Abstract
Bacterial cell-wall-associated fibronectin binding proteins A and B (FnBPA and FnBPB) form bonds with host fibronectin. This binding reaction is often the initial step in prosthetic device infections. Atomic force microscopy was used to evaluate binding interactions between a fibronectin-coated probe and laboratory-derived Staphylococcus aureus that are (i) defective in both FnBPA and FnBPB (fnbA fnbB double mutant, DU5883), (ii) capable of expressing only FnBPA (fnbA fnbB double mutant complemented with pFNBA4), or (iii) capable of expressing only FnBPB (fnbA fnbB double mutant complemented with pFNBB4). These experiments were repeated using Lactococcus lactis constructs expressing fnbA and fnbB genes from S. aureus. A distinct force signature was observed for those bacteria that expressed FnBPA or FnBPB. Analysis of this force signature with the biomechanical wormlike chain model suggests that parallel bonds form between fibronectin and FnBPs on a bacterium. The strength and covalence of bonds were evaluated via nonlinear regression of force profiles. Binding events were more frequent (p < 0.01) for S. aureus expressing FnBPA or FnBPB than for the S. aureus double mutant. The binding force, frequency, and profile were similar between the FnBPA and FnBPB expressing strains of S. aureus. The absence of both FnBPs from the surface of S. aureus removed its ability to form a detectable bond with fibronectin. By contrast, ectopic expression of FnBPA or FnBPB on the surface of L. lactis conferred fibronectin binding characteristics similar to those of S. aureus. These measurements demonstrate that fibronectin-binding adhesins FnBPA and FnBPB are necessary and sufficient for the binding of S. aureus to prosthetic devices that are coated with host fibronectin.
Introduction
Staphylococcus aureus is the leading cause of infections involving prosthetic medical implants.1,2 Although the morbidity, mortality, and cost associated with S. aureus device-associated infection are significant, our understanding of the pathogenesis of this infection is incomplete. A better understanding of the initial steps leading to device-associated S. aureus infection could ultimately lead to novel interventions to prevent this serious, common complication of medical progress.
Fibronectin binding proteins A and B (FnBPA and FnBPB) are integral components in the initial adhesion of S. aureus to host surfaces.3,4 These cell wall molecules, part of a family of microbial surface components recognizing adhesive matrix molecules (MSCRAMMs),5,6 form bonds with host ligands such as fibronectin (Fn), which coat prosthetic devices in vivo. The binding of the intrinsically disordered FnBPA to human Fn has recently been characterized with NMR, and it involves 11 discrete binding regions in FnBP which can form tandem β-zipper interactions with Fn, burying large surface areas.(3) Furthermore, it has also been shown that FnBPs account, at least in part, for the formation of biofilms, a hallmark feature of device-associated infections that limits the efficacy of traditional antibiotic therapies.(4)
Our group has recently measured the magnitude of the Fn−FnBP bond and shown that it could be used as an indicator of the S. aureus-related risk for patients who are candidates for implanted medical devices.(7) This work is part of an emerging body of research that utilizes atomic force microscopy (AFM) and optical tweezers to probe the nature of the bond between FnBP on S. aureus and a Fn-coated substrate,7−10 as well as related research that measures forces between inanimate surfaces and S. aureus(11,12) or S. epidermidis.(13−15) This form of “force spectroscopy” represents a time-tested, validated technique for evaluation of cell−substrate attractive forces. These investigations involve repeated characterization of binding forces between an AFM tip that is typically “baited” with a ligand, and receptors on the surface of a living bacterium.7,9,16−18
Our previous work with AFM(7) utilized isolates of S. aureus from clinical patients. We used PCR and Western blots to confirm the presence of fnbA and fnbB, as well as FnBPA and FnBPB, respectively, in the clinical isolates. However, the resulting AFM force spectra were not compared to laboratory-derived strains of S. aureus expressing well-characterized products of FnBPA and FnBPB. The goal of this study is to evaluate the force characteristics of the bond formed between host Fn and S. aureus FnBPA and FnBPB by using AFM on several elegant laboratory constructs developed by other investigators.
First, we used AFM on laboratory-derived S. aureus strains that either can or cannot express FnBPA and FnBPB.(19) Next, we used AFM on recombinant Lactococcus lactis that express S. aureus fnbA and fnbB, the genes for FnBP A and B.20,21L. lactis, like S. aureus, is a Gram-positive bacterium, but it does not have the MSCRAMM genes of S. aureus. Focusing upon the Fn binding properties of these laboratory strains will increase our understanding of the Fn−FnBP bond, and may provide a force-based model of the Fn−FnBP bond to complement recent structural-based models that have been determined with X-ray crystallography or NMR (e.g., see refs (22 and 23)).
Materials and Methods
Staphylococcus aureus and Lactococcus lactis Strains
Bacterial strains used in the AFM experiments are described in Table 1. The three S. aureus strains are detailed in ref (19). DU5883 is a fnbA fnbB double mutant that has lost the ability to attach to surface-bound Fn.(19) The shuttle plasmids pFNBA4 and pFNBB4 carry the fnbA and fnbB genes, respectively. These plasmids restore the adhesion-defective phenotype of DU5883 and also confer greater adhesion of bacterial cells to Fn-coated coverslips.(19)
Table 1. List of Bacterial Strains Used in Atomic Force Microscopy Experiments.
L. lactis was selected as a control bacterium because it is a noninvasive Gram-positive bacterium that does not naturally adhere to fibronectin or endothelium.(24) Three recombinant L. lactis strains were used in the AFM experiments: pOri23-fnbA, pOri23-fnbB, and pIL 253 (see Table 1). The pOri23 strains were created via insertion of fnbA or fnbB as well as antibiotic-resistance DNA fragments, as detailed in refs (20 and 21). Other studies have demonstrated functional expression of FnBP on the surface of the engineered L. lactis strains.20,21,24
Growth and Sample Preparation of S. aureus and L. lactis Isolates
Growth cultures for AFM analysis were started from cryogenically preserved samples. S. aureus was cultured to exponential phase (OD550 = 0.51−0.54) at 37 °C in tryptic soy broth containing 0.2% dextrose. Tetracycline (3 μg/mL) and/or erythromycin (10 μg/mL) were added to the broth according to ref (19). S. aureus is known to express MSCRAMMs including FnBP when cultured under these conditions.7,25L. lactis strains were grown to exponential phase at 30 °C in M17 medium supplemented with the appropriate antibiotics according to references (20), (21), and (24). Under such conditions, L. lactis pOri23-fnbA and pOri23-fnbB constitutively express FnBPA and FnBPB, respectively.20,21
Approximately 1 mL of cell suspension was harvested via centrifugation (5000g for 3 min). Cells were then washed three times in saline buffer. A small volume of washed cells was transferred onto a glass coverslip and allowed to sit for ∼5 min without drying. Nonadherent cells were washed away with 0.1 M phosphate-buffered saline (PBS; containing 0.85% NaCl at pH 7.2). The glass coverslip, with attached cells, was then transferred to the atomic force microscope.
Force Measurements Using the Atomic Force Microscope
Silicon nitride AFM cantilevers were cleaned in piranha solution, rinsed with Milli-Q water (18.2 M Ω cm), immersed in a 100 μg/mL fibronectin (Sigma-Aldrich) PBS solution for 45 min, and then rinsed four times in PBS.(7) This procedure creates a Fn-coated substrate (i.e., Fn-coated AFM tip or probe). For consistency of measurements, each Fn-coated tip was used across as many cells as possible, intermittently checking for tip degradation by probing a clean glass slide. A total of seven different Fn-coated tips were used in the AFM experiments.
Force measurements were performed in a buffer solution using an atomic force microscope (Veeco/Digital Instruments Bioscope AFM and NanoSCOPE IV controller) as described in ref (7). Briefly, an inverted optical microscope (Axiovert 200M, Zeiss) was used to position a Fn-coated probe (nominal tip radius 20 nm, spring constant 0.2 nN nm−1) over a binary fission pair or a small patch of four cells on the coverslip (Figure 1). The probe was brought into contact with a bacterium and pushed against the cell wall until the cantilever flexed 100 nm. The probe was then retracted away from the bacterium until the probe was completely separated from the cell. This process resulted in an approach force curve as well as a retraction force curve. The vertical travel distance of the z-piezoelectric scanner was 2.7 μm. A single approach−retraction cycle took 1−2 s (i.e., 0.5−1.0 Hz scan rate).
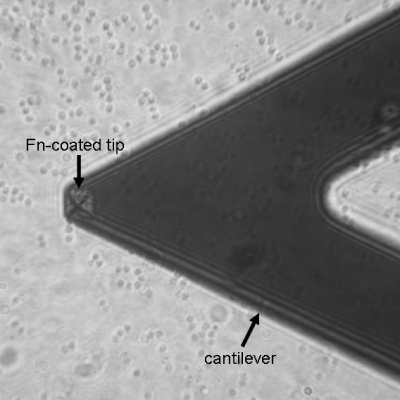
Figure 1.
Optical micrograph that shows the positions of the AFM cantilever and tip (square pyramid near end of cantilever) as well as cocci-shaped S. aureus cells on a coverslip. The bacteria are blurred because the plane of focus is on the AFM tip.
NanoSCOPE software (Veeco) was used to covert the raw data (voltage versus piezo displacement) into force−separation curves according to standard protocol.26,27 The retraction curves were analyzed for the presence of a binding force signature, a procedure often termed force spectroscopy.11,28 Peak detection was performed using the NanoSCOPE software with a minimum attractive force set at 0.1 nN, and a minimum separation distance of 5 nm to avoid potential nonspecific adhesion near the contact point between a cell and the AFM tip. This analysis is described in more detail below.
Results
Force Measurements with S. aureus Strains
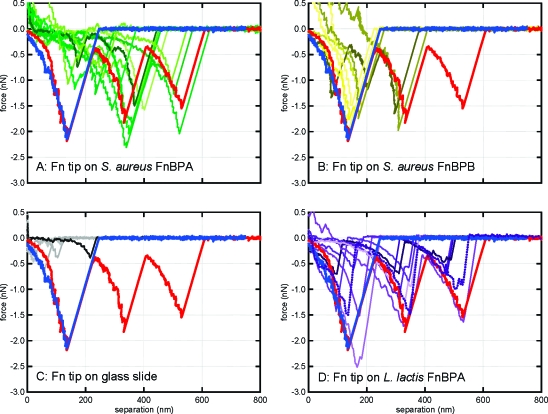
Approximately 3100 force curves were collected on samples prepared from multiple, monoculture growth flasks of the three different S. aureus strains. Figure 2 shows some representative force curves for the two S. aureus strains that express FnBP (i.e., DU5883+pFNBA4 and DU5883+pFNBB4). Those strains that expressed FnBP often exhibited a distinct, nonlinear, sawtooth-shaped force−distance relationship (see Figure 2). Such signatures have been attributed to specific binding events mediated by proteins (e.g., see refs (29−35)). For comparison, Figure 2 also shows force curves for a Fn-coated tip on the glass coverslip, as well as a Fn-coated tip on clinical isolates of S. aureus as reported in ref (7). The curves corresponding to the clinical isolates in Figure 2 (blue and red) are some of the exact same curves (blue and red) that are shown in Figure 1 of ref (7).
Figure 2.
Force spectra collected by atomic force microscopy in buffered saline. Shown are randomly selected retraction curves that exhibited a sawtooth-shaped binding event from a total of 4574 force profiles. A Fn-coated tip was used on (a) S. aureus that expresses only FnBPA (green); (b) S. aureus that expresses only FnBPB (yellow); (c) a glass slide (gray), and (d) recombinant L. lactis that expresses FnBPA or FnBPB (purple). The blue and red curves on each panel are taken directly from previous work with clinical isolates as reported in ref (7). The blue and red spectra are from nasal carriage isolates of S. aureus and invasive isolates of S. aureus, respectively.(7)
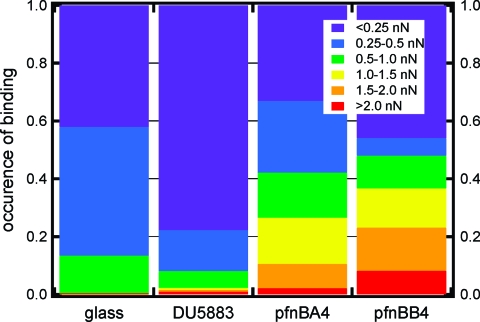
The force curves for all S. aureus strains were subsequently analyzed for the presence of a nonlinear, sawtooth-shaped binding profile. Some retraction curves exhibited more than one sawtooth waveform (e.g., two sawteeth in a single force−distance profile). Such profiles were only counted as a single event in order to determine the occurrence of specific binding between the cell and the Fn-coated probe. A total of 1654 force profiles were analyzed for 21 different cells from the DU5883 double mutant, which does not produce FnBPA or FnBPB. A total of 816 force profiles were analyzed from 6 different cells for the S. aureus that produces FnBPA (pFNBA4); while 543 force profiles were analyzed from 5 different cells for the S. aureus that produces FnBPB (pFNBB4). Approximately 100 curves (∼3% of the collected data) were discarded because of instrumental difficulties, such as excessive thermal drift or a damaged AFM tip. Figure 3 summarizes the results for each of the three S. aureus strains. It is worth noting that this figure includes analyses of all force curves collected using 5 different Fn tips on each of the 32 different S. aureus cells (21 DU5883, 6 pFNBA4, 5 pFNBB4) as well as 9 different bacteria-free spots on glass slides (1561 force curves).
Figure 3.
Occurrence of binding and binding force (in nN) for a Fn-coated substrate on bare glass and the three S. aureus mutants: (i) DU5883 is a double mutant that cannot produce either FnBP, (ii) DU5883 + pFNBA4 expresses only FnBPA, and (iii) DU5883 +pFNBB4 expresses only FnBPB. This figure includes analyses of 4574 force curves collected using 5 different Fn tips, 21 different cells of DU5883 (1654 force curves), 6 different cells of pFNBA4 (816 force curves), 5 different cells of pFNBB4 (543 force curves), and 9 different spots on glass slides (1561 force curves).
Specific binding events of a least 0.5 nN were observed with far greater frequency in the retraction curves associated with the pFNPA4 (frequency = 0.42 ± 0.21) and pFNPB4 (frequency = 0.48 ± 0.14) strains, and with far lower frequency in the retraction curves collected from the DU5883 double mutant (frequency = 0.08 ± 0.13) which does not express either gene (see Figure 3). Comparison of groups using Student’s t test demonstrates significant differences between the pFNBA4 and DU5883 mutants (p = 0.01) and the pFNBB4 and DU5883 mutants (p = 0.001), but not between the pFNPA4 and pFNBB4 strains (p = 0.59).
Force Measurements with Lactococcus lactis Strains
AFM experiments were also performed on recombinant strains of L. lactis (see Table 1). L. lactis is a surrogate Gram-positive bacterium that lacks specific staphylococcal-specific adhesins such as FnBP20,21 and does not bind to Fn-coated surfaces.(24) There were very few specific-binding events observed for pIL253 L. lactis (FnBPA−, FnBPB−), similar to the observations for the S. aureus DU5883 mutant (FnBPA−, FnBPB−). On the other hand, there were parallels in the force spectra of L. lactis and S. aureus that express FnBPA and FnBPB (see Figure 2).
Discussion
Bonds That Initiate S. aureus Infections of Medical Devices
S. aureus prosthetic device infections are a serious and growing medical problem.1,36 This type of infection is a multistep, biofilm-based process that begins with the attachment of bacteria to the surface of an indwelling device. A range of cell wall macromolecules are important in the adhesion of S. aureus to a substrate. These include teichoic acids(37) and polysaccharide intercellular adhesins.38,39 Arguably, the most important molecular adhesin on S. aureus is the so-called MSCRAMM family of proteins.5,6
MSCRAMMs are covalently anchored transmembrane proteins that bind to extracellular matrix molecules such as fibronectin (Fn), fibrinogen, and collagen.6,40 These matrix molecules are common constituents of the human bloodstream. This is significant because implanted materials become coated with host proteins, which in turn serve as attachment sites for S. aureus.19,41 Fn is the predominant ligand-promoting attachment molecule for implants such as cardiac devices, which remain in the body for extended periods of time.41,42 Therefore, S. aureus infections of implants are often attributed to bonds that form between FnBPs on S. aureus and host Fn molecules that coat the surface of indwelling devices.(43)
Fibronectin and Fibronectin-Binding Proteins
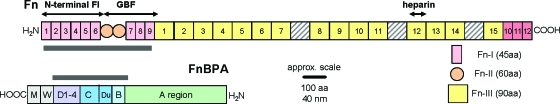
A great deal is known about the structures of Fn and S. aureus FnBP (e.g., see refs (22−24, 33, 44, and 45)). Figure 4 shows the structures of Fn and FnBPA. Note that the N- and C-termini are opposite for these two proteins to better illustrate the binding sites in each molecule. One binding site on Fn is believed to be the ∼29 kD string of FI modules at the N-terminus.5,46−48 Other regions of Fn, such as the GBF or FnIII heparin binding module, have also been shown to bind to FnBP.23,47
Figure 4.
Structures of Fn (top) and FnBPA (bottom) drawn to scale. Highlighted are regions on Fn (N-terminal Fn-I and Fn-II domains) and FnBP (A, B1, B2, Du, C, D1-D4) that are important in binding. Fn is approximately 2000 amino acids or 800 nm in length, and FnBPA is approximately 1000 amino acids or 400 nm in length.
In FnBPA, repeats of 35−40 amino acids in the C-terminal portion (D1-D4 in Figure 4) have been shown to bind to the 29 kD region of Fn.5,19,48 Others have identified regions encompassing Du and B1−B2 within FnBP as being able to adhere to Fn within either the N-terminal FI region(24) or the GBF region of Fn.(23) Some of the most recent experiments(22) provide evidence that FnBP of S. aureus contains 11 subdomains between the A and D regions (see Figure 4) that bind to Fn in a β-zipper type of arrangement.
Recently, we used AFM to probe the bond between Fn and FnBP as expressed on 15 different strains of S. aureus that were isolated from clinical patients.(7) This study was the first to suggest that the “force taxonomy” of an S. aureus isolate may serve as a fundamental and practical indicator of risk for patients who are considering implanted devices. The significance of our original paper was due, at least in part, to the use of isolates of S. aureus from an actual clinical setting, as opposed to type-strains obtained from a culture collection (e.g., the American Type Culture Collection). However, this approach has limitations, namely that binding proteins on the clinical isolates were not as well constrained as those on laboratory-derived strains of S. aureus.
Bonds between Fn and FnBP on S. aureus and L. lactis
Herein, we have used AFM on laboratory-derived strains of S. aureus and L. lactis, whose surface adhesins have already been well characterized in the literature. We hypothesized that if the force signature observed for the clinical isolates of S. aureus in ref (7) was also observed in the mutant strains used herein, then the bond observed in those clinical isolates would originate from attractive force interactions between FnBP on S. aureus and Fn on the substratum. Further, this approach could form the basis of a force model for the Fn−FnBP bond, which would complement recently published structure-based models for the Fn−FnBP bond.3,22,23
Regardless of species or strain, all bacteria exhibited similar repulsive forces in their AFM approach curves (data not shown). The repulsive interaction as S. aureus approaches a Fn-coated substrate is consistent with previously described electrostatic and/or steric forces.9,35,49 Retraction curves, however, often exhibited a strong attraction between the Fn-coated probe and the surface of the bacterium in the >50 nm range. Figure 2 shows attractive, nonlinear force−distance profiles (or waveforms) when a bond forms between a Fn-coated substrate and S. aureus that express either FnBPA or FnBPB on their cell wall. Sawtooth-shaped force spectra such as these have been attributed to the mechanical unraveling of protein molecules that have formed a bond between two surfaces (e.g., between a bacterium and a solid substrate).34,50,51 These force signatures are largely absent when AFM is used on S. aureus or L. lactis mutants that do not produce FnBPs (e.g., see DU5883 in Figure 3); whereas these signatures are present when AFM is used on L. lactis that express FnBP (see Figure 2). Furthermore, retraction curves demonstrating such nonlinear, force−distance profiles differ meaningfully from the profiles observed when a Fn probe is retracted from an inert surface (see Figure 2). Therefore, it can be concluded that this force signature is indicative of the bond that forms between Fn on a substrate and FnBP on the cell wall of a bacterium.
Similarities in force spectral characteristics and frequencies of binding events between S. aureus FnBP A and B expression mutants (DU5883+pFNBA4 and DU5883+pFNBB4), along with statistically significant differences between these strains and the S. aureus FnBP-deficient mutant DU5883 (see the Results section), suggest that Fn−FnBP bonds represent a primary means of adhesion for S. aureus, that the presence of this particular MSCRAMM is both necessary and sufficient to produce AFM-observable binding of the microbe to Fn-coated surfaces, and that FnBPA and FnBPB share similar structures and similar binding affinities for fibronectin, as proposed in refs (52) and (53). Comparison of the S. aureus data with data from the L. lactis strains demonstrates similarity of results (Figure 2) in terms of the characteristics of the observed force spectra. This indicates that adhesion characteristics conferred by these molecules may be transferred to a nonadherent species, and bolsters the conclusion that these molecules play an integral role in mediating S. aureus binding to Fn-coated surfaces.
Protein Binding Mechanics Explain the Shape of the Binding Signature
An important point that has yet to be fully addressed is the origin of the binding signature observed in Figure 2. This distinct feature in the force spectra can be explained with the so-called wormlike chain (WLC) model. The WLC model approximates the biomechanical force−extension relationship of folded polymers (e.g., proteins) that are mechanically extended (or unfolded) into their primary, linear form.(54) The WLC equation is given as
where F (in newtons) is the entropic restoring force generated when a protein is mechanically unfolded to distance x (in meters), kB is the Boltzmann constant (kB = 1.381 × 10−23 J K−1), and T is temperature (in kelvins). The adjustable parameters of the WLC model are the persistence length (p) and the contour length (L). The persistence length is a measure of the bending rigidity or stiffness of a polypeptide chain. For a single protein molecule, the persistence length is typically less than 2.0 nm,30−32,55,56 which is similar to the physical length of 0.4 nm between Cα atoms in the backbone of a protein.(57) The contour length is the extended length of either an entire protein molecule or a structural domain within a protein. A number of groups have studied the force−structure relationship of ligand−receptor pairs by comparing AFM force spectra to WLC models.16,29−31,33−35
Here, the WLC theory can be used to interpret the experimentally measured unbinding mechanics of the Fn−FnBP bond. If Fn on the tip forms a bond with a receptor on the outer cell wall of a bacterium, then an increasingly nonlinear force will be exerted on the tip as the tip is pulled away from the bacterium’s surface (i.e., the separation distance increases from left to right on a force−distance profile). This process causes the mechanical unfolding of the protein(s) that bridges the bacterium to the AFM tip. At some distance from the surface, the force exerted by the tip’s spring constant will exceed the tolerance of the ligand/receptor interaction, and the bond will break or the load-bearing domain will unravel. At this point, the tip will “snap” back to its index position, producing a sawtooth-shaped profile or waveform.
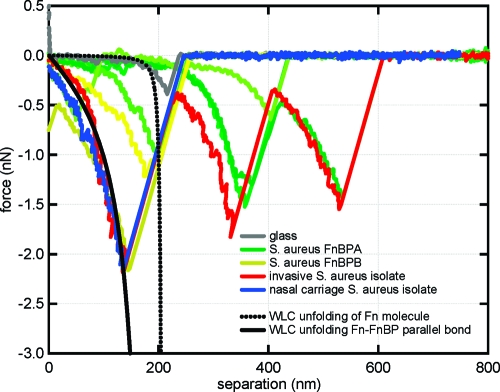
Figure 5 (dotted black curve) shows the force−extension relationship for the N-terminal FnI and FnII domains on Fn as predicted by the WLC model (see Figure 4 for reference). As noted above, this region of Fn has been shown to bind to FnBP from S. aureus. The WLC prediction compares well with the force spectra corresponding to a Fn-coated tip on a glass substrate (see Figure 5, gray curves). There is a nonlinear force−distance (or force−extension) relationship until the Fn breaks free of the glass slide at an extension distance of ∼200 nm, which corresponds to the extended length of the N-terminal FnI and FnII domains on Fn.
Figure 5.
Experimentally measured force spectra for Fn on glass (gray); Fn on S. aureus DU5883+pFNBA4 (FnBPA+ FnBPB−; green); Fn on S. aureus DU5883+pFNBB4 (FnBPA− FnBPB+; yellow); Fn on invasive S. aureus isolate from ref (7) (red); Fn on nasal carriage S. aureus isolate from ref (7) (blue). Theoretical profiles, predicted by the WLC model, are shown as black curves. The dotted black curve corresponds to the unfolding of the N-terminal FnI and FnII domains on Fn (p = 0.4 nm; L = 210 nm). The solid black curve corresponds to the unfolding of parallel bonded molecules of Fn and FnBP (p = 0.004 nm; L = 210 nm).
The WLC model can also be used to explain what happens when FnBP on S. aureus forms a bond with Fn molecules on the AFM tip. As shown in Figure 4, there are multiple binding sites along the lengths of Fn and FnBP. Parallel bonds that form along the length of Fn and FnBP would cause a stiffening (i.e., decreasing persistence length, p, in eq 1) of the protein−protein bond. Figure 5 (solid black curve) shows the hypothetical sawtooth-shaped binding profile for FnBP that forms parallel bonds with the N-terminal region of Fn. For this curve, the persistence length was set at 0.004 nm, which is significantly smaller than the physical dimension of an amino acid (∼0.4 nm). The small value for persistence length will be discussed in more detail below.
As shown in Figure 5, there is a strong correlation between the WLC model (solid black curve) and the force spectra collected for a Fn-coated tip on S. aureus. This indicates that the force signatures observed in the AFM curves are consistent with specific protein−ligand binding events and subsequent unfolding of the protein as the AFM tip is retracted from the cell wall. In this case, bacterial FnBP and the Fn on the tip represent the most likely MSCRAMM−ligand pair. The longer sawteeth observed in the force spectra can be explained by modeling the force−extension relationship of additional structural domains within Fn or FnBP. However, this is beyond the scope of the paper.
Very small values for persistence length have been reported by others as a result of multiple protein chains acting in parallel.55,58−62 For example, 10 protein chains in parallel, each with the same contour length, would exert a force 10 times that of a single chain. Fitting this response by a single chain model, such as the WLC model, would result in one-tenth the true persistence length value. This may be the situation for S. aureus FnBP, as other studies suggest that one FnBP has the capacity to bind to multiple copies (2−9) of Fn.23,48,63 This idea of multiple proteins acting in parallel is further supported by comparing the magnitude of forces shown herein (∼1 nN) to those in ref (64). In this reference, forces of ∼0.1 nN were measured between Fn and putative “fibronectin attachment proteins” on Mycobacterium bovis. The authors attributed their force measurement to single protein interactions, as fibronectin attachment proteins on M. bovis do not, apparently, have the capacity to bind to multiple copies of Fn like FnBPs in S. aureus.
Comparison to Prior Clinical Experiment
In our previous experiments with invasive isolates of S. aureus,(7) we commonly observed multiple sawteeth in a single AFM force curve. However, multiple sawtooth profiles were not commonly observed for any of the strains examined in this experiment. Instead, the S. aureus that expressed either FnBPA (DU5883 pFNBA4) or FnBPB (DU5883 pFNBB4) exhibited force profiles characterized by a single sawtooth, most similar to those from the control (nasal carriage) group in the previous clinical experiment.(7) A number of possible explanations might account for these differences, including (1) that each sawtooth in the multiple-sawtooth pattern was associated with a specific FnBP, (2) that other MSCRAMMs on the surface of S. aureus produced the other sawteeth, or (3) that structural differences resulting from single nucleotide polymorphisms (SNPs) in the fnbA and fnbB genes result in unusual binding characteristics that produce the observed force spectra.
The first of these possibilities seems unlikely given the degree of structural similarity between FnBPA and FnBPB, suggesting that their force spectra would likely overlap rather than appear as discrete sawteeth. However, as may be observed in Figure 2, FnBPA and FnBPB mutants did occasionally form discrete sawteeth that clustered at distances of less than or greater than ∼200 nm. Further investigation of this phenomenon is warranted.
With regard to the second possibility, although the presence of other Fn-binding proteins, including Emp,(65) Eap,(66) and Ebh,(67) might explain both the additional sawteeth (due to difference in size between these molecules and FnBPA and FnBPB) and the incidence of binding events (∼8%) in mutant strains not expressing FnBPA or FnBPB, the observation of similar patterns in species that do not express Emp or Ebh (L. lactis) suggest that this explanation is unlikely.
The third possibility seems most likely; that is, different S. aureus strains may express structurally different versions of the FnBPA and FnBPB proteins on their surfaces. Such differences might result in binding of variable numbers of Fn molecules by a single FnBP. Alternatively, these differences may result in domains along the length of FnBP that function as “structural elbows” that yield (i.e., unfold) rather than allow the breakage of a bond with Fn.(7) In other words, structural domains along the length of FnBP unravel such that the Fn-binding site(s) is not disturbed. This would permit S. aureus to remain attached to a Fn-coated substrate even when an external force (e.g., flow of blood) is placed on the Fn−FnBP bond.
Conclusions
The study suggests potential for differences in the nature of the Fn−FnBP bond due to specific alterations in the primary, secondary, and tertiary structures of FnBPA and FnBPB. Observable differences in the behaviors of in vivo S. aureus infections seem to be mediated by both host- and pathogen-specific factors.68,69 The ability of a bacterium to adhere to a common “scaffolding” molecule such as Fn is quite likely to reflect one such influential pathogen-specific factor. Further studies with truncated forms of Fn and FnBP are underway to increase our understanding of this complex interaction. Research should also continue with other MSCRAMMs (e.g., fibrinogen) and coagulase-negative staphylococci (e.g., see ref (13)) to determine the universality of these results.
In conclusion, these results may represent a key step toward understanding the diversity of clinical manifestations of S. aureus infection, and help to explain why S. aureus infection is lethal in some patients while other patients clear it with only a few days of antibiotics. In addition, classification of S. aureus by the force binding signature, which characterizes its interactions with Fn and thereby its means of adhesion, may represent a powerful clinical tool in the prediction and treatment of S. aureus infections of indwelling medical prostheses. An assay which directly assesses the ability of a specific strain of S. aureus to form bonds with a Fn-coated substrate may help predict whether the strain is capable of hematogenous seeding of a medical implant or formation of a biofilm, whether antibiotic treatment is warranted, and/or whether a patient whose nares are colonized by this strain requires preoperative nasal decolonization.
Acknowledgments
Timothy Foster kindly supplied the S. aureus strains used in these experiments. We thank the anonymous reviewers for their constructive comments. This work was supported by Grant HL086593 from the National Institutes of Health (NHLBI) and Grant 0745808 from the National Science Foundation. Y.-A.Q was supported by fellowships from the Swiss National Foundation (PASMP3-123226), the Foederatio Medicorum Helveticae, and the SICPA foundation. S.K.L. acknowledges the contributions of J. Tak.
Funding Statement
National Institutes of Health, United States
References
- Darouiche R. O. New Engl. J. Med. 2004, 350, 1422–1429. [DOI] [PubMed] [Google Scholar]
- Xiong Y. Q.; Fowler V. G.; Yeaman M. R.; Perdreau-Remington F.; Kreiswirth B. N.; Bayer A. S. J. Infect. Dis. 2009, 199, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham R. J.; Rudino-Pinera E.; Meenan N. A. G.; Schwarz-Linek U.; Turkenburg J. P.; Hook M.; Garman E. F.; Potts J. R. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 12254–12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E.; Pozzi C.; Houston P.; Humphreys H.; Robinson D. A.; Loughman A.; Foster T. J.; O'Gara J. P. J. Bacteriol. 2008, 190, 3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti J. M.; Allen B. L.; McGavin M. J.; Hook M. Annu. Rev. Microbiol. 1994, 48, 585–617. [DOI] [PubMed] [Google Scholar]
- Foster T. J.; Hook M. Trends Microbiol. 1998, 6, 484–488. [DOI] [PubMed] [Google Scholar]
- Yongsunthon R.; Fowler V. G.; Lower B. H.; Vellano F. P.; Alexander E.; Reller L. B.; Corey G. R.; Lower S. K. Langmuir 2007, 23, 2289–2292. [DOI] [PubMed] [Google Scholar]
- Simpson K. H.; Bowden G.; Hook M.; Anvari B. J. Bacteriol. 2003, 185, 2031–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongsunthon R.; Lower S. K. Adv. Appl. Microbiol. 2006, 58, 97–124. [PubMed] [Google Scholar]
- Mitchell G.; Lamontagne C.-A.; Brouillette E.; Grondin G.; Talbot B. G.; Grandbois M.; Malouin1 F. Mol. Microbiol. 2008, 70, 1540–1555. [DOI] [PubMed] [Google Scholar]
- Yongsunthon R.; Lower S. K. J. Electron Spectrosc. Relat. Phenom. 2006, 150, 228–234. [Google Scholar]
- Deupree S. M.; Schoenfisch M. H. Langmuir 2008, 24, 4700–4707. [DOI] [PubMed] [Google Scholar]
- Bustanji Y.; Arciola C. R.; Conti M.; Mandello E.; Montanaro L.; Samori B. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 13292–13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R. J.; Bergstrom T. S.; Liu Y. T.; Soto E. R.; Brown C. A.; McGimpsey W. G.; Camesano T. A. Langmuir 2006, 22, 11311–11321. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Strauss J.; Camesano T. A. Biomaterials 2008, 29, 4374–4382. [DOI] [PubMed] [Google Scholar]
- Lower B. H.; Shi L.; Yongsunthon R.; Droubay T. C.; McCready D. E.; Lower S. K. J. Bacteriol. 2007, 189, 4944–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhami A.; Jericho M. H.; Beveridge T. J. Langmuir 2007, 23, 2755–2760. [DOI] [PubMed] [Google Scholar]
- Lower B. H.; Yongsunthon R.; Shi L.; Wildling L.; Gruber H. J.; Wigginton N. S.; Reardon C. L.; Pinchuk G. E.; Droubay T. C.; Boily J. F.; Lower S. K. Appl. Environ. Microbiol. 2009, 75, 2931–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene C.; McDevitt D.; Francois P.; Vaudaux P. E.; Lew D. P.; Foster T. J. Mol. Microbiol. 1995, 17, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Que Y. A.; Haefliger J. A.; Francioli P.; Moreillon P. Infect. Immun. 2000, 68, 3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y. A.; Francois P.; Haefliger J. A.; Entenza J. M.; Vaudaux P.; Moreillon P. Infect. Immun. 2001, 69, 6296–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Linek U.; Werner J. M.; Pickford A. R.; Gurusiddappa S.; Kim J. H.; Pilka E. S.; Briggs J. A. G.; Gough T. S.; Hook M.; Campbell I. D.; Potts J. R. Nature 2003, 423, 177–181. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U.; Hook M.; Potts J. R. Mol. Microbiol. 2004, 52, 631–641. [DOI] [PubMed] [Google Scholar]
- Massey R. C.; Kantzanou M. N.; Fowler T.; Day N. P. J.; Schofield K.; Wann E. R.; Berendt A. R.; Hook M.; Peacock S. J. Cell. Microbiol. 2001, 3, 839–851. [DOI] [PubMed] [Google Scholar]
- Lowy F. D. New Engl. J. Med. 1998, 339, 520–532. [DOI] [PubMed] [Google Scholar]
- Ducker W. A.; Senden T. J.; Pashley R. M. Nature 1991, 353, 239–241. [Google Scholar]
- Lower S. K.; Tadanier C. J.; Hochella M. F. Jr. Geomicrobiol. J. 2001, 18, 63–76. [Google Scholar]
- Dupres V.; Menozzi F. D.; Locht C.; Clare B. H.; Abbott N. L.; Cuenot S.; Bompard C.; Raze D.; Dufrêne. Y. F. Nat. Methods 2005, 2, 515–520. [DOI] [PubMed] [Google Scholar]
- Rief M.; Gautel M.; Oesterhelt F.; Fernandez J. M.; Gaub H. E. Science 1997, 276, 1109–1112. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M.; Oberhauser A. F.; Fowler S. B.; Marszalek P. E.; Broedel S. E.; Clarke J.; Fernandez J. M. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. J.; Baumeister W.; Engel A. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 13170–13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser A. F.; Marszalek P. E.; Carrion-Vazquez M.; Fernandez J. M. Nat. Struct. Biol. 1999, 6, 1025–1028. [DOI] [PubMed] [Google Scholar]
- Oberdorfer Y.; Fuchs H.; Janshoff A. Langmuir 2000, 16, 9955–9958. [Google Scholar]
- Lower S. K.; Hochella M. F.; Beveridge T. J. Science 2001, 292, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Lower B. H.; Yongsunthon R.; Vellano F. P.; Lower S. K. J. Bacteriol. 2005, 187, 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V. H.; Crosslin D. R.; Friedman J. Y.; Reed S. D.; Cabell C. H.; Griffiths R. I.; Masselink L. E.; Kaye K. S.; Corey G. R.; Reller L. B.; Stryjewski M. E.; Schulman K. A.; Fowler V. G. Jr. Am. J. Med. 2005, 118, 1416. [DOI] [PubMed] [Google Scholar]
- Gross M.; Cramton S. E.; Gotz F.; Peschel A. Infect. Immun. 2001, 69, 3423–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton S. E.; Gerke C.; Schnell N. F.; Nichols W. W.; Gotz F. Infect. Immun. 1999, 67, 5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T.; Sau S.; Gomez M.; Lee J. C.; Lee C. Y. Infect. Immun. 2002, 70, 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. A.; Mosher D. F.; Olbrantz P. J. J. Biol. Chem. 1982, 257, 14788–14794. [PubMed] [Google Scholar]
- Foster T. J.Staphylococcus. In Medical Microbiology; Baron S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, 1996; p Ch. 2. [Google Scholar]
- Arrecubieta C.; Asai T.; Bayern M.; Loughman A.; Fitzgerald J. R.; Shelton C. E.; Baron H. M.; Dang N. C.; Deng M. C.; Naka Y.; Foster T. J.; Lowy F. D. J. Infect. Dis. 2006, 193, 1109–1119. [DOI] [PubMed] [Google Scholar]
- Menzies B. E. Curr. Opin. Infect. Dis. 2003, 16, 225–9. [DOI] [PubMed] [Google Scholar]
- Oberhauser A. F.; Badilla-Fernandez C.; Carrion-Vazquez M.; Fernandez J. M. J. Mol. Biol. 2002, 319, 433–447. [DOI] [PubMed] [Google Scholar]
- Craig D.; Gao M.; Schulten K.; Vogel V. Structure 2004, 12, 21–30. [DOI] [PubMed] [Google Scholar]
- Kuusela Nature 1978, 276, 718–720. [DOI] [PubMed] [Google Scholar]
- Bozzini S.; Visai L.; Pignatti P.; Petersen T. E.; Speziale P. Eur. J. Biochem. 1992, 207, 327–333. [DOI] [PubMed] [Google Scholar]
- Huff S.; Matsuka Y. V.; McGavin M. J.; Ingham K. C. J. Biol. Chem. 1994, 269, 15563–15570. [PubMed] [Google Scholar]
- Taylor E. S.; Lower S. K. Appl. Environ. Microbiol. 2008, 74, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower B. H.; Hochella M. F.; Lower S. K. Am. J. Sci. 2005, 305, 687–710. [Google Scholar]
- Lower S. K. Am. J. Sci. 2005, 305, 752–765. [Google Scholar]
- Jonsson K.; Signas C.; Muller H. P.; Lindberg M. Eur. J. Biochem. 1991, 202, 1041–1048. [DOI] [PubMed] [Google Scholar]
- Signas C.; Raucci G.; Jonsson K.; Lindgren P. E.; Anantharamaiah G. M.; Hook M.; Lindberg M. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory P. J.Statistical Mechanics of Chain Molecules; Hanser Publishers: New York, 1989; p 432. [Google Scholar]
- Kellermayer M. S. Z.; Smith S. B.; Granzier H. L.; Bustamante C. Science 1997, 276, 1112–1116. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L.; Trinick J.; Sleep J. A.; Simmons R. M. Nature 1997, 387, 308–312. [DOI] [PubMed] [Google Scholar]
- Mueller H.; Butt H. J.; Bamberg E. Biophys. J. 1999, 76, 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis J. E.; Akhremitchev B. B.; Walker G. C. Langmuir 1999, 15, 2799–2805. [Google Scholar]
- Higgins M. J.; Crawford S. A.; Mulvaney P.; Wetherbee R. Protist 2002, 153, 25–38. [DOI] [PubMed] [Google Scholar]
- Dugdale T. M.; Dagastine R.; Chiovitti A.; Mulvaney P.; Wetherbee R. Biophys. J. 2005, 89, 4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugdale T. M.; Dagastine R.; Chiovitti A.; Wetherbee R. Biophys. J. 2006, 90, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.; Abdi K.; Jiang Y.; Michaely P.; Bennett V.; Marszalek P. E. Nature 2006, 440, 246–249. [DOI] [PubMed] [Google Scholar]
- Froman G.; Switalski L. M.; Speziale P.; Hook M. J. Biol. Chem. 1987, 262, 6564–6571. [PubMed] [Google Scholar]
- Verbelen C.; Dufrene Y. F. Integr. Biol. 2009, 1, 296–300. [DOI] [PubMed] [Google Scholar]
- Hussain M.; Becker K.; von Eiff C.; Schrenzel J.; Peters G.; Herrmann M. J. Bacteriol. 2001, 183, 6778–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M.; Haggar A.; Peters G.; Chhatwal G. S.; Herrmann M.; Flock J.-I.; Sinha B. Infect. Immun. 2008, 76, 5615–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. R.; Harris L. G.; Richards R. G.; Foster S. J. Infect. Immun. 2002, 70, 6680–6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh H. S.; Hamburger J. B.; Ahn S. H.; McCafferty D. G.; Yang S. R.; Fowler V. G. Jr. Infect. Immun. 2009, 77, 1376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A.; Melles D. C.; Nouwen J.; van Leeuwen W. B.; van Wamel W.; Vos M. C.; Wertheim H. F.; Verbrugh H. A. Infect., Genet. Evol. 2009, 9, 32–47. [DOI] [PubMed] [Google Scholar]