Abstract
Biomechanical models simulating brain motion under loading and boundary conditions in the operating room (OR) are gaining attention as alternatives for brain shift compensation during open cranial neurosurgeries. Although the significance of brain–skull boundary conditions (BCs) in these models has been explored in dynamic simulations, it has not been fully investigated in models representing the quasi-static brain motion that prevails during neurosurgery. In this study, we extend the application of a brain–skull contact BC by incorporating it into an inversion estimation scheme for the deformation field using the steepest gradient descent (SGD) framework. The technique allows parenchymal surface motion normal to the skull while maintaining stress-free BCs at the craniotomy and minimizing the effect of measurement noise. Application of the algorithm in five clinical cases using sparse data generated at the tumor boundary confirms the significance of brain–skull BCs in the model response. Specifically, the results demonstrate that the contact BC enhances model flexibility and achieves improved or comparable performance at the tumor boundary (recovering about 85% of the deformation) relative to that obtained when normal motion of the parenchymal surface is not allowed. It also significantly improves model estimation accuracy at the craniotomy (1.6 mm on average), especially when the normal motion is large. The importance of the method is that model performance significantly improves when brain–skull contact influences the deformation field but does not degrade when the contact is less critical and simpler BCs would suffice. The computational cost of the technique is currently 3.9 min on average, but may be further reduced by applying an iterative solver to the linear systems of equations involved and/or by local refinement of the mesh in regions of interest.
Keywords: Brain shift, Brain–skull boundary condition, Finite element modeling, Image-guided surgery, Inverse method
1. Introduction
Dynamic (Nabavi et al., 2001; Sun et al., 2005a) and complex (Hartkens et al., 2003) brain shift resulting from surgical intervention during open cranial procedures often exceeds 20 mm at the cortical surface post-durotomy and can reach more than 3 mm at the tumor boundary (Nimsky et al., 2000). Brain motion of such magnitude degrades the registration accuracy of internal anatomical structures of interest (e.g., tumor), and thus, poses a significant challenge to image-guidance systems that solely depend on preoperative images (Buckner, 2003; Lacroix et al., 2001; Laws et al., 2003). To compensate for brain shift during these procedures, biomechanical models that estimate brain deformation under loading and boundary conditions applied in the operating room (OR) are being developed as alternatives to various intraoperative imaging techniques (e.g., Clatz et al., 2005; Dumpuri et al., 2007; Hu et al., 2007; Lunn et al., 2005, 2006; Skrinjar et al., 2002; Wittek et al., 2005), most notably, intraoperative magnetic resonance (iMR) and ultrasound (iUS).
The significance of brain–skull boundary conditions (BCs) in these models has been recognized in dynamic impact and inertial rotational injury computational simulations, where the results indicate that brain mechanical response is sensitive to the type of BCs applied (e.g., Bandak Eppinger, 1994; Kleiven and Hardy, 2002; Miller et al., 1998; Zhang et al., 2001). However, no consensus has been reached about the best way to represent the brain–skull interface (e.g., Bandak Eppinger, 1994; Kleiven and Hardy, 2002; Miller et al., 1998; Ruan et al., 1994; Takhounts et al., 2003; Zhang et al., 2001), in part because of the limited availability of experimental data. Consequently, the interface has been modeled as either a tied, frictionless or frictional sliding BC, with or without brain–skull separation (Ji et al., 2004; Ji and Margulies, 2007).
Modeling of the brain–skull interface in simulations of the quasi-static brain motion that prevails in neurosurgery has been similarly inconsistent. For example, Miller et al. (2000) fixed the bottom half of the brain surface in all directions, while allowing the top half to be free to deform. Using this BC, they were able to predict a force with a magnitude on the same order as measured in a confined compression experiment involving swine brain in vivo under moderate loading rates similar to those in the OR (1 mm/s up to 3.9 mm). More recently, Skrinjar et al. (2002) employed a contact algorithm to model the brain–skull interface to allow brain nodes to move freely unless they reached the skull, in which case they were stopped from further penetration.
Previously, we have assigned brain–skull BCs where all surface nodes, except those at the craniotomy and brainstem, are fixed in the direction normal to the skull, but allowed tangential movement (e.g., Lunn et al., 2005, 2006; Miga, 1998; Platenik et al., 2002; Sun et al., 2005b). Although this approach has successfully recovered 75–80% of the deformation in porcine brain retraction studies (Miga, 1998; Platenik et al., 2002), it prohibits parenchymal movement towards or away from the cranial wall in ways we frequently observe in the OR. A contact BC that we have recently implemented in a forced displacement method (FDM) enables modeling of this type of parenchymal motion, and thus, improves model flexibility (Ji et al., 2007). However, when applied in an FDM, it contributes to the generation of fictitious forces at otherwise stress-free locations and measurement noise in the applied data is directly incorporated into the model solution (Lunn et al., 2005, 2006), which compromises the accuracy of the overall model estimate.
In parallel, we have also developed an inversion scheme which is able to maintain stress-free BCs at the craniotomy site while fitting the model response to the measured data in a minimum-norm sense by adjusting the forcing conditions (Ji et al., accepted for publication; Lunn et al., 2005, 2006; Lynch, 2004). While this approach was found to improve the accuracy of model estimates (on average 33% in Lunn et al., 2006), its initial numerical implementation (based on the Representer algorithm) was too slow computationally to be used in the OR. More recently, we have been able to reduce the computational cost of the inversion scheme by a factor of 5–10 times (with an execution time of under 2 min being possible) by using the steepest gradient descent (SGD) algorithm, without compromising model performance (capturing 74–82% of the subsurface deformation in a series of clinical cases, Ji et al., accepted for publication).
These promising results initially obtained independently through studies of the contact BC applied in an FDM and the development of a computationally efficient, more accurate approach to data assimilation through inversion, have prompted us to incorporate the brain–skull contact BC constraint in our adjoint inverse algorithm computed efficiently with iterative SGD. Accomplishing this overall goal in the form of an effective, yet, efficient implementation is challenging because the iterative update of the forcing conditions must monitor the dynamically changing parenchymal surface in search of two-way breaches of the contact surface in order to adjust the penalty term that controls the allowable normal displacement of the brain surface. The validity of the implementation has been demonstrated with a simulated ground truth example, and the performance of the technique has been evaluated in five clinical cases (N = 5), where tumor displacement data was derived from iUS post-durotomy. Based on a contact BC assigned in the vicinity of the craniotomy and in regions opposite to the direction of gravity to allow for parenchymal motion normal to the skull, we show that the contact BC achieves comparable or improved tumor deformation estimates relative to those obtained with the fixed BCs that we have previously applied. Additionally, the contact BC significantly improves model estimation accuracy at the craniotomy, especially when the motion normal to the parenchymal surface is large. The importance of the approach we have implemented is that when motion occurs where brain–skull contact is critical, the BC acts to improve model estimation accuracy, whereas when no such motion is evident, the BC does not degrade the accuracy of the model estimates which can be achieved with fixed BCs.
2. Materials and methods
2.1. Finite element brain model formulation
Biot’s consolidation theory (Biot, 1941) was adopted to model the brain as a biphasic porous deformable medium where interstitial fluid pressure appears as a hydrodynamic driving force. The detailed mathematical formulation has been developed elsewhere (Miga, 1998; Paulsen et al., 1999), and is summarized by the following expressions, where Navier’s equations of linear elasticity are coupled with changes in fluid pressure from Darcian flow:
| (1) |
| (2) |
where u and p are the displacement and pore fluid pressure to be computed, respectively. Other material parameters and their values are listed in Table 1, and are identical to those in Ji et al. (2007) and Lunn et al. (2005). The utility of these material properties has been demonstrated in previous porcine brain retraction studies (Miga, 1998; Platenik et al., 2002). Discretizing these equations into the finite element matrix system results in:
| (3) |
where K is the stiffness matrix that embeds the discretized model equations, x is the nodal displacement and pore fluid pressure vector (4 degrees-of-freedom per node), and b includes the internal forces and boundary conditions resulting from surgical intervention, including interior forces due to pressure gradients in interstitial fluid and tissue buoyancy forces caused by gravitational effects.
Table 1.
Model parameters and their values used in this study.
| Parameter | Description | Value |
|---|---|---|
| G | Brain shear modulus | 2100 Pa |
| v | Brain Poisson’s ratio | 0.46 |
| ρt | Brain tissue density | 1000 kg/m3 |
| ρf,s | Surrounding fluid density for saturated nodes | 1000 kg/m3 |
| ρf,u | Surrounding fluid density for unsaturated nodes | 1.0 kg/m3 |
| k | Hydraulic conductivity | 1e−7 m3 s/kg |
| α | Ratio of fluid volume extracted to change in solid volume | 1 |
| 1/S | Fluid that can be forced into the tissue under constant volume | 0 |
| ψ | Pressure source strength | 0 Pa/s |
2.2. Adjoint equations inversion scheme
When incomplete measurements are available, a least squares inversion is suitable for minimizing the difference (ε) between the model estimate (x) and the measured data (d). This is achieved through the use of the well-known Lagrange method by introducing data error and forcing condition covariance models and a set of Lagrange multipliers (λ) to construct an objective function (Lunn et al., 2005; Lynch, 2004):
| (4) |
where A is the sampling matrix that generates computed values at given “sparse” measurement locations, while Wε and Wb (the inverse of the covariances of ε and b, respectively) describe the covariances of the data-model misfit between any pair of data points and the forcing conditions between any two locations. These covariances are controlled by the standard deviation (std) in the measurement error (σε) and the correlation length (L), respectively, through a “distance-based” model (Lynch, 2004). In this study, we have chosen their values based on previous investigations that yielded the best deformation estimates (σε = 0.2 mm and L = 5 mm; Lunn et al., 2006), similarly to those used in (Ji et al., submitted for publication). To minimize the scalar objective function, Ω, its derivatives with respect to x, b and λ are set to zero, generating the following set of adjoint equations after some further mathematical simplification (Lynch, 2004):
| (5) |
| (6) |
| (7) |
| (8) |
In the next two sections, we summarize the mathematical foundations of the contact BC constraint and introduce an algorithm that incorporates the contact solution process into the iterative SGD framework, so that the important features of both a contact BC and an efficient inversion scheme can be realized.
2.3. Contact BC formulation
Brain–skull contact under quasi-static loading conditions can be represented as a Signorini contact problem (Laursen, 2002). Initially, the closest point projection onto the master surface (i.e., contact point) is obtained for any slave node (s; Fig. 1) through a three-step contact search algorithm (Ji et al., 2007). Briefly, the slave node’s closest master node (m) and the neighboring master elements are first obtained. The slave node’s projection points on the resulting elements are computed and the one closest to s and within the elemental border is denoted as the closest point projection. When no valid projection is returned (e.g., due to nonconvexity), the closest element edge is considered. If no projection point is within the corresponding end points, the closest master node, itself, is used as the contact point. To alleviate the difficulty when searching for a contact point in nonconvex regions such as the basal region of the skull, the range of the neighboring master elements is expanded (a recursive depth of 3 was used in this study). The resulting contact point, y(0s), serves as the reference for determining slave node penetration, which, at time zero before deformation, is given by:
| (9) |
where N(0s) is the local master surface normal at the contact point prior to deformation. At time t after deformation, the penetration is updated to account for displacement of a slave node:
| (10) |
where sx and yx are displacements of the slave node and its corresponding contact point (yx = 0 when the master surface is fixed), respectively, and N(ts) is approximated by N(0s) assuming small changes occur in the master surface normal N(ts) = N(0s), when the master surface is fixed). The kinematical impenetrability condition for slave node, s (i.e., s does not penetrate through the master surface) is given by (Laursen, 2002; Zhong, 1993):
| (11) |
leading to the matrix form for assembling L (L ≥ 1) slave nodes:
| (12) |
where Q is a dim × L (dim equals 2 or 3 for a two- or three-dimensional problem, respectively) matrix of L diagonal submatrices with diagonal elements, qij, in each submatrix given as:
| (13) |
The subscripts in Eq. (13) denotes the jth local master surface normal component (j = 1, dim) for the ith slave node (i = 1, L).
Fig. 1.
Illustration of a one body frictionless contact against a fixed rigid-body formulated as a Signorini problem. The contact boundaries of the deformable (Ωs) and rigid body (Ωm) domains are defined as the slave (Γs) and master (Γm) surfaces, respectively. For a given slave node (s), its contact point on the master surface (y(s)) is determined through a three-step contact search algorithm (Ji et al., 2007), and the local master surface normal at the contact point (N(s)) is subsequently calculated.
When a slave node penetrates through the master surface, a force normal to the master surface is generated with a magnitude that is proportional to the penetration, which satisfies the kinematical impenetrability condition (Eq. (12)), and is applied at the contract point using a penalty method. Effectively, the total body potential energy of the system is rewritten to account for these contact forces:
| (14) |
where each term represents the component of total body potential energy due to internal, external and contact forces, respectively. The penalty coefficient, β, is a diagonal matrix (βij > 0; i = 1, dim × L) that controls the magnitude of the contact forces. The exact impenetrability condition is only achieved when β → ∞, which is not possible in practice because of matrix ill-conditioning (Laursen, 2002). If β is too large, the matrix solution becomes unstable, whereas if β is too small, slave nodes penetrate the master surface too far, which is also undesirable. In this paper, we have empirically chosen (βij = 5, i = 1, dim × L) for all computations (Ji et al., 2007). The total body potential energy is minimized when:
| (15) |
Combining Eqs. (12), (14), and (15) leads to the modified system of equations (setting the penetration, P, to zero in Eq. (12); Zhong, 1993):
| (16) |
where Kp = K + QTβQ, and bp = b – QTβ0P. Essentially, both sides of Eq. (3) are reformulated to include the penalty terms due to slave node penetration.
2.4. Incorporating contact BCs with SGD
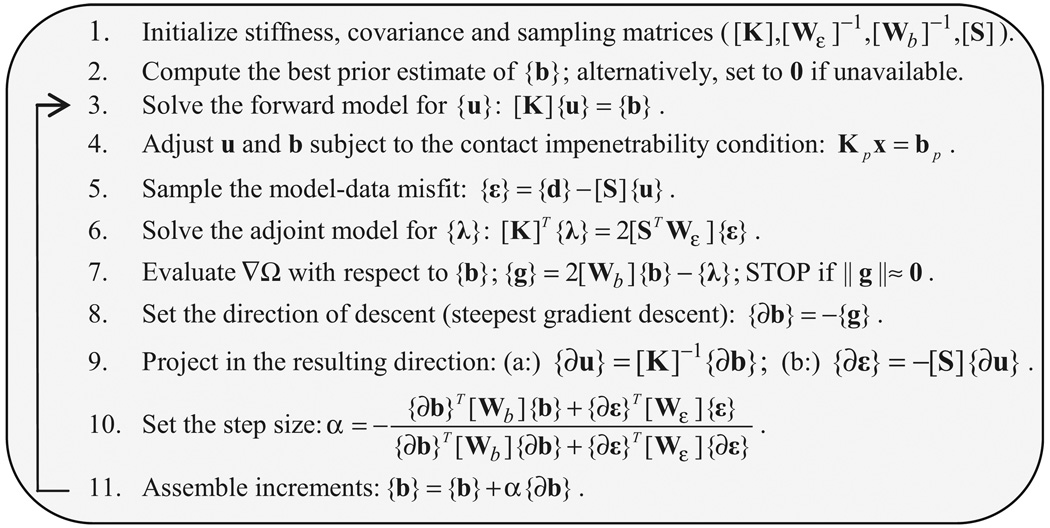
The difficulty in resolving the contact constraints (Eq. (12)) stems from the fact that the contact nodes are not generally known a priori. To ensure stability (Miga et al., 1998), we have adopted a fully implicit trial-and-error approach that iteratively solves the system of equations (Eq. (16)) until the set of contact nodes is stable (Le Tallec, 1994). We have found that a steady state in the contact system can be achieved in less than 10 iterations of an FDM when an empty initial set of contact nodes is used (Ji et al., 2007).
A similar iterative process occurs in the SGD for minimizing the model-data misfit (Ji et al., accepted for publication; Lynch, 2004). At the onset of each iteration, a forward model solution is computed to adjust the displacement estimate subject to the updated forcing conditions. With a contact BC, forces normal to the skull are generated when slave node penetration occurs. Essentially, these contact forces act as additional perturbations to the system forcing conditions obtained from SGD. Thus, a second forward model is computed with the reformulated system of equations (Eq. (16); Section 2.3). However, instead of using multiple iterations to stabilize the set of contact nodes within each SGD iteration, which would increase computational cost significantly and limit practical application in the OR, only one contact iteration is performed within each SGD iteration, and the slave node penetrations are resolved over the course of multiple SGD iterations. Consequently, the number of system solutions required to solve the model estimation problem with contact constraints is increased by one per SGD iteration, if contact nodes are detected. The detailed algorithm is described in Fig. 2 (note that x is replaced by u, i.e., only model displacement estimation is considered).
Fig. 2.
Flowchart of the contact solution process in the inversion scheme using the iterative SGD framework. The most computationally intensive part is the solution of full linear systems of equations (steps 3, 4, 6 and 9a). When slave node penetration is detected, an additional linear system of equations (step 4) is solved during SGD iteration.
When using an iterative algorithm such as SGD, a practical stopping criterion for achieving sufficient accuracy in the model estimate with minimum computational costs is required. In this study, we have empirically defined the minimum number of iterations (Itermin) needed to reach an approximate convergence of SGD to be the one where the change in the model-data misfit relative to the previous iteration is less than 0.1% of the initial model-data misfit (εinit), or formally:
| (17) |
where refers to εsprs achieved at the kth iteration (k > 1), and εinit is the initial model-data misfit. Model estimation performance was evaluated when Itermin was achieved for all computations in this study. To ensure that a sufficient number of iterations was used, we compared model response achieved at 2 × Itermin, and found that the difference in these solutions was less than 2% of εinit, indicating that Itermin was sufficient for estimating model performance.
A parallelized direct sparse solver (PARDISO; Schenk and Gartner, 2004) was adopted to compute all linear systems of equations in this work. Only one factorization is needed at the beginning of the solution process for fixed BCs, but factorization is required multiple times with contact BCs to account for the modification of the global stiffness matrix (every time slave node penetration occurs).
2.5. Clinical cases and brain model development
To demonstrate the applicability of the proposed technique, a total of five (N = 5) patients undergoing image-guided tumor resection were recruited at the Mary Hitchcock Memorial Hospital, under approval by the Institutional Review Board at the Dartmouth-Hitchcock Medical Center. All patients had well-defined tumor or cyst boundaries in both iUS and pMR, to allow for unbiased tumor segmentation. Patient age, gender, and type, location, and size of tumor are summarized in Table 2. For each patient, preoperative T1-weighted axial MRIs (pMR; N = 124; field of view: 240 mm; voxel size: 1.016 × 1.016 × 1.5 mm) were obtained from a 1.5 T GE MR scanner. The brain was segmented through a two-step level set scheme (Wu et al., 2005) and was uniformly meshed into tetrahedral elements (Sullivan et al., 1997) from which the corresponding triangular boundary elements were extracted. The average number of mesh nodes (in thousand) for all patients was 17.18 ± 3.05, while it was 94.98 ± 17.48 and 8.58 ± 0.59 for the number of tetrahedral and boundary elements, respectively.
Table 2.
Summary of patient cases, and type, location, and size of tumor
| Patient | Age (gender) | Type of tumor | Location of tumor | Size of tumor (cc) |
|---|---|---|---|---|
| 1 | 38 (M) | Low grade glioma | Left temporal | 4.7 (25.73)a |
| 2 | 62 (M) | Metastasis | Right posterior temporal | 10.99 |
| 3 | 49 (F) | Ganglioglioma | Right parietal | 0.20 (10.51)b |
| 4 | 62 (F) | Meningioma | Right parietal | 13.77 |
| 5 | 61 (F) | Meningioma | Right parietal | 0.62 |
Size of cyst in parenthesis when applicable.
Prior to surgery, the patient’s head was rigidly clamped, and the 3D locations of the fiducial markers attached to the scalp were identified in the OR with an optical tracking system (Polaris; The Northern Digital Inc., Canada). These locations were matched with their counterparts manually localized in pMR using an iterative closest point procedure (ICP; Besl and McKay, 1992) or a genetic algorithm (Hartov et al., 2007) to establish a rigid transformation. During surgery (specifically, before and after dural opening), a set of 2D B-mode iUS scans (typically N = 20–200; Siemens Sonoline Sienna, C8–5 transducer; image size: 640 × 480; 8-bit grayscale; image acquisition time: 50–60 ms) were digitized through a frame grabber (DT3155; Data Translations Inc., Marlboro, MA). The scan depth of the iUS images was 60 mm for all surgeries evaluated in this study (resulting in a pixel size of 0.15 × 0.16 mm).
The tumor (and cyst, when applicable) boundary from each pMR image was identified using isointensity contours at a level specified by an expert (typical intensity level of 4000–6000). A binary image stack was then constructed from these contours, and a triangulated surface was obtained to represent the undeformed tumor configuration. By contrast, tumor (cyst) segmentations in iUS images were performed manually with in-house software used to trace the boundaries on the monitor. An interpolation scheme was used to achieve a set of equally spaced points (0.2 mm, or approximately the iUS pixel size) for smooth representation of the tumor boundary. The number of resulting data points for each patient is reported in Table 3.
Table 3.
Summary of model estimation performance at the tumor boundary. Shown are the total number of tumor displacement data points (Ntot), εinit, εsprs, εcross and the percentage of subsurface deformation recovery achieved by applying the contact and fixed BCs.
| Patient | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| Ntot | 1180 | 3366 | 4014 | 1186 | 524 | |
| εinit | 4.8 ± 0.30 | 5.9 ± 2.4 | 6.1 ±1.4 | 4.0 ± 0.12 | 3.6 ± 0.56 | |
| εsprs | Contact BC | 0.40 ± 0.14 | 1.62 ± 0.96 | 1.57 ± 1.0 | 0.70 ± 0.33 | 0.12 ± 0.10 |
| Fixed BC | 0.57 ± 0.29 | 1.68 ± 0.85 | 1.62 ± 1.3 | 0.52 ± 0.30 | 0.14 ± 0.13 | |
| εcross | Contact BC | 0.4 ± 0.2 | 1.7 ± 0.8 | 1.5 ± 1.3 | 0.5 ± 0.3 | 0.13 ± 0.1 |
| Fixed BC | 0.72 ± 0.38 | 1.7 ± 0.8 | 1.5 ± 1.2 | 0.6 ± 0.2 | 0.12 ± 0.1 | |
| % of recovery | Contact BC | 92 | 72 | 75 | 88 | 96 |
| Fixed BC | 85 | 72 | 75 | 85 | 96 |
2.6. Simulated displacement with controlled measurement error
A simulated ground truth deformation field was produced for patient 2 to validate the implementation of the algorithm incorporating the brain–skull contact BC within the SGD inversion framework. This was achieved by generating a noise-free synthetic displacement field using a forward model solution, where the boundary nodes at the craniotomy (N = 68) and brainstem (N = 38) were stress-free. The size of the craniotomy was approximately 45 × 24 mm, which was defined as the maximum distance between any pair of craniotomy nodes (long axis) and the cross-sectional length of the craniotomy elements intersected by a plane orthogonal to the long axis but passing through the center of the craniotomy (short axis). Fluid pressure at the craniotomy nodes was fixed (zero pressure), while free otherwise. A fluid plane passing through the “lowest” craniotomy node but perpendicular to gravity was defined. Elements below/above the fluid plane were saturated/unsaturated, and the surrounding fluid density was assigned accordingly (1000 and 1 kg/m3 for saturated and unsaturated regions, respectively, Ji et al. (2007), Lunn et al. (2005). A second plane was determined by moving the fluid plane along the direction of gravity by 20 mm. Boundary nodes found above the second plane were assigned to be slave nodes (N = 1829) potentially moving towards or away from the inner-surface of the skull, while the remaining boundary nodes were fixed in the normal direction but allowed to move tangentially (i.e., fixed BC). The inner-surface of the skull was obtained by projecting the brain boundary nodes along the average nodal normal by 1 mm to simulate the cerebrospinal fluid (CSF) gap. These BCs remained stress-free at the craniotomy site, and allowed normal motion of the parenchyma in the vicinity of the craniotomy as well as in regions contralateral to gravity, as reported in the literature (e.g., Hartkens et al., 2003; Hu et al., 2007; Maurer et al., 1998; Nabavi et al., 2001; Nimsky et al., 2000).
To simulate the measurements, displacements at the undeformed tumor surface nodes generated from pMR (Section 2.5) were interpolated from the noise-free deformation field. These nodes and their displacements were used as a pooled sample (N = 2541) of displacement data, from which 200 points were randomly selected as sparse data (εsprs) for model guidance, and the remaining sample was defined as cross-validation locations. The displacements forming the sparse measurement data were superimposed with noise having a normally distributed magnitude (zero mean and a std of half a pixel or 0.51 mm) and a uniformly distributed direction, to simulate measurement error (Ji et al., accepted for publication).
When using the SGD inversion scheme to recover the deformation, the BCs assigned were identical to those used in the forward model. Model response at the cross-validation locations (εcross) achieved at iteration Itermin (Section 2.4) was evaluated against the noise-free ground truth. In addition, displacements of the parenchymal surface at the craniotomy were also utilized for cross-validation purposes.
2.7. Clinical tumor displacement
Clinical tumor displacements post-durotomy were measured from the spatially merged tumor boundaries segmented from iUS and pMR images using the transformation determined between iUS pre-durotomy and pMR. Proper transformation is critical for accurate measurement of tumor displacements post-durotomy, as any misalignment between tumor boundary in iUS pre-durotomy and pMR (due to errors from the fiducial-based registration, limited feature localization accuracy in iUS and even possible brain shift at this very early stage of surgery) could accumulate and compromise the accuracy obtained in subsequent brain shift compensations. To achieve a more faithful reference for displacement measurement, a mutual-information-based (MI-based) patient re-registration scheme was employed to minimize the tumor boundary misalignment pre-durotomy (average misalignment of 2.5 mm, maximum misalignment of up to 5 mm was reduced to 1.0 mm; Ji et al., 2008) before determining tumor displacements post-durotomy.
The ICP algorithm was applied to generate a rigid-body transformation that minimized the overall distance between the transformed iUS tumor boundary points (deformed configuration) and the pMR tumor surface (undeformed configuration). Tumor displacements were essentially the difference between the original coordinates of the iUS tumor boundary points and their new coordinates after applying the transformation (Lunn et al., 2005). The rigid-body transformation generated a rigid displacement field locally (both translation and rotation) around the tumor to approximate the brain deformation in this region and obtain the initial model-data misfit (i.e., εinit; Table 3 in the results). Here, we have used the rigid tumor displacement to guide the model deformation that generates a non-rigid displacement field in the tumor region.
Similarly to the simulated data, a total of 200 sparse data points were randomly selected for model guidance. Model performance was evaluated using the residual model-data misfit at the remainder of the pooled sample (i.e., εcross). In addition, the 3D parenchymal surface at the craniotomy after dural opening recorded with stereopsis independently of the measured tumor displacement was employed for further unbiased cross-validations (except for patient 5 whose stereopsis data was unavailable; The details of the stereopsis technique appear in Sun et al., 2005a, 2005b.) using a “closest point projection distance” (CPPD) analysis (Ji et al., 2007). The accuracy of the stereopsis system was found to be within 1 mm using a brain–shaped phantom (Sun et al., 2005a) or iUS (Sun et al., 2005b). This approach is similar to that used in the contact search (Section 2.3), where the closest point projection distance from each craniotomy node relative to the triangulated parenchymal surface measured from stereopsis was defined positive/negative when the vector from the corresponding surface projection to the node was opposite/along the out-ward surface normal defined at the projection point. The mean absolute value of the distances from all craniotomy nodes was used to assess the mismatch between the two surfaces.
Model performance with both contact and the fixed BCs was evaluated. Fluid density and BC assignments with the contact BC were analogous to those used with the simulated data (Section 2.6). When applying the fixed BCs, slave nodes assigned in the contact BCs were fixed in the direction normal to the skull, but allowed to move tangentially.
2.8. Data analysis
With the simulated ground truth data, εcross and CPPD of the parenchymal surface at iteration Itermin (the minimum iteration number that satisfies Eq. (17)) were examined to assess the model estimation performance using the contact BC. In addition, we report the computational cost and the difference between model solution and the ground truth throughout the entire deformation domain to investigate the effect of the locally generated sparse data used in model guidance. Whole-brain deformation was also produced using the fixed BC for comparison purposes. In addition, we report the principal direction of model deformation relative to the gravitational axis, using an approach similar to that described in Hartkens et al. (2003) with both types of BCs.
With the clinical cases, similarly, we report εsprs, εcross and CPPD achieved at iteration Itermin and the resulting computational cost when either the contact or the fixed BCs were applied. Paired-t tests were employed to evaluate the significance of the differences in model estimation accuracy due to the two BCs. Statistical significance was defined at 95%. The angular difference between the principal direction of computed whole-brain deformation and gravity was also evaluated.
All deformation computations were executed on an eight-processor Linux cluster (2.6 GHz, 8G RAM of shared-memory running Ubuntu 6.10) using the PARDISO sparse direct solver for the solution of all linear systems of equations, and all data analysis was performed in Matlab 7.3 (The Mathworks, Natick, MA).
3. Results
3.1. Simulated data with measurement noise
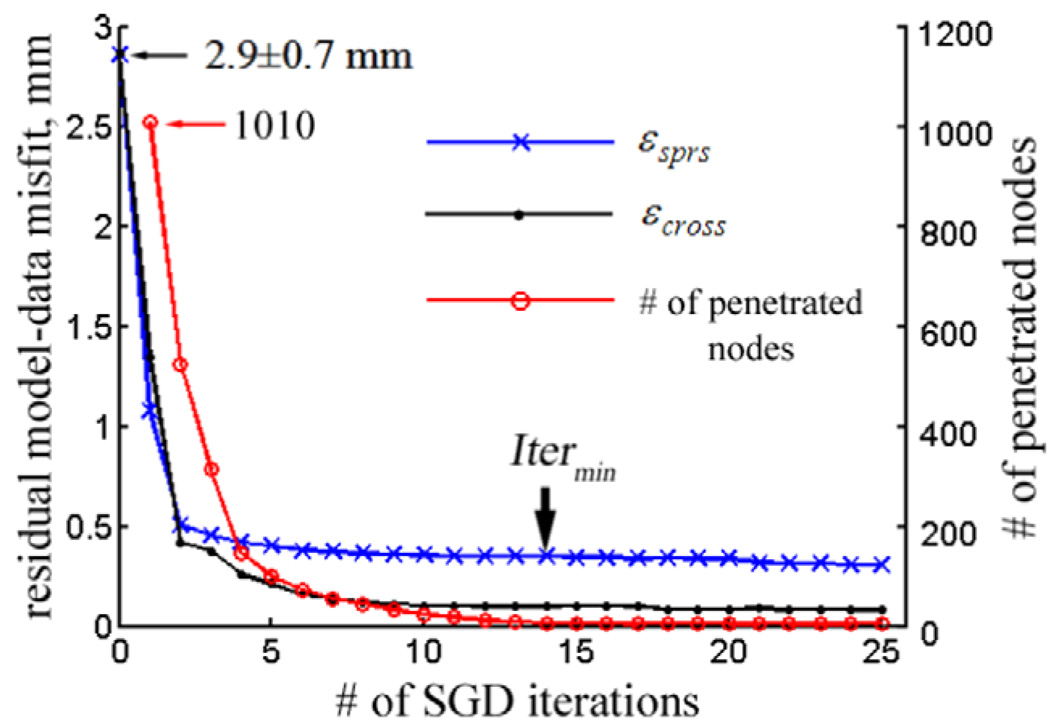
The residual model-data misfit, εsprs and εcross, as well as the number of brain surface nodes penetrating the inner-surface of the skull are plotted against the SGD iteration. Itermin of 14 was found (Fig. 3), resulting in a total computational cost of 186 s. The number of nodes penetrating the skull virtually diminished to zero at Itermin (the remaining penetrated nodes (N = 8) were due to mesh irregularities in the basal region of the skull that caused ambiguity in calculating surface normals), demonstrating the fulfillment of the contact impenetrability condition (Eq. (12)).
Fig. 3.
The residual model-data misfit εsprs and εcross, as well as the number of brain surface nodes penetrating the skull as a function of SGD iteration for the simulated ground truth case. Itermin of 14 was found, resulting in εsprs and εcross of 0.40 ± 0.20 and 0.11 ± 0.07 mm, respectively. The number of nodes penetrating the skull surface virtually diminished to zero after Itermin iterations.
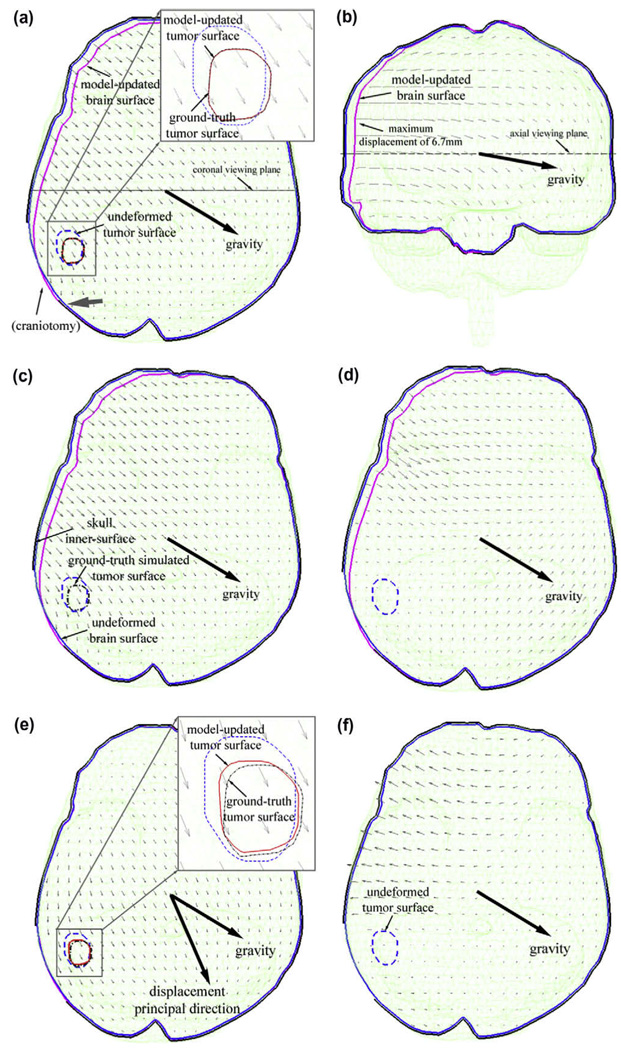
Model estimation accuracy reached 0.11 ± 0.07 mm at the tumor cross-validation locations, much improved from that at the sparse data points used to drive the model (0.40 ± 0.20 mm, about equal to the noise level in the data). The smaller cross-validation error resulted from the fact that noise was initially superimposed on the sparse data used to drive the model estimation to simulate measurement error, while noise-free displacements were used to evaluate accuracy at the cross-validation locations. These results demonstrate the inversion algorithm’s tolerance to measurement error, which is important in clinical settings where measurement error is generally less predictable or unknown (Ji et al., accepted for publication). Fig. 4a and b shows the model-estimated displacement projections on the axial (coronal) plane passing through the center of the craniotomy (centroid of all mesh nodes), which recovered about 96% of tumor deformation (εinitof 2.9 ± 0.67 mm). The CPPD between the model-updated and the ground truth parenchymal surfaces at the exposed cortex (found in the same manner as the cross-validation error at the tumor boundary) was reduced from 1.3 ± 0.82 to 0.07 ± 0.06 mm, indicating that about 95% of the parenchymal surface deformation was recovered.
Fig. 4.
Model response simulating brain sagging due to gravitational forces using either the contact (a–d) or fixed (e and f) BC with simulated data. Shown are the projections of the model-estimated displacements (in mm) on the axial plane through the center of the craniotomy (a and e) and on the coronal plane through the centroid of mesh nodes (b), the axial projections of the ground truth displacements (c), the displacement difference relative to the ground truth (d; magnified five times for visualization), and the displacement difference between model estimates produced by the two types of BCs (f). A substantial angular difference between the principal direction of the whole-brain deformation and gravity was evident when the fixed BC was used (e; 36.2°), which was not present when the contact BC was otherwise used (3.1°; principal direction not shown). The tumor surface estimated with the inversion algorithm is visually compared with the ground truth for the contact BC in (a) where the two surfaces closely matched with each other, and for the fixed BC in (e) where the two surfaces were markedly different.
To compare the model-estimated solution with the ground truth (Fig. 4c) over the entire deformation domain, the difference in the resulting displacements was projected onto an axial plane through the center of the craniotomy (Fig. 4d). The majority of the displacement differences (>99%) were less than 5% when normalized by the maximum ground truth displacement of 6.7 mm (which occurred in the right parietal region of the brain opposite to gravity; Fig. 4b), and were larger in regions far away from the tumor where the sparse data were generated. The maximum difference occurred in the nonconvex basal region of the skull due to mesh irregularities (up to 10%; not shown).
For comparison, model estimation was also produced with the fixed BC using the identical set of sparse data for model guidance. As expected, parenchymal displacements normal to the skull were not permissible with this type of BC (Fig. 4e), which resulted in large displacement differences compared with the ground truth, especially in regions where the parenchyma moved away from the skull using the contact BC (Fig. 4f). The fixed BC generated larger model-data residual errors around the tumor (1.05 ± 0.73 mm at sparse data points and 0.53 ± 0.32 mm at the cross-validation points). A much more substantial angular difference was also present between the principal direction of the computed whole-brain deformation and gravity for the fixed BC (36.2°; Fig. 4e) relative to the contact BC (3.1°). The angular difference between the two principal directions as a result of the two types of BCs was 34.6°.
3.2. Clinical datasets
3.2.1. Model performance at the tumor boundary
The number of measured tumor boundary displacements and the initial model-data misfit (εinit) are listed in Table 3, along with εsprsand εcrossgenerated by both the contact and fixed BCs at their respective Itermin (see Table 5). When the contact BC was applied, model estimation accuracy based on εcrosswas improved for patients 1 and 4 relative to when the fixed BC was used (p < 0.01), whereas no significant difference was found for patients 2, 3, and 5 (p > 0.05). On average, the BCs generated comparable model estimation accuracies at the tumor boundary, recovering 85% and 83% of the subsurface deformation when the contact and fixed BCs were applied, respectively.
Table 5.
Summary of the number of SGD iterations needed to reach an approximate convergence of the inversion scheme and the resulting computational cost with the contact and fixed BCs.
| Patient | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| Itermin | Contact BC | 19 | 13 | 10 | 17 | 12 |
| Fixed BC | 23 | 19 | 75 | 36 | 16 | |
| Time(s) | Contact BC | 265 | 142 | 289 | 215 | 250 |
| Fixed BC | 252 | 73 | 255 | 107 | 91 |
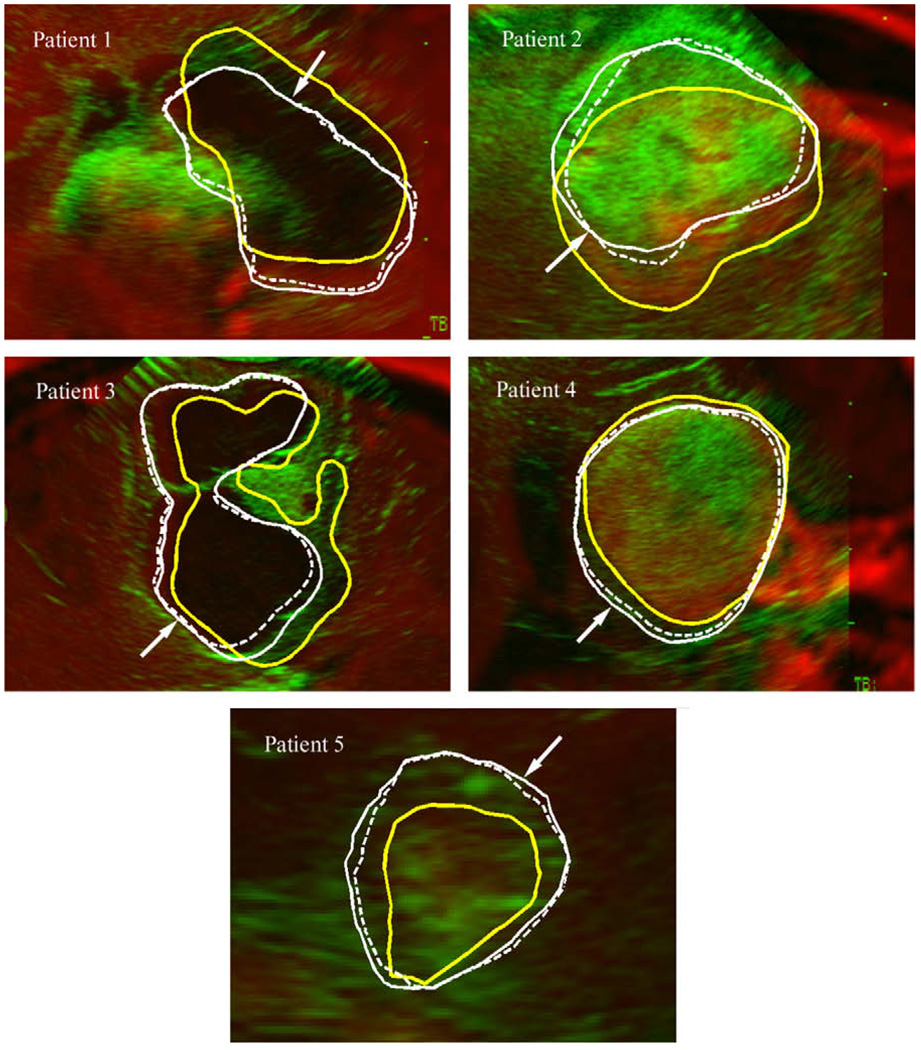
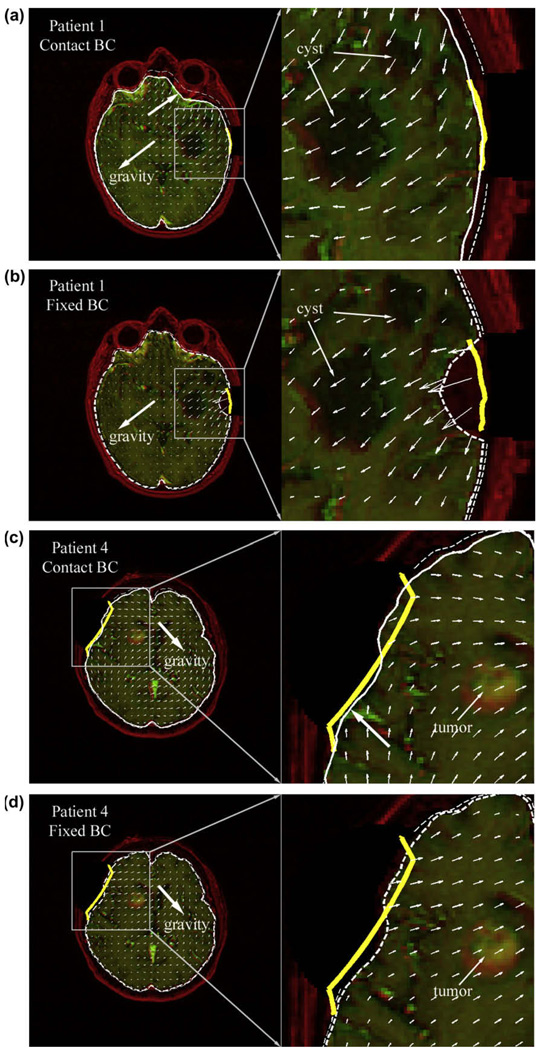
To provide visual assessment of model estimation accuracy at the tumor boundary, typical overlays of iUS images post-durotomy against the corresponding oblique pMR images are shown for each patient (Fig. 5).
Fig. 5.
Typical overlays of iUS post-durotomy (green) on the corresponding oblique pMR (red) using the transformation between iUS pre-durotomy and pMR. For all patients, misalignments between the cross-sections of the undeformed pMR tumor surface (thick yellow lines; intersected by the iUS imaging plane) and the tumor boundaries present in iUS indicate tumor motion post-durotomy. The model-updated tumor cross-sections (solid/dashed thin lines generated by the contact/fixed BC) improved the match with the tumor boundary present in iUS in all patients (indicated by arrows), regardless of the type of BCs used. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.2. Model performance at the craniotomy
Model response at the craniotomy surface varied between cases. For patient 1 whose parenchyma fully contracted, the model-updated parenchymal surface closely matched the measured surface, producing a CPPD approximately equal to the accuracy of the stereopsis data when the contact BC was applied (Table 4; Fig. 6a). By contrast, a much more localized deformation was apparent when the fixed BC was used (Fig. 6b), because it did not allow the parenchymal surface to move away from the skull other than at the craniotomy (its CPPD was about eight times that achieved with the contact BC; p ≪ 0.01). The improvement in model flexibility was also evident when the parenchyma partially sagged (patient 4; approximately 40% of the measured surface sagged; Fig. 6c and d), where movements both towards and away from the skull were present when the contact BC was applied, while only the latter was achieved using the fixed BC.
Table 4.
Summary of the size of craniotomy and model estimation performance in this region.
| Patient | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Size of craniotomy, mma | 28 × 18 | 45 × 24 | 42 × 33 | 59 × 56 |
| Initial CPPD, mmb | 4.7 ± 1.1 | 5.0 ± 1.6 | 0.74 ± 0.67 | 2.1 ± 1.0 |
| CPPD, mm; Contact BC | 1.1 ± 0.8 | 1.1 ± 0.96 | 1.2 ± 0.57 | 1.9 ± 1.0 |
| CPPD, mm; Fixed BC | 7.8 ± 2.8 | 2.3 ± 1.1 | 1.1 ± 0.64 | 2.3 ± 0.9 |
Size of craniotomy for patient 5 (50 × 40 mm) was approximated using a ruler in the OR; initial CPPD for patient 5 was 0.57 ± 0.27 mm, as estimated from the initial mesh configuration relative to the model-estimated surface at Itermin (not shown).
Using the undeformed craniotomy nodes relative to the measured parenchymal surface post-durotomy.
Fig. 6.
Model responses generated by applying the contact and fixed BCs for two patients whose exposed parenchymal surface contracted either fully (patient 1) or partially (patient 4). The undeformed (red) and model-updated (green) axial MR images at the center of the craniotomy are overlaid with the corresponding displacement projections (unit in pixels). Thick solid (ac) or dashed (bd) white lines represent the cross-sections of model-updated brain surface generated by the contact or fixed BCs, respectively, while thin dashed white lines represent the skull inner-surface. Thick yellow lines are the cross-sections of the parenchymal surface at the craniotomy measured from stereopsis. Thick arrows in (ac) indicate a sagging or bulging response generated by the contact BC that was lacking with the fixed BC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
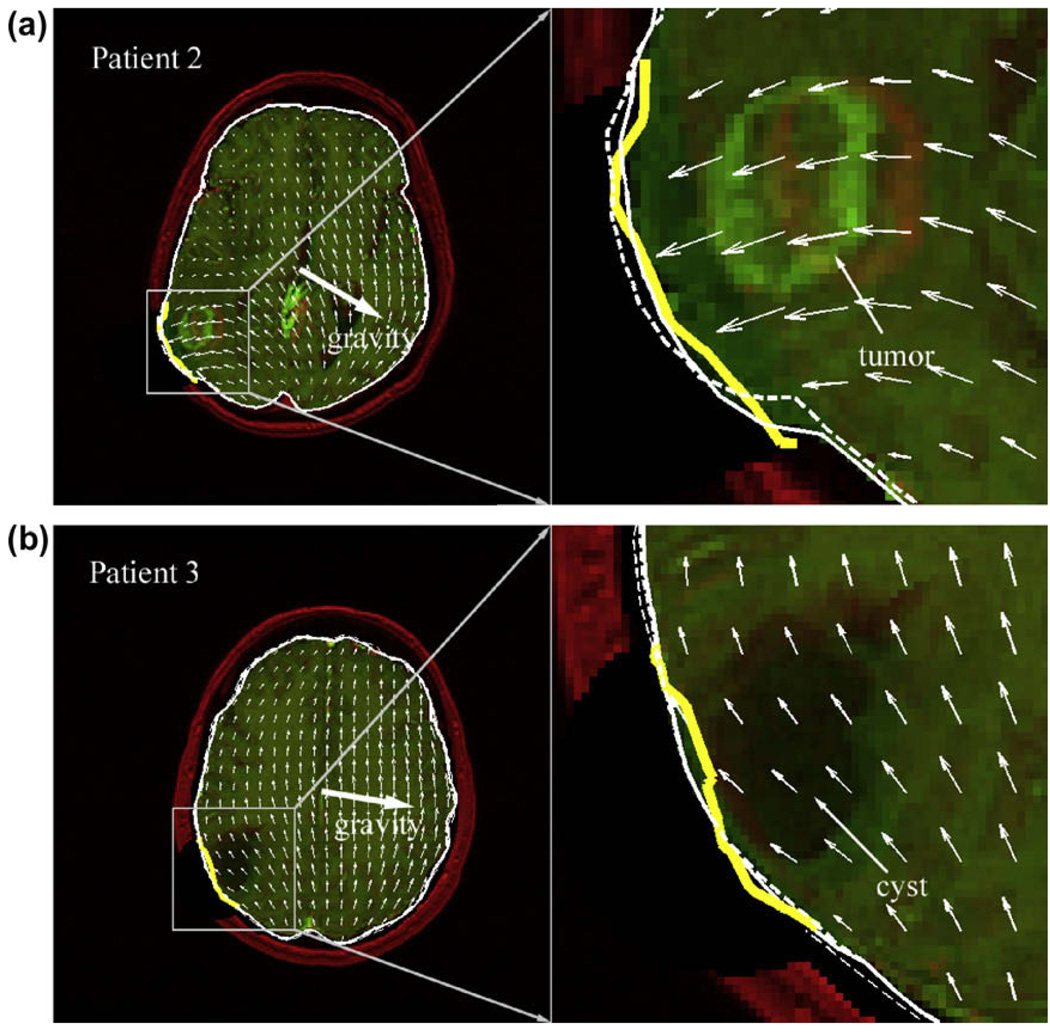
Similar localized parenchymal deformation around the craniotomy was evident when applying the fixed BC to patient 2 whose parenchyma surface fully distended at the craniotomy (Fig. 7a). By allowing the parenchymal surface to move towards the skull, the contact BC reduced the amount of distension compared with the response produced by the fixed BC, thereby improving the match with the surface measured from stereopsis. By comparison, both the contact and the fixed BCs produced similar deformation near the craniotomy in patient 3, where dominant tangential movement of the parenchymal surface (as opposed to distention or contraction as in other patient cases) was apparent (Fig. 7b).
Fig. 7.
Overlays of model-updated and undeformed axial MR images for 2 patients with displacement projections generated by the contact BC where the parenchyma distended either fully (patient 2) or partially (patient 3) at the craniotomy. All other labels are identical to those in Fig. 6.
3.2.3. Computational efficiency
The number of iterations to attain approximate SGD convergence (Itermin) resulting from the contact BC was consistently less than that needed with the fixed BC for all patients (by 43% on average; Table 5), demonstrating the improved model flexibility when the parenchymal surface is allowed to move normal to the skull. However, because of the extra factorization required in the solution process to account for the modified global system matrix when slave node penetration is detected, model solutions using the contact BC resulted in increased computational cost (about 49% more than needed for the fixed BC).
Representative curves of εcrossgenerated from the contact and fixed BCs in relation to SGD iterations are plotted for two patients, whose parenchymal surface at the craniotomy either fully contracted (patient 1) or distended (patient 2).
After Itermin iterations, the number of penetrated nodes virtually diminished to zero for both patients using the contact BC, suggesting fulfillment of the contact impenetrability condition (Eq. (12); the residual number of penetrated nodes was 13 and 10, respectively, due to mesh irregularities in the nonconvex basal region of the skull; Fig. 8b and d). Oscillation was evident in the distension case, suggesting that dynamic out-ward motion of the brain surface nodes normal to the skull existed during the contact solution process, which was less evident in the contraction case (Fig. 8b and d). The magnitude of the residual penetrations in both cases was similar (<0.1 mm) (Fig. 8).
Fig. 8.
Representative curves of εcross using the contact and fixed BCs, and the number of nodes penetrating the inner-surface of the skull and the average penetration magnitude with the contact BC as a function of SGD iterations for patient 1 (a and b) and 2 (c and d), whose parenchyma either fully contracted or distended at the craniotomy. Itermin for each patient using the two BCs are identified (thick arrows; see also Table 3).
3.2.4. Displacement principal direction
Finally, the principal direction of the model-estimated displacement field was extracted using principal component analysis (PCA; Hyvärinen et al., 2001). All nodal displacement vectors, starting from the origin in a Cartesian coordinate system, were represented by the corresponding end points, and the principal direction was determined as the vector emanating from the origin along which the variance of the points was maximized, similar to the approach in Hartkens et al. (2003). The angular difference between the resulting principal direction and gravity was tabulated, along with the difference in the principal directions produced by the contact and fixed BCs (Table 6). Except for patient 1 whose parenchyma virtually moved along the direction of gravity, model displacements in general did not correspond to gravity, and nodes even moved in nearly opposite directions (patient 2 and 5), regardless of which BCs were applied (Fig. 6a and b). The angular difference in the principal directions produced by the two BCs was small when the parenchymal surface at the craniotomy largely contracted (patients 1 and 4), while it was relatively large otherwise (patients 2 and 5; Table 6).
Table 6.
Summary of the angular differences between the gravitational direction and the principal direction of model-estimated brain motion produced by the contact and fixed BCs, as well as the angular differences between the principal directions generated by the two BCs (in degrees).
| Patient | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Contact BC; with gravity | 10 | 140 | 105 | 93 | 141 |
| Fixed BC; with gravity | 9 | 164 | 117 | 88 | 168 |
| Between BCs | 13 | 55 | 14 | 3 | 28 |
4. Discussion
We have incorporated a brain–skull contact BC into a data assimilation scheme using an SGD inversion framework to simulate movement of the parenchymal surface towards or away from the skull. This type of motion frequently occurs in the OR (e.g., Hartkens et al., 2003; Maurer et al., 1998; Nabavi et al., 2001; Nimsky et al., 2000), but is not well represented by the type of fixed BCs we have assigned previously (e.g., Lunn et al., 2005, 2006; Miga, 1998; Platenik et al., 2002; Sun et al., 2005b). The validity of the algorithm implementation was demonstrated with a model solution guided by sparse data generated locally from simulated ground truth where the majority of displacement differences between the model estimation and the ground truth were less than 5%. The technique was further applied to five clinical datasets where sparse data at the tumor boundary post-durotomy were randomly selected for model guidance. Model solutions produced at the tumor boundary were compared against the measured displacements to assess model performance (with either the brain–skull contact BCs or fixed BCs). In addition, stereopsis data for four cases were utilized to further cross-examine the validity of the model response. The accuracy of the stereopsis system has been assessed previously to be approximately within 1 mm (Sun et al., 2005a,b), and was not re-evaluated in this study.
Our results indicate that on average, these BCs achieve comparable model estimation performance at the tumor boundary where the sparse data were generated for model guidance, recovering about 83%–85% of the subsurface deformation, but statistically significant differences can occur in individual cases with the contact BC performance being better. Model response at the craniotomy varied significantly when the two types of BCs were applied. Specifically, a more localized parenchymal deformation was present with the fixed BC, especially when the parenchymal surface fully contracted (i.e, patient 1) because of its inability (other than that at the craniotomy) to move away from the cranial wall in concert with the tumor, which significantly degraded model estimation accuracy in this region (CPPD of up to 8 mm). In contrast, the contact BC significantly improved the model estimation in this region, achieving CPPDs on the order of the stereopsis data accuracy. The difference in model responses was less evident when parenchymal distension occurred (patients 2 through 5, as well as in the simulated data where the parenchyma partially distended; Fig. 4), likely due to the small brain–skull gap simulated in this study (1 mm), which limited the extent of brain motion towards the skull. However, the principal directions of the model deformation varied significantly with the two BCs in this group of patients, especially when the magnitude of distension was large (patient 2; Table 6). These observations suggest that the contact BC performs best when large parenchymal contraction is present. However, the unpredictable magnitude and pattern of parenchymal deformation in the OR make selection of the simpler fixed BCs, in situations where they perform well, difficult to anticipate. Importantly, the contact BCs do not degrade model performance even when the fixed BCs are adequate, thus, they can be activated in every case without negative consequences.
The computational efficiency (3.9 min on average) of applying the contact BC within the SGD inversion framework establishes its feasibility for use in clinical applications in the OR and may be further improved by applying iterative algorithms to solve the linear systems of equations (e.g., Algebraic Multigrid algorithms; Stüben, 2001), which do not involve factorization of the system stiffness matrix. In addition, utilizing a locally (instead of globally, as in this study) refined mesh in regions of interest (e.g., around the tumor and craniotomy) is also expected to further reduce the computational cost without sacrificing accuracy in the model estimates.
The model estimation accuracy at the craniotomy surface varied across the patients in this study (up to 77% for patient 1, but no improvement for patient 3), in part because we have only used sparse data generated at the tumor boundary for model guidance (the measurements of parenchymal surface displacement from stereopsis being utilized for evaluation of the different types of BCs applied). Incorporating intraoperatively acquired displacement data both at the tumor boundary and parenchymal surface at the craniotomy is expected to produce excellent model performance in both regions.
One limitation with the study is that we used rigid-body displacements produced by ICP to validate the accuracy of non-rigid tumor deformation generated from model estimates for clinical patients. Although it may be seemingly desirable to use non-rigid tumor displacements generated directly from image-based registration (e.g., via mutual information) for more faithful validation, the high computational cost (Holden, 2008) of this method would compromise its feasibility for clinical applications in the OR as a means of obtaining displacement data for model assimilation. For example, an average computational cost of 1–2 min is necessary for rigid (as opposed to non-rigid) registration between pMR and iUS based on normalized mutual information (Ji et al., 2008), and the computational cost for non-rigid registrations is expected to be much greater, easily surpassing that of the computational model updates (3.9 min in this study using contact BCs, while 2 min in Ji et al. (accepted for publication) using BCs). In addition, we also compared the model response by assimilating sparse data generated from ICP registration with that produced by assimilating the ground truth displacements for the simulated case. At the same iteration (Itermin of 14; Fig. 3), the model-data misfit at the cross-validation locations was 0.23 ± 0.11 mm when sparse data generated from ICP registration were used, capturing 92% of tumor deformation which was only slightly degraded from the 96% recovery when the ground truth displacements were otherwise used. These results indicate that even though the ICP algorithm produced rigid tumor displacements, non-rigid tumor deformation was achieved through data assimilation, which is an added benefit of the inversion approach used in this work.
Nevertheless, we have demonstrated that a contact BC improves model flexibility and leads to more rapid SGD convergence than the fixed BC, especially when the motion of the parenchymal surface is large (e.g., full contraction). However, it is also important to realize that this type of BC is not able to transmit tension and does not represent the connective tissues that may restrict brain–skull separation (e.g, dura and arachnoidal trabecular). Therefore, the magnitude and pattern of parenchymal surface collapse and distention obtained in this study requires further investigation (e.g., brain–skull separation is likely exaggerated in the left frontal region of the brain in Fig. 6a). Additionally, we have simulated the brain–skull separation as a uniform 1 mm gap to limit the maximum amount of parenchymal surface distension that is allowed. Clinically, however, the thickness of the CSF layer varies across patients and regions of the brain. In principle, a patient-specific brain–skull gap can be generated and inclusion of a contact surface that more accurately reflects the inner-surface of the skull warrants further investigation of its effect on brain deformation (but is likely to improve upon the results presented here). Nonetheless, we have shown that intraoperative brain shift compensation through biomechanical modeling is feasible with the contact BC, and further validation of a patient-specific version of this type of BC will be reported in the future.
5. Conclusion
We have incorporated a brain–skull contact BC within a computationally efficient inverse method for intraoperative data assimilation. The technique captures the important features of both the contact BC (which improves model flexibility by allowing two-way parenchymal surface motion normal to the skull) and displacement estimates from an inverse solution scheme (which maintains stress-free BCs at the craniotomy while minimizing the effect of measurement noise in the data). In the five patient cases evaluated where sparse data was generated locally at the tumor boundary, the contact BC recovered about 85% of subsurface tumor deformation on average, which was slightly better than the fixed BC (83% on average), whereas it significantly improved the accuracy in the model estimates at the craniotomy (1.6 mm on average) compared with the fixed BC (3.4 mm on average). The contact scheme is particularly attractive for OR use where the extent and pattern of parenchymal displacement is difficult to anticipate because it improves model performance in cases where brain–skull contact significantly influences the deformation field but does not degrade model estimates when the simpler fixed BCs would have been adequate. These results may be further improved by incorporating the stereopsis data at the craniotomy into the data assimilation itself. While the technique presently requires more computations relative to the fixed BC, the average computational cost was still under 3.9 min on average (2.6 min for the fixed BC), and can be further reduced by employing a fast iterative solver for the solution of the linear systems of equations and by applying locally refined meshes in regions of interest to reduce the number of degrees-of-freedom without sacrificing model accuracy. These characteristics of the proposed technique make it appear to be feasible for routine application in the OR as a model-based brain shift compensation scheme in the near future.
Acknowledgement
Funding from the National Institutes of Health grant R01 EB002082-11 is acknowledged.
References
- Bandak F, Eppinger R. A three-dimensional finite element analysis of the human brain under combined rotational and translational accelerations; Proceedings of 38th Stapp Car Crash Conference; 1994. [Google Scholar]
- Besl PJ, McKay ND. A method for registration of 3-d shapes. IEEE Trans. Pattern Anal. Machine Intell. 1992;14(2):239–256. [Google Scholar]
- Biot M. General theory of three-dimensional consolidation. J. Appl. Phys. 1941;12:155–164. [Google Scholar]
- Buckner JC. Factors influencing survival in high grade gliomas. Semin. Oncol. 2003;30:10–14. doi: 10.1053/j.seminoncol.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Clatz O, Delingette H, Talos IF, Golby AJ, Kikinis R, Jolesz FA, Ayache N, Warfield SK. Robust nonrigid registration to capture brain shift from intraoperative MRI. IEEE Trans. Med. Imaging. 2005;24(11):1417–1427. doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpuri P, Thompson RC, Dawant BM, Cao A, Miga MI. An atlas-based method to compensate for brain shift: preliminary results. Med. Image Anal. 2007;11:128–145. doi: 10.1016/j.media.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkens T, Hill DLG, Castellano-Smith AD, Hawkes DJ, Jr, Maurer CR, Martin MJ, Hall WA, Liu H, Truwit CL. Measurement and analysis of brain deformation during neurosurgery. IEEE Trans. Med. Imaging. 2003;22(1):82–92. doi: 10.1109/TMI.2002.806596. [DOI] [PubMed] [Google Scholar]
- Hartov A, Roberts DW, Paulsen KD. A comparative analysis of co-registered ultrasound and MRI in neurosurgery. Neurosurgery. 2007;62 Suppl. 1:91–101. doi: 10.1227/01.neu.0000317377.15196.45. [DOI] [PubMed] [Google Scholar]
- Holden M. A review of geometric transformations for nonrigid body registration. IEEE Trans. Med. Imaging. 2008;27(1):111–128. doi: 10.1109/TMI.2007.904691. [DOI] [PubMed] [Google Scholar]
- Hu J, Jin X, Lee JB, Zhang L, Chaudhary V, Guthikonda M, Yang KH, King AI. Intraoperative brain shift prediction using a 3D inhomogeneous patient-specific finite element model. J. Neurosurg. 2007;106:164–169. doi: 10.3171/jns.2007.106.1.164. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A, Karhunen J, Oja E. Independent Component Analysis. New York: John Willey and Sons, Inc; 2001. [Google Scholar]
- Ji S, Zhu Q, Dougherty L, Margulies S. In vivo measurements of human brain displacement. Stapp Car Crash J. 2004;48:527–539. doi: 10.4271/2004-22-0010. [DOI] [PubMed] [Google Scholar]
- Ji S, Margulies S. Brainstem motion within the skull: measurement of the pons displacement in vivo. J. Biomech. 2007;40(1):92–99. doi: 10.1016/j.jbiomech.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Ji S, Liu F, Hartov A, Roberts DW, Paulsen KD. Brain–skull boundary conditions in a computational deformation model; Medical Imaging 2007: Visualization, Display and Image-Guided Procedures, Proceedings of SPIE; San Diego, USA: 2007. [Google Scholar]
- Ji S, Hartov A, Roberts DW, Paulsen KD. Data assimilation using a gradient descent method for estimation of intraoperative brain deformation. Med. Image Anal. doi: 10.1016/j.media.2009.07.002. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Wu Z, Hartov A, Roberts DW, Paulsen KD. Mutual-information-based patient re-registration using intraoperative ultrasound for image-guided neurosurgery. Med. Phys. 2008;35(10):4612–4624. doi: 10.1118/1.2977728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiven S, Hardy W. Correlation of an FE model of the human head with local brain motion - consequences for injury prediction. Stapp Car Crash J. 2002;46:123–144. doi: 10.4271/2002-22-0007. [DOI] [PubMed] [Google Scholar]
- Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, Lang FF, McCutcheon IE, Hassenbush SJ, Holland E, Hess K, Michael C, Miller D, Sawaya R. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extend of resection and survival. J. Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- Laursen TA. Computational Contact and Impact Mechanics: Fundamentals of Modeling Interfacial Phenomena in Nonlinear Finite Element Analysis. Springer-Verlag; 2002. [Google Scholar]
- Laws ER, Shaffrey ME, Morns A, Anderson FA., Jr Surgical management of intracranial gliomas – does radical resection improve outcome? Acta Neurochir. 2003 Suppl. 85:47–53. doi: 10.1007/978-3-7091-6043-5_7. [DOI] [PubMed] [Google Scholar]
- Le Tallec P. In: Numerical methods for solids. Ciarlet PG, Lions JL, editors. North-Holland: Handbook of Numerical Analysis; 1994. [Google Scholar]
- Lunn KE, Paulsen KD, Lynch DR, Roberts DW, Kennedy FE, Hartov A. Assimilating intraoperative data with brain shift modeling using the adjoint equations. IEEE Trans. Med. Image Anal. 2005;9:281–293. doi: 10.1016/j.media.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Lunn KE, Paulsen KD, Liu F, Kennedy FE, Hartov A, Roberts DW. Data-guided brain deformation modeling: evaluation of a 3-D adjoint inversion method in porcine studies. IEEE Trans. Biomed. Eng. 2006;53(10):1893–1900. doi: 10.1109/TBME.2006.881771. [DOI] [PubMed] [Google Scholar]
- Lynch D. Numerical Partial Differential Equations for Environmental Scientists and Engineers. Berlin: Springer; 2004. [Google Scholar]
- Maurer CR, Jr, Hill DLG, Martin AJ, Liu H, McCube M, Rueckert D, Lloret D, Hall WA, Maxwell RE, Hawkes DJ, Truwit CL. Investigation of intraoperative brain deformation using a 1. 5-T interventional MR system: preliminary results. IEEE Trans. Med. Imaging. 1998;17(5):817–825. doi: 10.1109/42.736050. [DOI] [PubMed] [Google Scholar]
- Miga MI. Ph.D. Dissertation. Hanover, NH: Dartmouth College, Thayer School of Engineering; 1998. Development and Quantification of a 3D Brain Deformation Model for Model-Updated Image-Guided Stereotactic Neurosurgery. [Google Scholar]
- Miga MI, Paulsen KD, Kennedy FE. Von Neumann stability analysis of Biot’s general two-dimensional theory of consolidation. Int. J. Numer. Meth. Eng. 1998;43:955–974. [Google Scholar]
- Miller K, Chinzei K, Orssengo G, Bednarz P. Mechanical properties of brain tissue in-vivo: experiment and computer simulation. J.Biomech. 2000;33:1369–1376. doi: 10.1016/s0021-9290(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Miller RT, Margulies SS, Leoni M, Nonaka M, Chen X, Smith DH, Meaney DF. Finite element modeling approaches for predicting injury in an experimental model of severe diffuse axonal injury; Proceedings of 42nd Stapp Car Crash Conference; Tempe, AZ, SAE: 1998. [Google Scholar]
- Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS, Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB, Wells WM, III, Kikinis R, Jolesz FA. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001;48(4):787–798. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- Nimsky C, Ganslandt O, Cerny S, Hastreiter P, Greiner G, Falbusch R. Quantification of, visualization of and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery. 2000;47(5):1070–1080. doi: 10.1097/00006123-200011000-00008. [DOI] [PubMed] [Google Scholar]
- Paulsen KD, Miga MI, Kennedy FE, Hoopes PJ, Hartov A, Roberts DW. A computational model for tracking subsurface tissue deformation during streotactic neurosurgery. IEEE Trans. Biomed. Eng. 1999;46:213–225. doi: 10.1109/10.740884. [DOI] [PubMed] [Google Scholar]
- Platenik LA, Miga MI, Roberts DW, Lunn KE, Kennedy FE, Hartov A, Paulsen KD. In vivo quantification of retraction deformation modeling for updated image-guidance during neurosurgery. IEEE Trans. Biomed. Eng. 2002;49(8):823–835. doi: 10.1109/TBME.2002.800760. [DOI] [PubMed] [Google Scholar]
- Ruan JS, Khalil T, King AI. Dynamic response of the human head to impact by three-dimensional finite element analysis. J. Biomech. Eng. 1994;116(1):44–50. doi: 10.1115/1.2895703. [DOI] [PubMed] [Google Scholar]
- Schenk O, Gartner K. Solving unsymmetric sparse systems of linear equations with PARDISO. J. Future Gen. Compu. Syst. 2004;20(3):475–487. [Google Scholar]
- Skrinjar O, Nabavi A, Duncan J. Model-driven brain shift compensation. Med. Image Anal. 2002;6:361–373. doi: 10.1016/s1361-8415(02)00062-2. [DOI] [PubMed] [Google Scholar]
- Stüben K. A review of algebraic multigrid. J. Comput. Appl. Math. (JCAM) 2001;128:281–309. Vol. 7 of JCAM Millenium Issue. [Google Scholar]
- Sullivan JM, Jr, Charron G, Paulsen KD. A three dimensional mesh generator for arbitrary multiple material domains. Finite Element Anal. Des. 1997;25(3):219–241. [Google Scholar]
- Sun H, Roberts DW, Farid H, Wu Z, Hartov A, Paulsen KD. Cortical surface tracking using an operating microscope. Neurosurgery. 2005a;56(1):86–97. doi: 10.1227/01.neu.0000146263.98583.cc. [DOI] [PubMed] [Google Scholar]
- Sun H, Lunn KE, Faird H, Wu Z, Roberts DW, Hartov A, Paulsen KD. Stereopsis - guided brain shift compensation. IEEE Trans. Med. Imaging. 2005b;24(8):1039–1052. doi: 10.1109/TMI.2005.852075. [DOI] [PubMed] [Google Scholar]
- Takhounts EG, Eppinger RH, Campbell JQ, Tannous RE, Power ED, Shook LS. On the development of the SIMon finite element head model. Stapp Car Crash J. 2003;47:107–133. doi: 10.4271/2003-22-0007. [DOI] [PubMed] [Google Scholar]
- Wittek A, Kikinis R, Warfield SK, Miller K. Brain shift computation using a fully nonlinear biomechanical model. Med. Image Comput. Comput. Assist. Interv. Int. Conf. Med. Image Comput. Comput. Assist. Interv. 2005;8(Pt. 2):583–590. doi: 10.1007/11566489_72. [DOI] [PubMed] [Google Scholar]
- Wu Z, Paulsen KD, Sullivan JM., Jr Model initialization and deformation for automatic segmentation of T1-weighted brain MRI data. IEEE Trans. Biomed. Eng. 2005;52:1128–1131. doi: 10.1109/TBME.2005.846709. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang KH, Dwarampudi R, Omori K, Li T, Chang K, Hardy WN, Khalil TB, King AI. Recent advances in brain injury research: a new human head model development and validation. Stapp Car Crash J. 2001;45:369–393. doi: 10.4271/2001-22-0017. [DOI] [PubMed] [Google Scholar]
- Zhong ZH. Finite Element Procedures for Contact–Impact Problems. Oxford Science Publications; 1993. [Google Scholar]