Abstract
The prion hypothesis posits that a misfolded form of prion protein (PrP) is responsible for the infectivity of prion disease. Using recombinant murine PrP purified from Escherichia coli, we created a recombinant prion with the hallmarks of the pathogenic PrP isoform: aggregated, protease-resistant, and self-perpetuating. After intracerebral injection of the recombinant prion, wild-type mice developed neurological signs in ~130 days and reached the terminal stage of disease in ~150 days. Characterization of diseased mice revealed classic neuropathology of prion disease, the presence of protease-resistant PrP, and the capability of serially transmitting the disease, confirming that these mice succumbed to prion disease. Thus, as postulated by the prion hypothesis, the infectivity in mammalian prion disease results from an altered conformation of PrP.
Transmissible spongiform encephalopathies (TSEs or prion disease) are infectious neurodegenerative disorders. The prion hypothesis (1) proposes that the infectious agent is an aberrant conformational isoform of the normal PrP (PrPC), a GPI (glycosylphosphatidylinositol)-anchored glycoprotein. By virtue of its self-perpetuating characteristic, the aberrant isoform (PrPSc) converts host PrPC into the PrPSc conformation and leads to neurodegeneration (2–4). Despite strong supporting evidence (5–11), a crucial prediction derived from the prion hypothesis, that an infectious prion can be generated with bacterially expressed recombinant PrP (recPrP), remains unfulfilled (2, 12), leaving lingering doubts about the prion hypothesis (13).
Recombinant PrP has been folded into various forms similar to PrPSc, but none of them fully recapitulates the characteristics of the infectious agent (2, 12). The amyloid fiber of a recPrP fragment (recPrP89–230) causes prion disease in transgenic mice over-expressing PrP89–231 (10), but a prolonged incubation time in mice over-expressing PrP has led to uncertainty about whether the infectivity is indeed derived from recPrP89–230 amyloid fibers (2, 12). The difficulty in creating a recombinant prion is likely due to the lack of proper facilitating factors (14). Polyanions, particularly RNA, have been found to facilitate PrP conversion and promote de novo prion formation (9, 15–17). We investigated lipid as a potential facilitating factor because GPI-anchored PrPC is in the vicinity of lipid membranes and the interfacial lipid bilayer region strongly influences protein structure (18). Encouraged by the findings that lipid interaction converts recPrP to a PrPSc-like form (19), we applied protein misfolding cyclic amplification (PMCA) (8) to study recPrP conversion in the presence of both lipid and RNA.
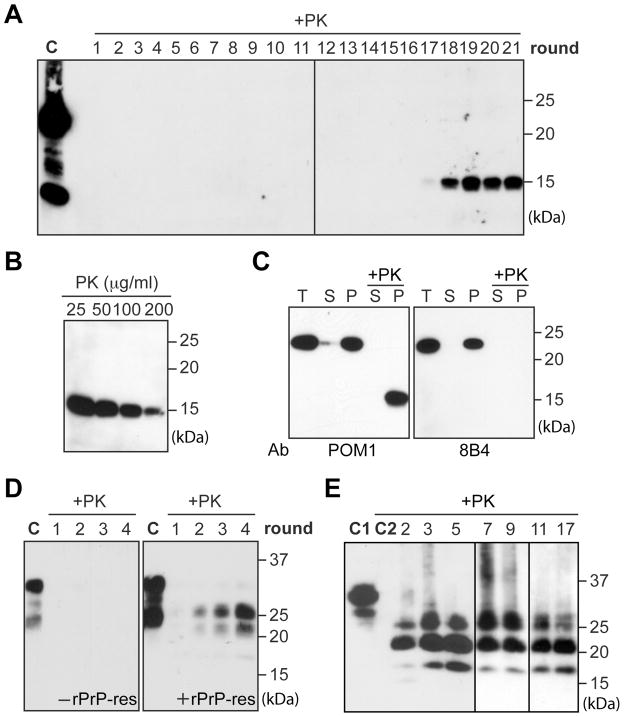
Using a serial PMCA protocol (20), we tested 16 different conditions in which recPrP was mixed with various combinations of lipids and/or total RNA isolated from normal mouse liver. In the presence of synthetic anionic phospholipid POPG (1-palmitoyl-2-oleoylphosphatidylglycerol) and RNA, a 15 kDa proteinase K (PK)-resistant band was detected after 17 rounds of PMCA (Fig. 1A). Once formed, the PK-resistant recPrP (rPrP-res) was able to serially propagate (Fig. 1A and fig. S1). The same procedure was repeated several times and, in our hands, the overall efficiency of de novo rPrP-res formation was around 20% (fig. S2).
Fig. 1.
In vitro generated rPrP-res. (A) One round PMCA consisted of 48 cycles of sonication (0.5 minute) and incubation (29.5 minute). At the end, 1/10 reaction mixture was transferred to fresh substrate mixture to start a new round (20). PMCA products were digested with 25 μg/ml PK. C, undigested recPrP. (B) Serial PK digestion of PMCA products. (C) PMCA product was separated into supernatant(s) and pellet(p) by a 1-hour 100,000g centrifugation at 4°C. T, total input; +PK, 25 μg/ml PK digested. (D) With normal mouse brain homogenate (NMBH) as substrate, PMCA was performed with or without rPrP-res seed. Product was digested with 100 μg/ml PK. C, undigested NMBH. (E) After rPrP-res infection, SN56 cells were lysed, digested with 25 μg/ml PK, and centrifuged. The PK-resistant PrP in the pellet was detected by immunoblot analysis. Numbers indicates cell passages. C1, undigested SN56 cell lysate. C2, pellet of PK-digested, uninfected SN56 cell lysates. In all panels, PrP was detected by immunoblot analysis with POM1 anti-PrP antibody except for the right panel in (C). PK digestion was carried out at 37°C for 30 minutes (A, B, and C) or 1 hour (D and E).
Serial PK-digestion of rPrP-res revealed that the PK-resistant band was detectable after 200 μg/ml PK digestion (PK:recPrP molar ratio > 50:1) (Fig. 1B). After centrifugation, the rPrP-res was only detected in the pellet fraction (Fig 1C), indicating that rPrP-res was aggregated. Moreover, the 15 kDa PK-resistant band was not recognized by the 8B4 antibody that detects an N-terminal epitope of PrP (21) (Fig. 1C), revealing that the rPrP-res contained a C-terminal PK-resistant core. Thus, similar to PrPSc, rPrP-res is aggregated, PK-resistant, and contains a C-terminal PK-resistant core.
Next, we performed PMCA and cell culture analyses to determine whether rPrP-res could seed glycosylated and GPI-anchored endogenous PrPC. With normal mouse brain homogenate as substrate, PMCA was carried out with or without rPrP-res seed. The PK-resistant endogenous PrP, demonstrated by higher molecular weights of glycosylated PrP, was detected in samples seeded with rPrP-res (Fig. 1D). Whereas in reactions without rPrP-res seed, no PK-resistant PrP was detected, ruling out de novo PrP-res formation or insufficient PK digestion. The cell infection assay was performed on SN56 cells, a murine neuronal cell line susceptible to prion infection (22). Endogenous PrPC in SN56 cells was glycosylated and sensitive to PK digestion (Fig. 1E). After rPrP-res infection, the PK-resistant endogenous PrP was detected in passage 2 cells and remained detectable after 17 passages (Fig. 1E). A similar experiment revealed that the rPrP-res converted normal mouse brain homogenate (Fig. 1D) could infect SN56 cells as well (fig. S3). Thus, rPrP-res is able to propagate its PK-resistant conformation to endogenous PrPC.
To determine whether rPrP-res was capable of causing bona fide prion disease, we infected 8-week-old female CD-1 mice by intracerebral injection. The rPrP-res (inoculum 4) was prepared by propagating rPrP-res through 24 rounds of PMCA. All PMCA products were pooled together and centrifuged through a sucrose cushion. The pellet was washed, resuspended, and used for inoculation. Three control inocula were used for animal study (Table 1). Inoculum 1, consisting of all the components used for rPrP-res propagation except for recPrP and rPrP-res seed, was subjected to the same treatments as inoculum 4. Inoculum 2, consisting of all the components of rPrP-res propagation except for rPrP-res seed, was incubated at 37°C for 24 days without sonication, and subjected to the same pelleting and washing treatments. Omitting the sonication step prevented the de novo rPrP-res formation in this control sample, which was confirmed by the PK digestion analysis described below (Fig. 2A). Inoculum 3 was prepared by directly mixing recPrP, POPG, and RNA in the inoculum diluent. The amount of each component was equal to the total amount in the final pool of inoculum 4, ensuring that the result was not influenced by insufficient dosage of recPrP or any other component. The inoculation and animal care were carried out in an animal vivarium that had never been exposed to animals with prion disease.
Table 1.
Intracerebral inoculation of rPrP-res
| Inoculum | Component | Processing | Preparation for injection | Diseased/Inoculated | Survival Time(dpi)† |
|---|---|---|---|---|---|
| 1 | Buffer + POPG + RNA The amount of each component equaled to that in the rPrP-res propagation reaction. |
Serial PMCA | Pelleting through a sucrose cushion and washing twice with PBS | 0/15 | > 360 |
| 2 | Buffer + POPG + RNA + recPrP The amount of each component equaled to that in the rPrP-res propagation reaction. |
Incubated at37°C without sonication | Pelleting through a sucrose cushion and washing twice with PBS | 1*/14 | > 360 (286*) |
| 3 | POPG + RNA+ recPrP The amount of each component equaled to that in the final pool of inoculum 4. |
No processing | No preparation | 0/5 | > 360 |
| 4 (rPrP-res) | Buffer + POPG +RNA+ recPrP+ rPrP-res seed | Serial PMCA | Pelleting through a sucrose cushion and washing twice with PBS | 15/15 | 150 ± 2.2(mean ± SEM) |
One mouse died from an unrelated disease at 286 dpi. It had no neurological sign or weight loss.
One mouse from each control groups was sacrificed at 275 dpi to serve as controls.
Fig. 2.
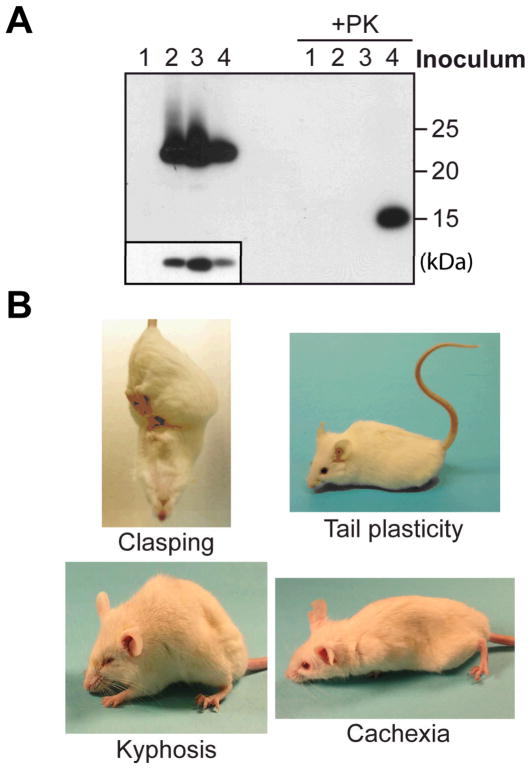
Intracerebral rPrP-res inoculation. (A) After injection, the remaining inocula with or without PK digestion were electrophoresed and PrP was detected by immunoblot analysis with POM1 anti-PrP antibody. The insert is a lighter exposure of undigested inocula. (B) Mice injected with rPrP-res showed indicated signs.
After the injection, the remaining inocula were analyzed (Fig. 2A). Inoculum 1 did not contain PrP and, accordingly, no PrP was detected. Of three inocula that contained recPrP, inoculum 4 had the lowest amount of recPrP (Fig. 2A). However, the 15 kDa PK-resistant band was only detected in inoculum 4, verifying that it was the only inoculum containing rPrP-res.
Around 130 days post inoculation (dpi), all 15 rPrP-res inoculated mice developed clinical signs of prion disease. The earliest sign was clasping, an indication of neurological dysfunction (Fig. 2B). Soon after that, mice developed tail plasticity and akinesia, that is, mice remained stationary in response to external stimuli (Fig. 2B and video S1). The disease progressed rapidly and mice developed kyphosis, head twitching, mild ataxia, and eventually became cachexic and lethargic (Fig. 2B and video S2). The rPrP-res inoculated mice reached the terminal stage at 136–161 dpi and the average survival time was 150 ± 2.2 days (mean ± SEM) (Table 1 and fig. S4). None of the mice injected with control inocula developed prion disease for more than 360 days and are still alive. Thus, intracerebral rPrP-res injection caused neurodegenerative disorders in wild-type mice with infectivity specifically associated with the rPrP-res conformation.
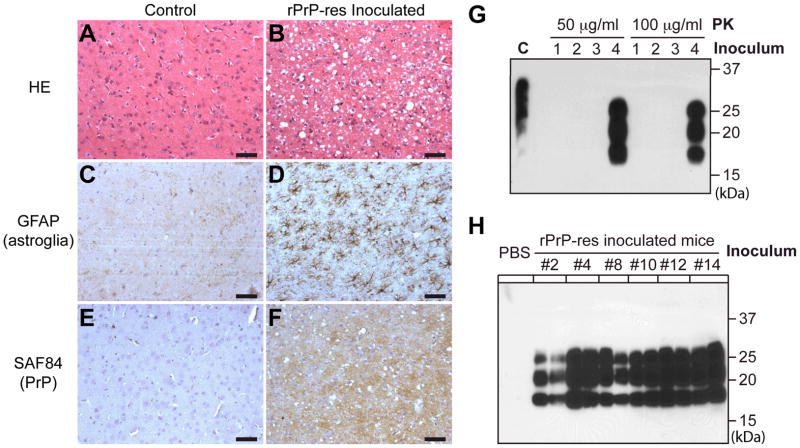
To ensure that every rPrP-res inoculated mouse received both pathological and biochemical analyses, each mouse brain was bisected sagittally and the half brain was subjected to histological or biochemical analysis. Severe spongiosis was detected in multiple brain regions (fig. S5 and S6). Dense small vacuoles were observed in the frontal cortex and caudate nucleus (Fig. 3B and fig. S6), while larger vacuoles were detected in the pons, midbrain (areas around raphe nuclei and periaqueductal gray) and cerebellar white matter (areas around cerebellar dentate and fastigial nuclei). Moderate spongiosis was present in the occipital cortex, thalamus, medulla, and hippocampus, whereas little spongiosis was detected in the superior or inferior colliculus, hypothalamus, or olfactory bulb (fig. S6 and S7). Prominent astrogliosis and microgliosis were detected in rPrP-res inoculated mouse brains (Fig. 3D and fig. S8). PrP immunohistochemistry revealed abnormal PrP deposition in a pattern similar to the diffuse synaptic accumulation (Fig. 3F). The densest PrP deposition was in thalamus (fig. S8), which was supported by the paraffin-embedded tissue blot (PET blot) analysis (fig. S9). Collectively, rPrP-res inoculated mouse brains exhibited the classic neuropathological features of prion disease: spongiosis, astrogliosis, microgliosis, and abnormal PrP deposition.
Fig. 3.
Characterization of rPrP-res caused prion disease. Histological analyses of age-and sex-matched control mice (A, C, and E) and rPrP-res injected mice (B, D, and F). Brain sections were stained by hematoxylin and eosin (A) and (B), an anti-GFAP (glial fibrillary acidic protein) antibody (C) and (D), and SAF84 anti-PrP antibody (E) and (F). Immunohistochemical stains were counterstained with hematoxylin. Scale bar represents 50 μm. (G) Brain homogenates of an rPrP-res inoculated mouse or mice inoculated with control inocula 1–3 were digested at 37°C for 1 hour with indicated concentrations of PK. PrP was detected by immunoblot analysis with M20 anti-PrP antibody. (H) Brain homogenates of mice that received second round transmission or control mice inoculated with inoculum diluent (PBS) were digested by 50 μg/ml PK at 37°C for 1 hour. PrP was detected by immunoblot analysis with POM1 antibody. Number indicates the mouse from which the inocula were prepared.
To determine whether PrPSc was specifically present in rPrP-res inoculated mice, we sacrificed one mouse from each control group at 275 dpi. PrPSc was detected in rPrP-res inoculated mouse brain but not in any other control brains (Fig. 3G). Histological analysis confirmed that there was no spongiosis in the control mice. The glycosylation and the electrophoretic pattern of PrPSc were similar among all 15 mice (fig. S10), in agreement with the similar neuropathology and relatively synchronized disease onset observed among these mice.
To determine whether the rPrP-res induced disease could be serially transmitted, 1% brain homogenates were prepared from 6 diseased mice and each sample was inoculated intracerebrally into 4 or 5 wild-type CD-1 mice. Around 130 dpi, all mice (n = 29) developed disease and the behavior phenotypes were essentially the same as those of the rPrP-res inoculated mice (video S3). These mice reached the terminal stage of disease at 151–180 dpi and the average survival time was 166 ± 1.5 days (fig. S11). The marginal increase in the survival time of second round transmission could be due to the reported variation among inoculation experiments (23), or the influence of other components in the brain homogenate used in second round transmission. Nonetheless, PrPSc was detected in all groups of mice inoculated with diseased mouse brain homogenates, but not in control mice (Fig. 3H). The spongiosis pattern remained similar to that of rPrP-res inoculated mice (fig. S12). Thus, similar to natural prion disease, the rPrP-res caused disease can be serially transmitted.
Inadvertent contamination is always a concern for PMCA. The only naturally occurring prion used in our lab was the RML strain, which was used only three times in our failed attempts to convert recPrP. During the last two years while we were working with rPrP-res, absolutely no naturally occurring prion was used. Our latest de novo rPrP-res formation (fig. S2) was achieved in a new sonicator and the substrate was prepared in a lab that has never been exposed to prion. Furthermore, both behavioral and pathological phenotypes of rPrP-res inoculated mice were clearly different from those reported for RML infected mice (24). Therefore, it is highly unlikely that rPrP-res formation was due to an inadvertent contamination. It is also important to note that, before the inoculum was prepared, the rPrP-res had been propagated for more than 35 rounds of PMCA. Thus, even if the initial rPrP-res formation were due to contamination, the >1035 dilution ensures that recPrP was the only PrP in the inoculum (Fig. 2A). Collectively, we conclude that the disease-causing agent was rPrP-res.
The three main components in our system were recPrP, POPG, and RNA. The purity of recPrP was verified by silver staining and recPrP was the only protein detected (fig. S13). The mouse liver RNA was carefully chosen because PrP is not normally expressed in liver and ectopic PrP expression in the liver of PrP null mice does not support prion propagation (25). Because synthetic polyanions that do not encode protein can replace RNA in cell-free prion formation and propagation (9, 16, 17), the role of RNA in generating infectious prion is likely to facilitate PrP conversion rather than encoding an infectious protein. Supporting this notion, rPrP-res was successfully propagated using synthetic poly(A) RNA (fig. S14), revealing that rPrP-res can be generated with virtually completely defined components. The requirement of lipid is in accordance with previous reports of higher prion infectivity in lipid membrane associated PrPSc (22, 26). Notably, the purified GPI-anchored PrPC, which was used to produce infectious prion de novo (9), contained stoichiometric amounts of co-purified lipids, supporting a general role of lipid in PrP conversion. Finally, the POPG and RNA used here may simply mimic an unknown in vivo facilitating factor(s). Further studies are required to identify these factors.
Here we provide direct evidence supporting the prion hypothesis. First, rPrP-res is in a conformational state similar to the pathogenic PrPSc isoform. Second, rPrP-res possesses the self-perpetuating characteristic of a prion. Third, rPrP-res causes bona fide prion disease in wild-type mice. The fact that only rPrP-res inoculated mice developed prion disease establishes that prion disease is caused by the altered conformational form of PrP.
Supplementary Material
Acknowledgments
We thank Drs. A. Aguzzi, M.-S. Sy, and B. Wainer for reagents, A. Steele for comments on the manuscript. This work was supported by the Ellison Medical Foundation, NIH R01NS060729, and research support from ECNU.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
One-sentence summaries
Recombinant prion protein purified from E. Coli was converted into an infectious prion that recapitulates the characteristics of infectious agent in prion disease.
Materials and Methods
References
References and Notes
- 1.Prusiner SB. Science. 1982;216:136. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Caughey B, Baron GS, Chesebro B, Jeffrey M. Annu Rev Biochem. 2009;78:177. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguzzi A, Baumann F, Bremer J. Annu Rev Neurosci. 2008;31:439. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Clarke AR. Science. 2007;318:930. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 5.Bueler H, et al. Cell. 1993;73:1339. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 6.Kocisko DA, et al. Nature. 1994;370:471. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 7.Bessen RA, et al. Nature. 1995;375:698. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 8.Castilla J, Saa P, Hetz C, Soto C. Cell. 2005;121:195. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Deleault NR, Harris BT, Rees JR, Supattapone S. Proc Natl Acad Sci U S A. 2007;104:9741. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legname G, et al. Science. 2004;305:673. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 11.Telling GC, et al. Science. 1996;274:2079. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 12.Weissmann C. Cell. 2005;122:165. doi: 10.1016/j.cell.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Manuelidis L. J Cell Biochem. 2007;100:897. doi: 10.1002/jcb.21090. [DOI] [PubMed] [Google Scholar]
- 14.Caughey B, Baron GS. Nature. 2006;443:803. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 15.Deleault NR, Lucassen RW, Supattapone S. Nature. 2003;425:717. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 16.Deleault NR, et al. J Biol Chem. 2005;280:26873. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 17.Geoghegan JC, et al. J Biol Chem. 2007;282:36341. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White SH, Ladokhin AS, Jayasinghe S, Hristova K. J Biol Chem. 2001;276:32395. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, et al. Biochemistry. 2007;46:7045. doi: 10.1021/bi700299h. [DOI] [PubMed] [Google Scholar]
- 20.Materials and methods are available as supporting material on Science Online.
- 21.Pan T, et al. J Virol. 2005;79:12355. doi: 10.1128/JVI.79.19.12355-12364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron GS, Magalhaes AC, Prado MA, Caughey B. J Virol. 2006;80:2106. doi: 10.1128/JVI.80.5.2106-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamguney G, et al. J Gen Virol. 2008;89:1777. doi: 10.1099/vir.0.2008/001255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castilla J, et al. Embo J. 2008;27:2557. doi: 10.1038/emboj.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raeber AJ, et al. Proc Natl Acad Sci U S A. 1999;96:3987. doi: 10.1073/pnas.96.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabizon R, McKinley MP, Prusiner SB. Proc Natl Acad Sci U S A. 1987;84:4017. doi: 10.1073/pnas.84.12.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.