Abstract
Flor strains of Saccharomyces cerevisiae form a biofilm on the surface of wine at the end of fermentation, when sugar is depleted and growth on ethanol becomes dependent on oxygen. Here, we report greater biofilm formation on glycerol and ethyl acetate and inconsistent formation on succinic, lactic, and acetic acids.
Flor or velum formation by certain wine strains of Saccharomyces cerevisiae (flor strains) is a form of cellular aggregation observed as an air-liquid interfacial biofilm at the end of the alcoholic fermentation. Formation of the biofilm appears to be an adaptive mechanism because it ensures access to oxygen and therefore permits continued growth on nonfermentable ethanol. In general, nonbuoyant cells cease growth at the end of completed wine fermentations not for lack of carbon but for lack of oxygen. Biofilm cells have been found to have an elevated and/or altered lipid content and increased surface hydrophobicity (3, 5, 8, 9, 11). While both Hsp12, a small heat shock protein (13), and Muc1 (also known as Flo11), a hydrophobic cell wall mannoprotein (4, 6), have been shown to be required for the flor biofilm (10, 12, 14), other genetic or environmental requirements, other than an absence of glucose and the presence of ethanol and oxygen, have not been demonstrated. Here, we asked whether flor formation could be induced during growth on nonfermentable substrates other than ethanol. On the basis of dry weight of biofilm formed per mg of available carbon, the best carbon sources were found to be glycerol, ethyl acetate, and ethanol, in descending order. While subsurface growth occurred on acetic, dl-lactic, and succinic acids, an air-liquid interfacial biofilm did not always form. Microarray analysis of cells shifted from growth on glucose to growth on ethanol did not detect significant changes in expression of known biofilm formation-associated genes.
Yeast strain, growth conditions, and quantitation of biofilm.
A flor strain of S. cerevisiae, 3238-32 MATa leu2-Δ1 lys2-801 ura3-52 (12), was grown for 24 h in YEPD (2% Bacto peptone, 1% yeast extract, 2% glucose) at 30°C and 200 rpm. Cells were harvested and washed twice in sterile distilled water by centrifugation, resuspended in sterile distilled water, and diluted 500-fold into sterile-filtered YNB (Bacto yeast nitrogen base without amino acids) plus 30 μg/ml Leu, 30 μg/ml Lys, and 10 μg/ml Ura and supplemented with 4% (vol/vol) ethanol (YNB-EtOH), 3% (vol/vol) glycerol (YNB-Glyc), 1% (wt/vol) potassium acetate (YNB-Acet), 2% (wt/vol) dl-lactic acid (YNB-Lact), 2% (wt/vol) succinic acid (YNB-Succ), or 2% (vol/vol) ethyl acetate (YNB-EtAc). All YNB-based media were adjusted to pH 5.4. Four replicate 1-ml/well cultures were grown in a 24-well polystyrene microtiter plate (Becton Dickinson Labware, NJ). Biofilms were harvested by aspiration after 5 days of static incubation at 30°C by collecting the 1-ml cultures and an additional 1 ml of sterile distilled water used to rinse each well. The 2-ml samples were transferred to a cuvette and read at A600. Biofilm biomass (dry weight) was determined from a standard curve that related A600 values to dry weights of biofilm formed by 3238-32 grown in an independent 200-ml culture of YNB-EtOH from which the biofilm had been collected, washed with sterile distilled water, and dried under vacuum. The standard curve was generated by suspending about 60 mg of dried biofilm in 10 ml of distilled water from which a number of dilutions were prepared and read at A600. The mean slope of two such curves (coefficient of variation, <20%) was used as the conversion factor.
Biofilm formation not limited to growth on ethanol.
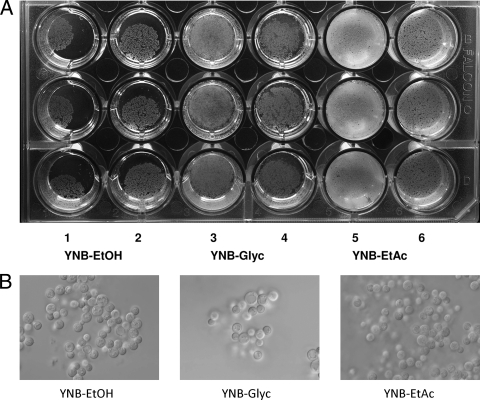
Figure 1A is a representative photograph of air-liquid interfacial biofilms formed in a 24-well microtiter plate containing 2 or 3 ml of medium per well. Biofilms consistently formed in YNB-EtOH, YNB-Glyc, and YNB-EtAc but were not always observed in YNB-Succ, YNB-Lact, or YNB-Acet, in spite of subsurface growth in the latter three media. The greatest amount of biofilm formed in YNB-Glyc, followed by YNB-EtAc and then by YNB-EtOH (Table 1). Per mg of available carbon, the amounts of biofilm formed in YNB-Glyc and in YNB-EtAc were about 3.8 and 2.7 times greater, respectively, than in YNB-EtOH. Per ml of medium, the amounts formed in YNB-Glyc and in YNB-EtAc were about 3.4 and 1.8 times greater, respectively, than in YNB-EtOH. Consistent with catabolism of glycerol and ethyl acetate by flor yeasts, a decrease in both compounds during biological aging of sherry wine has been reported (2). While biofilm formation in the other media was inconsistent, subsurface growth was always observed. Cell yields in YNB-Succ, YNB-Lact, and YNB-Acet were mostly from bulk growth (Table 1). One interpretation of these data is that biofilm formation requires an energy input not needed for growth in bulk culture and growth on the more reduced substrates—glycerol, ethyl acetate, and ethanol—provided this additional energy.
FIG. 1.
Representative air-liquid interfacial biofilms formed by 3238-32 in a 24-well microtiter plate. (A) YNB-EtOH (columns 1 and 2), YNB-Glyc (columns 3 and 4), and YNB-EtAc (columns 5 and 6). Wells in the odd-numbered columns contained 3 ml of medium, and wells in the even-numbered columns contained 2 ml. (B) Photomicrographs of unfixed biofilm cells taken at ×1,000 using differential interference contrast microscopy and an Olympus BX61 microscope. Images were acquired with a XM10 high-resolution CCD camera using CELL software.
TABLE 1.
Biofilms formed as a function of carbon sourcea
| Medium | Yield (mean ± SD) of biofilmsb expressed as: |
|
|---|---|---|
| μg dry wt/ml medium | μg dry wt/mg available C | |
| YNB-EtOH | 114 ± 10 † | 6.9 ± 1 ¶ |
| YNB-EtAc | 205 ± 25 ‡ | 18.8 ± 2 $ |
| YNB-Glyc | 385 ± 40 § | 26 ± 3 # |
While subsurface growth occurred in YNB-Succ, YNB-Lact, and YNB-Acet, a substantial air-liquid interfacial biofilm was not consistently observed. Cell yields in YNB-Succ, YNB-Lact, and YNB-Acet were 185 ± 25, 107 ± 11, and 29 ± 5 μg dry weight cells/ml medium, respectively, and 22.8 ± 3, 13.4 ± 1, and 12.1 ± 2 μg dry weight cells/mg available C, respectively. Yields were based on harvest of nonbiofilm cells and cells in a biofilm when present.
A one-way analysis of variance (ANOVA) was performed to assess the effect of carbon source on biofilm formation using Tukey's honestly significant difference (HSD) test as a post-hoc comparison of means (P < 0.05). The analysis was performed independently for the data in columns one and two. Means within the same column that are followed by different symbols are significantly different (Statgraphics Centurion XV; StatPoint Technologies, Inc., Warrenton, VA).
Microarray analysis.
To determine if biofilm-specific changes in gene expression accompanied the shift from growth in bulk culture in YNB plus 2% glucose to growth as a biofilm in YNB-EtOH, a microarray analysis was undertaken. We exploited a previous observation made when growing 3238-32. Namely, when a large inoculum (∼10%) of glucose-grown cells was shifted to YNB-EtOH, cells initially settled to the bottom of the vessel to form a visible layer that remained qualitatively unchanged in thickness over the course of 2 to 3 days while a biofilm became evident at the air-liquid interface after about 1 to 2 days. We reasoned that at a first approximation, the biofilm and nonbiofilm populations should have been genetically identical and that environmental factors were likely to explain the differential growth response. Briefly, cells grown 24 h at 30°C in YEPD to a density of 5 × 108 cells/ml were pelleted, washed twice in distilled water, diluted 10-fold into 100 ml of YNB-EtOH in triplicate 250-ml beakers, and grown statically at 27°C. After 48 h, the visible biofilm covering the entire liquid surface was collected by aspiration. Once removed, cells at the bottom were collected similarly. Cells from both populations were washed once in sterile distilled water prior to RNA isolation and microarray analysis (GEO series accession number GSE19156 [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19156]).
Initially, a gene ontology (GO) process analysis was performed on genes upregulated in both biofilm and nonbiofilm cells (data not shown). Categories for which gene frequencies differed ≥2-fold between the two populations are listed in Table 2. Based on the centrality of these processes to cell growth (e.g., ribosome biogenesis and assembly, translation, cytokinesis), these data suggest that cells in the biofilm were growing faster than the nonbiofilm cells or that the latter had stopped growing. In support of the no-growth possibility, half of the 653 genes found to be upregulated in a stationary-phase yeast culture harvested after 5 days in YEPD (7) were also upregulated ≥2-fold in the nonbiofilm cells. Three genes whose overexpression was associated with the “quiescent” fraction of stationary-phase cells (1) were found to be highly overexpressed in the nonbiofilm cells: SIP18, 37.7-fold; GRE1, 10.7-fold; BCY1, 8.1-fold. A significant increase in expression of known biofilm formation-associated genes (FLO8, MUC1, NRG1, NRG2, SNF1, HSP12) was not observed. Expression of agglutinin-encoding genes (AGA1, AGA2, FIG2, FLO1, FLO5, FLO10, SAG1) in the biofilm cells was either the same or >2-fold greater than in the nonbiofilm cells. While expression of two genes involved in protein mannosylation was >2-fold greater than in the biofilm cells (MNN11 and MNN9), expression of others was either the same or >2-fold greater than in nonbiofilm cells (CSG2, PMI40, DPM1, VAN1, OCH1, ALG3). It is possible that because RNA was harvested well after the biofilm began to form, the primary biofilm-specific transcriptional response was missed. Alternatively, the shift to biofilm growth may not be accompanied by major shifts in transcription of known biofilm formation-related genes, or the key genes in the process may be unrecognized.
TABLE 2.
Ratios of GO process frequencies for genes expressed in biofilm vs nonbiofilm cellsa
| GO process category | Ratio of GO process frequencies for biofilm vs nonbiofilm cells |
|---|---|
| Ribosome biogenesis and assembly | 11.1 |
| Biological process unknown | 5.4 |
| Nuclear organization and biogenesis | 4.7 |
| Amino acid and derivative metabolic process | 4.5 |
| Translation | 4.3 |
| Cytokinesis | 4.1 |
| Heterocycle metabolic process | 3.6 |
| Cell budding | 3.5 |
| Aromatic compound metabolic process | 2.8 |
| Organelle organization and biogenesis | 2.8 |
| Anatomical structure morphogenesis | 2.3 |
| DNA metabolic process | 2.2 |
| RNA metabolic process | 2.2 |
| Protein folding | 2.2 |
Initially, a GO process analysis was performed on genes upregulated in both biofilm and nonbiofilm cells. The GO process categories listed here are those for which the frequency for biofilm cells was at least 2-fold higher than the frequency for nonbiofilm cells. The GO category of known biofilm formation-related genes (FLO8, MUC1, NRG1, NRG2, SNF1) was not a category for which the frequency of expression for biofilm cells was at least 2-fold higher than for nonbiofilm cells.
In summary, formation of an air-liquid interfacial biofilm by S. cerevisiae is not limited to aerobic growth on ethanol but occurs on other reduced nonfermentable carbon sources, glycerol and ethyl acetate. Its variable formation on less reduced substrates (succinic, acetic, and lactic acids) is consistent with an additional energy input that is not needed for growth in bulk culture.
Acknowledgments
We thank Anne-Marie Girard for technical support for the microarray analysis and Lynn G. Ketchum for photographic assistance (Fig. 1A).
Footnotes
Published ahead of print on 30 April 2010.
REFERENCES
- 1.Allen, C., S. Büttner, A. D. Aragon, J. A. Thomas, O. Meirelles, J. E. Jaetao, D. Benn, S. W. Ruby, M. Veenuis, F. Madeo, and M. Werner-Washburne. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlanga, T. M., R. Peinado, C. Millán, J. C. Mauricio, and J. M. Ortega. 2004. Influence of blending on the content of different compounds in the biological aging of sherry dry wines. J. Sci. Food Agric. 52:2577-2581. [DOI] [PubMed] [Google Scholar]
- 3.Cantarelli, C., and A. Martini. 1969. On the pellicle formation by “flor” yeast. Antoine Van Leeuwenhoek 35:F35-F36. [PubMed] [Google Scholar]
- 4.Dranginis, A. M., J. M. Rauceo, J. E. Coronado, and P. N. Lipke. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farris, G. A., M. Sinigaglia, M. Budroni, and M. E. Guerzoni. 1993. Cellular fatty acid composition in film-forming strains of two physiological races of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 17:215-219. [Google Scholar]
- 6.Fidalgo, M., R. R. Barrales, J. I. Ibeas, and J. Jimenez. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. U. S. A. 103:11228-11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iimura, Y., S. Hara, and K. Otsuka. 1980. Cell surface hydrophobicity as a pellicle formation factor in a film strain of Saccharomyces. Agric. Biol. Chem. 44:1215-1222. [Google Scholar]
- 9.Iimura, Y., S. Hara, and K. Otsuka. 1980. Fatty acids as hydrophobic substances on the cell surface of a film strain of Saccharomyces. Agric. Biol. Chem. 44:1223-1229. [Google Scholar]
- 10.Ishigami, M., Y. Nakagawa, M. Harakawa, and Y. Iimura. 2004. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 237:425-430. [DOI] [PubMed] [Google Scholar]
- 11.Martínez, P., L. Perez, and T. Benítez. 1997. Mechanism of velum formation by flor yeast isolated from sherry wine. Am. J. Enol. Vitic. 48:55-62. [Google Scholar]
- 12.Zara, S., A. T. Bakalinsky, G. Zara, G. Pirino, M. A. Demontis, and M. Budroni. 2005. FLO11-based model for air-liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zara, S., G. A. Farris, M. Budroni, and A. T. Bakalinsky. 2002. HSP12 is essential for biofilm formation by a Sardinian sherry strain of S. cerevisiae. Yeast 19:269-276. [DOI] [PubMed] [Google Scholar]
- 14.Zara, G., S. Zara, C. Pinna, S. Marceddu, and M. Budroni. 2009. FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae. Microbiology 155:3838-3846. [DOI] [PubMed] [Google Scholar]