following the microbe names, the numbers in parentheses are the number of molecular probes for each microbe. Blue diamonds represent the background-subtracted fluorescence for each of the 122 probes (compared to that for six control experiments in which no DNA was included in the reaction mixture). The amount of WGA target DNA is on the right ordinate and shown in red. The horizontal red lines represent the mass (in ng) of each WGA DNA included in sample E, as follows: 0.45 ng E. coli DNA, 1.6 ng L. gasseri DNA, and 10 ng T. pallidum DNA. Organisms not mentioned in the text are listed in the Fig. 3 legend. The horizontal red line at 0 represents zero fluorescence.
While there was excellent qualitative agreement, quantitative agreement was elusive. For example, in simulated sample E (Fig. 6), there were 0.45 ng (0.15 attomol) of WGA E. coli DNA and 10 ng (13 attomol) of WGA T. pallidum DNA. The range of fluorescence achieved for the five T. pallidum probes extends both higher and lower than the range of fluorescence of the three E. coli probes (Fig. 6), despite the ∼100-fold difference in the molar amount of target DNA. The explanation probably lies in the well-known facts that standard PCR is not quantitative and fluorescence intensity is not a linear function of mass.
DISCUSSION
We have described a method that combines molecular probes with molecular barcode array technology and is capable of detecting any microbe for which adequate DNA sequence information is available. Custom software (which we have made publicly available) was used to identify the 40 contiguous bases of unique target DNA sequence that is essential for the specificity of each molecular probe. We verified the specificity of our molecular probes by testing them against DNA from many other microbial species and against human genomic DNA. Finally, we successfully applied this method to identify microbes in simulated clinical samples.
There are several important benefits in this technology. A key benefit of the molecular probe method is that it does not require growth of the microbe. Another benefit is that the molecular probes are 96 bases in length and, therefore, can be purchased commercially. The molecular probe method employs a commercially available oligonucleotide array. Quality control has been built into the Tag4 array, so that results are reproducible from scientist to scientist and from laboratory to laboratory (17).
Another key benefit is that the method is designed to identify microbes in multiplex. The Tag4 array contains 16,000 features (also known as 20-mers or barcodes). In this work, we have used fewer than 200 features. Thus, molecular probes for a nearly unlimited number of microbes can be added at any time (9). Sequence data produced by the ongoing Human Microbiome Project will only increase the number of organisms that can be interrogated by this method. In the near future, it should be possible to interrogate the entire human microbiome on one Tag4 array. It is widely recognized that the complex population of microbes living in or on humans can reflect the health of the individual (for examples, see references 12, 13, 18, and 20). Therefore, the method presented herein has potential applications for both basic research and clinical medicine.
Supplementary Material
Acknowledgments
We have benefited from discussions with Johan Baner, Lisa Diamond, Simon Fredrickson, Sujatha Krishnakumar, Jochen Kumm, Michael Mindrinos, Curtis Palm, and Nader Pourmand.
We are deeply grateful to 47 generous colleagues who sent us gifts of a combined 87 genomic DNAs: C. Arvidson, A. Blanchard, F. Biville, E. Carretto, T. Chang, J. Christensen, J.-A. Dillon, L. Drago, D. Edmonson, E. Eribe, H. Falentin, M. Ferris, D. Forster, S. Harvey, N. Jones, T. Klaenhammer, S. Kleinsteuber, S. Lau, M. Muller, R. Nichols, H. Nojiri, S. Norris, S. Patrick, B. Peyton, W. Picking, S. Pournaras, J. Ramos, V. Rodwell, G. Rossolini, T. Sandrin, M. Schneegurt, V. Sivadon, Y. Song, G. Spear, T. Stanton, B. Stark, J. Steele, R. Stephens, H. Suzuki, C. Vadeboncoeur, A. Viale, W. Wade, G. Welling, J. West, K. Williams, E. Worobec, and K. Yuen.
This work was supported by National Human Genome Research Institute grant PO1 HG000205.
R.W.H. designed the experiments. R.W.H. and R.P.S. wrote the manuscript. E.A.A. wrote the custom software Blaster.rb. M.F. entered the sequences into the custom software and retrieved the results. R.W.H. designed the molecular probes and performed the molecular probe reactions. M.M. hybridized the molecular probes to the arrays. A.M.A. washed and scanned the arrays. R.P.S. and R.W.H. analyzed the array data. R.P.S. and M.F. produced the graphs thereof. R.W.D. provided the intellectual, physical, and financial milieu for these experiments.
The authors declare no competing financial interests. Affymetrix (San Carlos, CA) currently holds the commercial license to molecular inversion probe technology.
Footnotes
Published ahead of print on 23 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baner, J., P. Marits, M. Nilsson, O. Winqvist, and U. Landegren. 2005. Analysis of T-cell receptor V beta gene repertoires after immune stimulation and in malignancy by use of padlock probes and microarrays. Clin. Chem. 51:768-775. [DOI] [PubMed] [Google Scholar]
- 2.Curtin, C. 2009. Humans as host. Genome Tech. October:38-43.
- 3.DeSantis, T. Z., E. L. Brodie, J. P. Moberg, I. X. Zubieta, Y. M. Piceno, and G. L. Andersen. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371-383. [DOI] [PubMed] [Google Scholar]
- 4.Dumonceaux, T. J., J. Schellenberg, V. Goleski, J. E. Hill, W. Jaoko, J. Kimani, D. Money, T. B. Ball, F. A. Plummer, and A. Severini. 30 September 2009. Multiplex detection of bacteria associated with normal microbiota and with bacterial vaginosis in vaginal swabs by use of oligonucleotide-coupled fluorescent microspheres. J. Clin. Microbiol. doi: 10.1128/JCM.00112-09. [DOI] [PMC free article] [PubMed]
- 5.Friedrich, M. J. 2008. Microbiome project seeks to understand human body's microscopic residents. JAMA 300:777-778. [DOI] [PubMed] [Google Scholar]
- 6.Gao, Z., C. H. Tseng, B. E. Strober, Z. Pei, and M. J. Blaser. 2008. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 3:e2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, J. A. Segre, and the NISC Comparative Sequencing Program. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardenbol, P., J. Baner, M. Jain, M. Nilsson, E. A. Namsaraev, G. A. Karlin-Neumann, H. Fakhrai-Rad, M. Ronaghi, T. D. Willis, U. Landegren, and R. W. Davis. 2003. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21:673-678. [DOI] [PubMed] [Google Scholar]
- 9.Hardenbol, P., F. Yu, J. Belmont, J. Mackenzie, C. Bruckner, T. Brundage, A. Boudreau, S. Chow, J. Eberle, A. Erbilgin, M. Falkowski, R. Fitzgerald, S. Ghose, O. Lartchouk, M. Jain, G. Karlin-Neumann, X. Lu, X. Miao, B. Moore, M. Moorhead, E. Namsaraev, S. Pasternak, E. Prakash, K. Tran, Z. Wang, H. B. Jones, R. W. Davis, T. D. Willis, and R. A. Gibbs. 2005. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 15:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonasson, J., M. Olafsson, and H. J. Monstein. 2002. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. APMIS 110:263-272. [DOI] [PubMed] [Google Scholar]
- 12.Lampe, J. W. 2008. The Human Microbiome Project: getting to the guts of the matter in cancer epidemiology. Cancer Epidemiol. Biomarkers Prev. 10:2523-2524. [DOI] [PubMed] [Google Scholar]
- 13.Mai, V., and P. V. Draganov. 2009. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J. Gastroenterol. 15:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolaitchouk, N., B. Andersch, E. Falsen, L. Strömbeck, and I. Mattsby-Baltzer. 2008. The lower genital tract microbiota in relation to cytokine-, SLPI- and endotoxin levels: application of checkerboard DNA-DNA hybridization (CDH). APMIS 116:263-277. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson, M., H. Malmgren, M. Samiotaki, M. Kwiatkowski, B. P. Chowdhary, and U. Landegren. 1994. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265:2085-2088. [DOI] [PubMed] [Google Scholar]
- 16.Pierce, S. E., E. L. Fung, D. F. Jaramillo, A. M. Chu, R. W. Davis, C. Nislow, and G. Giaever. 2006. A unique and universal molecular barcode array. Nat. Methods 3:601-603. [DOI] [PubMed] [Google Scholar]
- 17.Pierce, S. E., R. W. Davis, C. Nislow, and G. Giaever. 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2:2958-2974. [DOI] [PubMed] [Google Scholar]
- 18.Proal, A. D., P. J. Albert, and T. Marshall. 2009. Autoimmune disease in the era of the metagenome. Autoimmun. Rev. 8:677-681. [DOI] [PubMed] [Google Scholar]
- 19.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 20.Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313-323. (Erratum, 9:600.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt, T. M., and D. A. Relman. 1994. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 235:205-222. [DOI] [PubMed] [Google Scholar]
- 23.Socransky, S. S., C. Smith, L. Martin, B. J. Paster, F. E. Dewhirst, and A. E. Levin. 1994. “Checkerboard” DNA-DNA hybridization. Biotechniques 17:788-792. [PubMed] [Google Scholar]
- 24.Stone, M. 2009. NIH builds substantial Human Microbiome Project. Microbe 4:451-456. [Google Scholar]
- 25.Sundquist, A., S. Bigdeli, R. Jalili, M. L. Druzin, S. Waller, K. M. Pullen, Y. Y. El-Sayed, M. M. Taslimi, S. Batzoglou, and M. Ronaghi. 2007. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol. 7:108-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarnberg, M., T. Jakobsson, J. Jonasson, and U. Forsum. 2002. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS 110:802-810. [DOI] [PubMed] [Google Scholar]
- 27.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The Human Microbiome Project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.