Abstract
Vesicular transport to and from the lysosome and late endosome is defective in patients with Chediak–Higashi syndrome (CHS) and in mutant beige (bg) mice1–4. CHS and bg cells have giant, perinuclear vesicles with characterises of late endosomes and lysosomes that arise from dysregulated homotypic fusion3–5. CHS and bg lysosomes also exhibit compartmental missorting of proteins, such as elastase, glucuronidase and cathepsin G2,3,6,7. Lyst, a candidate gene for bg, was identified by direct complementary DNA selection from a yeast artificial chromosome (YAC) clone containing a 650-kilobase segment of the bg-critical region on mouse chromosome 13. Lyst is disrupted by a 5-kilobase deletion in bg11J mice, and Lyst messenger RNA is markedly reduced in bg2J homozygotes. The homologous human gene, LYST, is highly conserved with mouse Lyst, and contains a frame-shift mutation at nucleotides 117–118 of the coding domain in a CHS patient. Thus bg mice and human CHS patients have homologous disorders associated with Lyst mutations. Lyst encodes a protein with a carboxy-terminal prenylation motif and multiple potential phosphorylation sites. Lyst protein is predicted to form extended helical domains, and has a region of sequence similar to stathmin, a coiled-coil phosphoprotein thought to act as a relay integrating cellular signal response coupling8–10.
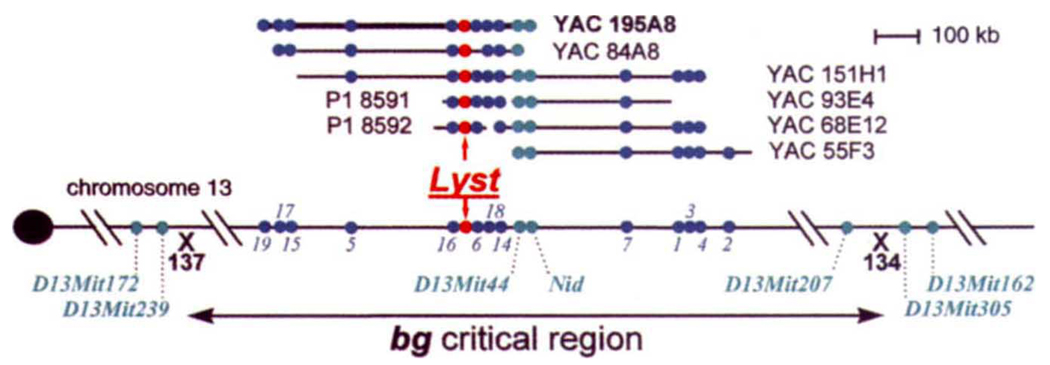
We have previously localized the bg locus within a 0.24-centimorgan interval on mouse chromosome 13, and isolated contiguous arrays of YACs that cover 2,400 kb of this interval11–13. Candidate cDNAs for bg were isolated from YAC 195A8, which contains 650 kb of the bg non-recombinant interval using direct cDNA selection with mouse spleen cDNA 13 (Fig. 1). Of 56 candidate cDNA clones analysed from a direct-selection experiment, evidence for causality in bg was found in one (see below), and this gene was designated Lyst (lysosomal trafficking regulator). As this clone was 132 nucleotides long, additional Lyst sequences were sought by screening three mouse cDNA libraries and performing polymerase chain reaction (PCR) amplification of cDNA ends14. Ten overlapping Lyst clones were identified, representing ~7kb (Genbank accession number, L77889). These were physically assigned to mouse chromosome 13 with pulsed field gel electrophoresis (PFGE) Southern blots, confirming that they were all derived from a single gene (mouse genome database accession number, MGD-PMEX-14). The Lyst probes identified the same polymorphic PFGE restriction fragments as nidogen (Nid)12, indicating that Lyst and Nid are clustered within 650 kb. Lyst was also mapped genetically in 504 [C57BL/6J-bgJ × (C57BL/6J-bgJ × CAST/EiJ)F1] backcross mice12 by means of three TaqI restriction fragment length polymorphisms (RFLPs). The Lyst RFLPs cosegregated with bg (and Nid), confirming their colocalization on proximal mouse chromosome 13 (MGD accession number, MGD-CREX-615).
FIG. 1.
Genetic and physical map of the bg non-recombinant interval on mouse chromosome 13 showing the location of Lyst. Mouse chromosome 13 is shown by a horizontal line with the centromere on the left. The bg critical region is delineated by chromosome crossovers (denoted with an X) in animals 134 and 137 of an intersub-specific mouse backcross12 [C57BL/6J – bgJ × (C57BL/6J – bgJ × CAST/EiJ)F1]. Microsatellite markers D13MM172 and D13Mit239 flank bg proximally; D13Mitl62 and D13Mit305 lie distal to bg (indicated by turquoise circles). YAC and P1 clones identified by PCR screening25,26 with oligo-nucleotides corresponding to Nid or D13Sfkl3 are shown above the chromosome. Novel sequence-tagged sites (STS, indicated by dark blue circles), generated by inverse repetitive element PCR or direct cDNA selection13, were used to order clones within the contiguous array. Novel mouse chromosome 13 STSs are numbered 1–18, corresponding to D13Sfkl to D13Sfkl8, respectively13. Lyst was isolated from YAC 195A8, a 650-kb clone, by using direct cDNA selection13. The physical location of Lyst-associated STSs on YAC and P1 clones are shown in red (MGD accession number MGD-PMEX-13).
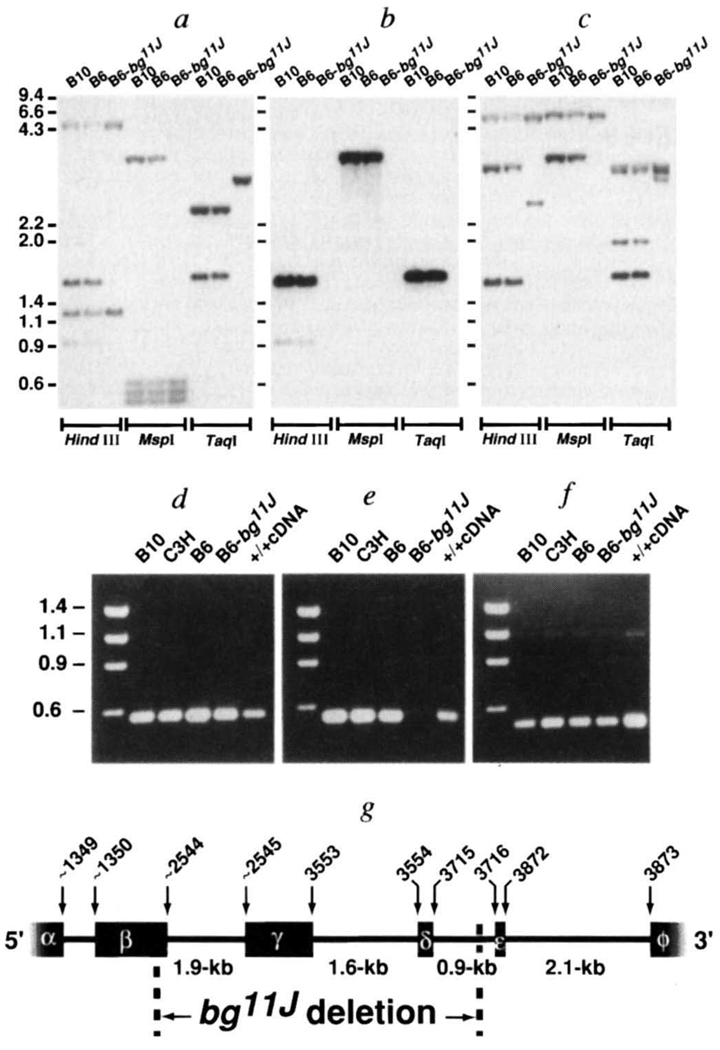
Evidence for Lyst mutations was found in two bg alleles. A 5-kb genomic deletion that contained the 3′ end of Lyst exon β, and exons γ and δ, was identified in bg11J DNA (Fig. 2). The bg11J deletion corresponds to loss of ~400 internal amino acids of the predicted Lyst peptide. Furthermore, whereas the 5′ end of the bg11J deletion occurs within Lyst exon β, the 3′ end is intronic. Therefore the truncated Lyst mRNA in bg11J mice is also anticipated to splice incorrectly, terminate prematurely, and lack polyadenylation.
FIG. 2.
Intragenic deletion of Lyst in bg11J DNA. a–c, Southern blot identification of an intragenic Lyst deletion in bg11J DNA. A Southern blot was sequentially hybridized11 with 3 Lyst probes: a, the probe (nucleotides 1,262–3,433 of Lyst cDNA) extends upstream from the bg11J deletion; b, the probe (nucleotides 2,835–3,433 of Lyst cDNA) is completely deleted; c, the probe (nucleotides 3,594–4,237 of Lyst cDNA) extends downstream from the bg11J deletion. Restriction endonucleases are indicated at the bottom of each panel, and molecular size standards (in kb) are shown to the left. Similar results were obtained with 3 additional restriction endonucleases (data not shown). The bg11J mutation was discovered in 1992 at The Jackson Laboratory in a C57BL/6J-jb mouse at generation N4 after transfer from B6C3Fe-a/a. The mutation jb had, in turn, been discovered 14 generations earlier in B6C3Fe-a/a-hyh mice at generation N3 after transfer form C57BL/10J. The hyh mutation arose in C57BL710J mice, and was maintained in that strain until transfer at F15. Thus the possible contributors of genetic information to bg11J include C57BL/6J, C3HeB/FeJ and C57BL/10J. Southern blots were prepared from genomic DNA of all potential progenitor mouse strains, but only C57B/V10J, C57BL/6J and C57BI/6J-bg11J are shown, d–f, PCR analysis of the bg11J deletion. C57BL/10J, C3HeB/FeJ, C57BL/6J and C57BL/6J-bg11J genomic DNA and Lyst cDNA were used as templates in the PCR reactions. Amplicons illustrated correspond to: d, Lyst cDNA nucleotides 1,337–1,837, which represent exon β and are upstream from the deletion; e, nucleotides 2,670–3,210, which represent exon γ, which is deleted in bg11J DNA; and f, nucleotides 4,913–5,433, which represents an exon downstream from the deletion. No amplicon was observed in control PCR reactions performed without template. More than 30 other STSs that had been localized within the bg non-recombinant interval PCR amplified normally from bg11J DNA. g, Genomic structure of Lyst in the vicinity of the bg11J deletion. Lyst exons (α, β, γ, δ, ε and φ) are depicted by black boxes, and intervening introns by a solid line. Nucleotides of the mouse Lyst cDNA that correspond to exonic boundaries are indicated above the boxes. The 3′ end of exon β, and all of exons γ and δ, are deleted in bg11J DNA. The region of Lyst protein that is deleted in bg11J contains a pair of helices with N-terminal phosphorylation sites. Genomic structure and intronic sequences were ascertained by sequence analysis of nested PCR products, performed with exonic primers and P1 clone DNA as template14. Boundaries of the bg11J deletion were determined by PCR of genomic DNA.
Quantitative reverse transcription (RT)–PCR demonstrated a moderate decrease in Lyst mRNA in bg and bgJ liver, and a gross reduction in bg2J (Lyst ΔOD after normalization for β-actin mRNA: +/+, 1.00; bg2J/bg2J 0.19; bg/bg, 0.28; bgJ/bgJ, 0.40). A commensurate reduction in bg2J transcript abundance was noted by using several primer pairs derived from different regions of the Lyst cDNA. Aberrant Lyst RT–PCR products were not observed. The particularly striking (more than fivefold) reduction in Lyst expression evident in bg2J homozygotes suggested the existence of a mutation in bg2J Lyst that results in decreased transcription or mRNA instability. The molecular basis of the decrease in Lyst mRNA in bg2J is not yet known, but it is reminiscent of the leaky ablation of mature message associated with an intronic retro-transposition event14.
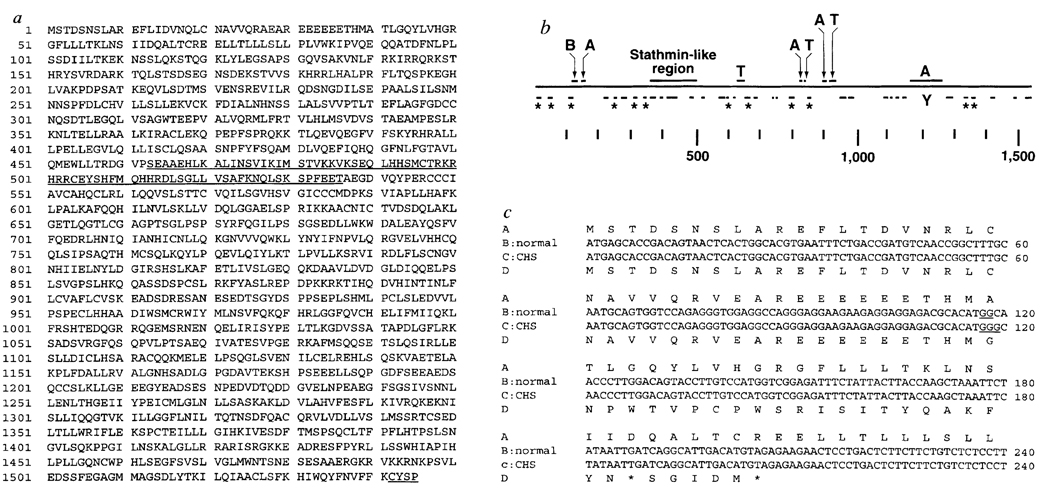
The predicted open reading frame (ORF) of Lyst was 4,635 nucleotides, encoding a protein of 1,545 amino acids and relative molecular mass 172,500 (Mr 172.5K) (Fig. 3a). Nucleotides 51–74 are rich in CG dinucleotides, a common feature of the 5′ region of housekeeping genes. Comparison with DNA databases indicated that Lyst is novel, and resembles only uncharacterized human expressed sequence tags (ESTs) (L. Hillier et al, unpublished data; J. Macke et al, unpublished data). The sequence of a cDNA clone corresponding to one such human EST (Genbank accession number L77889) matched the 5′ region of mouse Lyst (nucleotide identity was 76% in the 5′ untranslated region (UTR), 91% in the ORF, and amino-acid identity was 97%; Fig. 3c); another human EST matched the 3′ region of the mouse Lyst coding domain (Genbank accession number W26957). On hybridization to PFGE Southern blots of mouse DNA12, the human clones identified restriction fragments that were indistinguishable from mouse Lyst; physical mapping of the human clones to the same region of the mouse genome as Lyst indicates that they are indeed homologous to Lyst.
FIG. 3.
Amino-acid sequence and predicted peptide structure of mouse and human Lyst cDNAs. a, Predicted amino-acid sequence of the mouse Lyst cDNA. The region (residues 463–536) with sequence similarity to stathmin is underlined, as is a C-terminal prenylation motif, b, Predicted secondary structure27–30 and motifs of mouse Lyst protein. The solid line denotes the protein sequence. Above the line are the stathmin-like region and areas rich in acid (A; Asp, Glu), basic (B; His, Arg, Lys), and turn (T; Pro, Gly, Ser, Thr, Asp) residues, the latter, especially at regions 600–625 and 875–895, are too long to be a simple turn between secondary structural elements, and are either very flexible or very rigid (in a manner reminiscent of collagen, although they do not fit the collagen consensus motifs). Helices predicted with >80% confidence are shown as solid lines below the protein sequence, and asterisks identify helices with N-terminal phosphorylation motifs. A tyrosine phosphorylation motif (residue 1,214) is designated Y. c, Nuceiotide and predicted amino-acid sequence of the 5′ end of the human LYST coding domain showing a frame-shift mutation in a patient with CHS. Line A illustrates the predicted normal human amino-acid sequence; line B shows the normal human nucleotide sequence; line C shows the nucleotide sequence from a patient with CHS; line D shows the predicted amino-acid sequence from the CHS patient. The CHS nucleotide sequence contains an insertional mutation at nucleotide 117–118 that results in a frame shift and premature termination (asterisk).
It has been suggested that CHS and bg represent homologous disorders, as their clinical features1 and defects in lysosomal transport 4 are identical. Homology of bg and CHS is supported by genetic complementation studies: fusion of fibroblasts from bg mice and CHS patients fails to reverse lysosomal abnormalities, in contrast to fusions with normal cells15. Furthermore, recent genetic linkage studies have shown that CHS maps within a linkage group conserved between human Chromosome 1q43 and the bg region on mouse Chromosome 13 (F. J. Barrat et al., manuscript submitted). Therefore LYST mutations in CHS patients were sought by sequencing LYST lymphoblast and fibroblast cDNAs corresponding to these ESTs from 10 CHS patients. In one patient, a single-base insertional mutation was found at nucleotides 117–118 of the LYST coding domain, resulting in a frame shift and termination after amino acid 62 (Fig. 3c).
Previous studies showing spontaneous aggregation of membrane-bound concanavalin A (capping) suggest that there is a defect in microtubule dynamics in bg cells16,17. In a search of the SWISSPROT database, using BLITZ and BLASTP, a similarity was found between a domain in Lyst and stathmin (oncoprotein 18), a phosphoprotein that may regulate polymerization of micro-tubules10 (27% identity from residues 463 to 536; best expected occurrence by chance, 4.36 × 10−6). The domain in stathmin that matches Lyst is helical and has heptad repeats that participate in coiled-coil interactions with other proteins8–9. The stathmin-like region of Lyst is also predicted to be helical and form coiled coils. However, it is the charged residues, rather than the hydrophobic ones, that are conserved between Lyst and stathmin, suggesting that the sequence similarity is not primarily due to conserved secondary structure. Thus this region of Lyst potentially encodes a coiled-coil protein-interaction domain that may regulate microtubule-mediated lysosome transport. Although Lyst is not predicted to have transmembrane helices, the C-terminal tetrapeptide (CYSP; amino acids 1,542–1,545) is strikingly similar to known prenylation sites18, which could provide attachment to lysosomal/late endosomal membranes through thioester linkage with the cysteine.
Previous studies of bg leukocytes have shown correction of microtubule function (as assessed by Concanavalin A capping) and natural killer activity when treated with inhibitors of protein kinase C (PKC) breakdown19,20, suggesting that bg might be regulated by phosphorylation. Lyst contains 25 sites of potential phosphorylation by PKC, 36 by casein kinase II (CKII) (many of which overlap those of PKC), two by cAMP-dependent protein kinase, and one by tyrosine kinase (Fig. 3b). Almost half of the predicted helices outside the stathmin-like region (14 of 30) have a PKC- or CKII-phosphorylation signal at their amino terminus, and eight of them form consecutive helical pairs. Thus Lyst seems to contain helical bundles with clusters of phosphorylation sites at either end. Stathmin also has an N-terminal phosphorylation site and helix motif, and these Lyst domains may have a similar ‘signal-relaying’ function to stathmin8,9. Furthermore, phosphorylation of these positions could provide a control mechanism by causing a conformational shift in the bundles, thereby affecting interactions with other molecules.
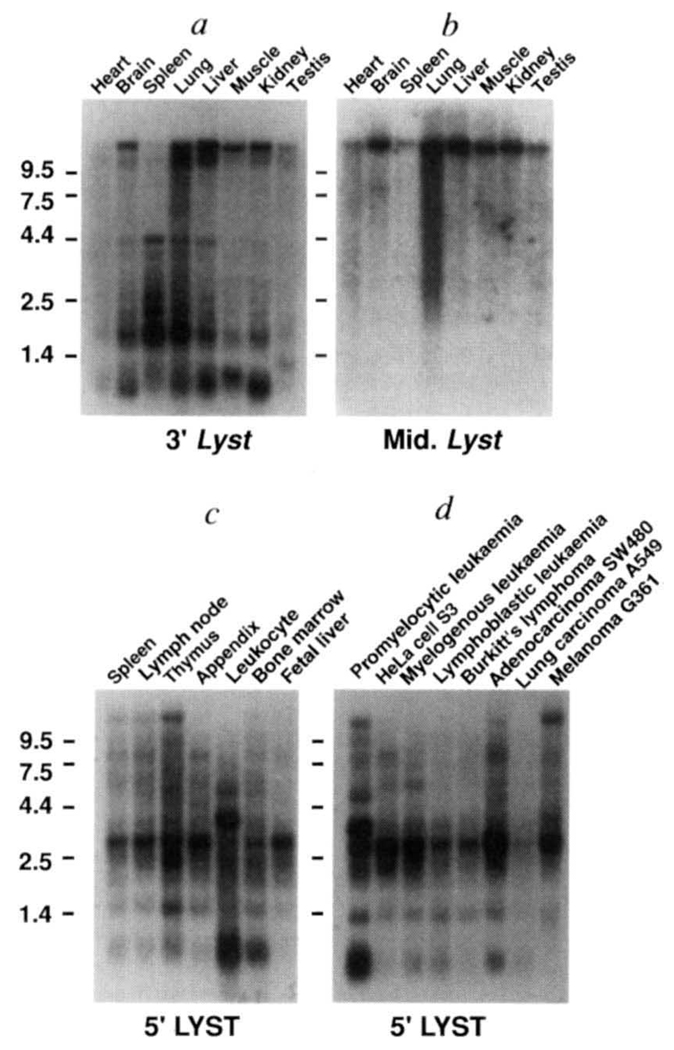
Northern analysis and RT–PCR indicated that Lyst is ubiquitously transcribed, both temporally and spatially, in mouse and human tissues (Fig. 4). Northern blot analysis also revealed complex alternative splicing of Lyst mRNA, with both constitutive and anatomically restricted Lyst mRNA isoforms. The largest Lyst transcript in human and mouse was 12–14 kb, but this transcript was not constitutively expressed. In mRNA from mouse spleen, human peripheral blood leukocytes, promyelocytic leukaemia HL-60, and several leukaemia lines, the 12–14-kb isoform was either undetectable or barely detectable, but smaller Lyst transcripts were abundant (Fig. 4). Given the significance for bg mice and CHS patients of defects in the lysosomal and late-endosomal compartments of granulocytes, NK cells and cytolytic T lymphocytes21–24, it is likely that these Lyst mRNAs of ~3 kb and 4 kb represent the transcripts of primary functional significance. Probes derived form the 5′ or 3′ ends of the Lyst ORF detected multiple bands on northern blots (Fig. 4a, c), whereas probes that corresponded to more central exons (β and γ) identified only the 12–14-kb transcript (Fig. 4b). Assuming that mature, cytoplasmic Lyst transcripts were detected, these results suggest modular assembly of Lyst mRNAs, with obligatory usage of the exons that contain start and termination codons, and adaptability in retention of central exons. The possible coexistence of several Lyst protein isoforms within cells would allow precise control of functions.
FIG. 4.
Northern blot analysis of mouse and human Lyst. a, b, Northern blots of 2 µg poly(A)+ RNA from various mouse tissues (Clontech) hybridized with probes that correspond to a, nucleotides 4,423–4,631, and b, nucleotides 1,430–2,457 (exon β) of mouse Lyst cDNA. c, d, Northern blot of 2µg poly(A)+ RNA from c, various human lymphoid tissues, or d, human cancer cell lines, hybridized with a probe that corresponds to nucleotides 357–800 of human LYST cDNA. Molecular size standards (in kb) are shown to the left. Hybridization of mouse mRNA with probes from mouse Lyst exons α and γ gave identical results to those shown with exon β (b), whereas probes from exons δ, ε, and ϕ gave results identical to those shown in a.
ACKNOWLEDGEMENTS
We thank H. Sweet for investigating the origin of bg11J; K. Achey for technical assistance; and S. Segal for inspiration. M.L is supported by the National Center for Human Genome Research. S.J.B. is supported by the National Institutes of Health and the Vanderbilt Cancer Center. S.F.K. is supported by the American Cancer Society, the Arthritis Foundation, Glaxo-Wellcome Research and Development, and the National Institutes of Health.
References
- 1.Blume RS, Wolff SM. Med. Baltimore. 1972;51:247–280. [PubMed] [Google Scholar]
- 2.Zhao H, et al. Lab. Invest. 1994;71:25–34. [PubMed] [Google Scholar]
- 3.Brandt EJ, Elliott RW, Swank RT. J. Cell Biol. 1975;67:774–788. doi: 10.1083/jcb.67.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhardt JK, Wiebel FA, Hester S, Argon Y. J. exp. Med. 1993;178:1845–1856. doi: 10.1084/jem.178.6.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willingham MC, Spicer SS, Vincent RA. Expl Cell Res. 1981;136:157–168. doi: 10.1016/0014-4827(81)90047-1. [DOI] [PubMed] [Google Scholar]
- 6.Holcombe RF, Jones KL, Stewart RM. Immunodeficiency. 1994;5:131–140. [PubMed] [Google Scholar]
- 7.Swank RT, Brandt E. J. Am. J. Path. 1978;92:755–769. [PMC free article] [PubMed] [Google Scholar]
- 8.Sobel A. Trends biochem. Sci. 1991;16:301–305. doi: 10.1016/0968-0004(91)90123-d. [DOI] [PubMed] [Google Scholar]
- 9.Maucuer A, Camonis JH, Sobel A. Proc. natn. Acad. Sci. U.S.A. 1995;92:3100–3104. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belmont LD, Mitchison T. J. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 11.Barbosa MDFS, et al. Genomics. 1995;30:439–444. doi: 10.1006/geno.1995.1262. [DOI] [PubMed] [Google Scholar]
- 12.Kingsmore SF, et al. J. invest. Med. in the press. [Google Scholar]
- 13.Kingsmore SF, et al. Mamm. Genome. in the press. [Google Scholar]
- 14.Kingsmore SF, et al. Nature Genet. 1994;7:136–142. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- 15.Perou CM, Kaplan J. Somat. Cell molec. Genet. 1993;19:459–468. doi: 10.1007/BF01233251. [DOI] [PubMed] [Google Scholar]
- 16.Oliver JM, Zurier RB, Berlin RD. Nature. 1975;253:471–473. doi: 10.1038/253471a0. [DOI] [PubMed] [Google Scholar]
- 17.Oliver JM, Zurier RB. J. Clin. Invest. 1976;57:1239–1247. doi: 10.1172/JCI108392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke SA. Rev. Biochem. :355–386. [Google Scholar]
- 19.Sato A, et al. J. Leuk. Biol. 1990;48:377–381. doi: 10.1002/jlb.48.5.377. [DOI] [PubMed] [Google Scholar]
- 20.Ito M, et al. Biochem. biophys. Res. Commun. 1989;160:433. doi: 10.1016/0006-291x(89)92451-0. [DOI] [PubMed] [Google Scholar]
- 21.Gallin JI, Bujak JS, Patten E, Sheldon MW. Blood. 1974;43:201–206. [PubMed] [Google Scholar]
- 22.Roder J, Duwe A. Nature. 1979;278:451–453. doi: 10.1038/278451a0. [DOI] [PubMed] [Google Scholar]
- 23.Saxena RK, Saxena QB, Adler WH. Nature. 1982;295:240–241. doi: 10.1038/295240a0. [DOI] [PubMed] [Google Scholar]
- 24.Baetz K, Isaaz S, Griffiths M. J. Immun. 1995;154:6122–6131. [PubMed] [Google Scholar]
- 25.Kusumi K, et al. Mamm. Genome. 1993;4:391–392. doi: 10.1007/BF00360591. [DOI] [PubMed] [Google Scholar]
- 26.Pierce JC, Sauer B, Stemberg N. Proc. natn. Acad. Sci. U.S.A. 1992;89:2056–2060. doi: 10.1073/pnas.89.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gribrat JF, Gamier J, Robson B. J. molec. Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 28.Deleage G, Roux B. Protein Engng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 29.Geourjon C, Deleage G. Protein Engng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 30.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]