Abstract
MicroRNAs (miRNA), small noncoding RNAs, are potential diagnostic and prognostic markers, as well as therapeutic targets. miRNA profiles of colorectal carcinomas have not been studied extensively in the context of microsatellite instability (MSI) status. We therefore evaluated 55 paired colorectal adenocarcinomas (CRC) and non-neoplastic mucosa samples using a panel of 24 miRNAs selected by literature review and prior studies in our laboratory. Stem-loop reverse transcriptase quantitative (real-time) polymerase chain reaction assays were done on RNA extracted from formalin-fixed, paraffin-embedded tissue of resection specimens. When miRNA expression was compared with clinicopathologic features and MSI status, eleven miRNAs (miR-183, -31, -20, -25, -92, -93, -17, -135a, -203, -133b, and -223) were over-expressed in CRC relative to mucosa, and nine (miR-192, -215, -26b, -143, -145, -191, -196a, -16, and let-7a) were under-expressed in CRC. Relative expression of miR-92, -223, -155, -196a, -31, and -26b were significantly different among MSI subgroups, and miR-31 and miR-223 were overexpressed in CRC of patients with hereditary non-polyposis colorectal cancer syndrome (Lynch syndrome). Our findings indicate that miRNA expression in CRC is associated with MSI subgroups, including low MSI and HNPCC-associated cancers, and that miRNAs may have posttranscriptional gene regulatory roles in these MSI subgroups and possible effects on the clinicopathologic and biomarker characteristics.
Colorectal adenocarcinoma (CRC) is the fourth most common cancer and the second most common cause of cancer deaths in the United States.1 CRC can be divided broadly into two groups: those exhibiting complex chromosomal abnormalities (chromosomal instability) and those exhibiting microsatellite instability (MSI).2 The 10 to 15% of CRC exhibiting high levels of MSI (MSI-H) is a complex and heterogeneous group, the vast majority of cases being sporadic due to acquired somatic methylation of the MLH1 mismatch repair gene. The remaining MSI-H CRCs include those resulting from germline mutation in a mismatch repair gene, usually MSH2 or MLH1, that causes hereditary nonpolyposis colorectal cancer syndrome (HNPCC, Lynch syndrome, Warthin-Lynch syndrome),3,4 and an approximately equal number of MSI-H non-CpG island methylator phenotype-high CRCs.5,6,7 Owing to biological heterogeneity in CRC, biomarkers may be useful in classification and therapeutic evaluation in individual patients. MSI status has been repeatedly validated in this context, both in HNPCC and in sporadic carcinomas.8,9,10,11 MSI-H evaluation contributes to the diagnosis of patients who have HNPCC. The presence of MSI-H also indicates a better prognosis and lack of improved survival after adjuvant chemotherapy, possibly leading to different treatment strategy.8,9,10,11,12 Recent data suggest that patients with MSI-H colon cancer may have better survival in the postoperative adjuvant setting after treatment with a 5-fluorouracil and irinotecan regimen than after 5-fluorouracil alone.13
Recently, evaluation of the predictive and prognostic capability of microRNAs (miRNA) in cancer, including CRC, has been the focus of several studies.14,15,16 miRNA are short 21 to 25 nucleotide noncoding RNA molecules that regulate gene expression at the posttranscriptional level by repressing translation or cleaving RNA transcripts. miRNAs are implicated as crucial factors in diverse regulatory pathways in human tissues, including cell differentiation, proliferation, apoptosis, and stress response.17,18,19,20,21,22 Emerging data implicate miRNA in oncogenesis and suggest that miRNA function both as tumor suppressor genes as well as oncogenes.23,24,25,26 Predicted targets for differentially expressed miRNA are enriched for genes that encode tumor suppressors and oncogenes.27
miRNA expression levels have been associated with prognosis and treatment response in cancer24,28 and have the potential to be exploited as specific therapeutic targets, or even used as therapeutic agents.29,30 Though studies have implicated specific miRNA in colorectal carcinogenesis through their role as gene expression modifiers,15,31,32,33 only recently have the first attempts been made to correlate miRNA levels with MSI status.16 In our study, expression of a selected panel of miRNA was evaluated by stem-loop reverse transcriptase quantitative (real-time) polymerase chain reaction (RT-qPCR) assay of formalin-fixed paraffin-embedded (FFPE) samples obtained from microsatellite-stable (MSS), microsatellite instability low (MSI-L), and microsatellite instability high (MSI-H) CRC and compared with clinicopathologic characteristics. We provide new insights into miRNA expression in relation to MSI status and other clinicopathologic features.
Materials and Methods
Patient Selection
Fifty-five CRC samples with paired non-neoplastic colorectal mucosa were identified from 54 patients, including one patient with synchronous CRC, in pre-existing specimen sets at The University of Texas M. D. Anderson Cancer Center. The specimen sets consisted of MSI-H, MSI-L and MSS colorectal adenocarcinomas evaluated for MSI status for clinical purposes for which FFPE tissue blocks, applicable patient consent, and institutional review board approval were available. Cases were accessioned between December 2001 and August 2007.
The 55 specimens included 22 adenocarcinomas with MSI-H, 8 with MSI-L, and 25 MSS CRC. Six of the patients with MSI-H CRC had the clinical diagnosis of HNPCC. Routine immunohistochemistry for mismatch repair gene products and/or molecular tests for MSI (see below) had been performed at the request of the attending physicians in accordance with University of Texas M. D. Anderson Cancer Center clinical care guidelines. In some cases, MLH1 promoter methylation (n = 8) and BRAF mutation analysis (exon 15 codons 595 to 600, n = 3) were also done. Additionally, nine patients with MSI-H CRC were tested at reference laboratories for germline mutations in MLH1 or MSH2.
The study included 30 females and 24 males with a mean age of 51.3 years. Clinicopathologic characteristics, including age, gender, primary tumor site, stage at the time of resection, MSI status, and HNPCC diagnosis were recorded. The characteristics of the patients and tumors are summarized in Table 1 and presented in detail in Supplemental Table S1 (see http://jmd.amjpathol.org).
Table 1.
Summary of Characteristics of Patients and Colorectal Adenocarcinomas
| Tumor category* | MSI method for CRC† | Mean age and range | Sex | Tumor side‡ | Tumor stage§ |
|---|---|---|---|---|---|
| All tumors, n = 55 | |||||
| PCR 1 | 51.3 | M 25 | Right colon 35 | I 5 | |
| IHC 9 | 25–85 | F 30 | Left colon 15 | II 23 | |
| Both 45 | Indeterminate 5 | III 18 | |||
| IV 1 | |||||
| MSI-H CRC, n = 22 | |||||
| PCR 1 | 57.2 | M 10 | Right colon 16 | I 3 | |
| IHC 3 | 28–85 | F 12 | Left colon 2 | II 12 | |
| Both 18 | Indeterminate 4 | III 5 | |||
| IV 2 | |||||
| MSI-L CRC, n = 8 | |||||
| PCR 0 | 47.5 | M 5 | Right colon 4 | I 0 | |
| IHC 0 | 25–77 | F 3 | Left colon 4 | II 3 | |
| Both 8 | Indeterminate 0 | III 3 | |||
| IV 2 | |||||
| MSS CRC, n = 25 | |||||
| PCR 0 | 47.4 | M 10 | Right colon 15 | I 2 | |
| IHC 6 | 28–66 | F 15 | Left colon 9 | II 8 | |
| Both 19 | Indeterminate 1 | III 10 | |||
| IV 5 |
One patient had synchronous MSI-L and MSI-H tumors.
See Supplemental Table 1 at http://jmd.amjpathol.org for characteristics of each case.
MSI-H CRC = colorectal adenocarcinoma with high levels of MSI, MSI-L = low levels of MSI, and MSS = microsatellite-stable.
PCR = polymerase chain reaction analysis of panel of mono- and dinucleotide repeat sequences only, IHC = immunohistochemistry for mismatch repair gene products only.
The tumors were assigned to “left” and “right” as follows: Right—from cecum to hepatic flexure; Left—from splenic flexure to rectum; Indeterminate—transverse colon and late recurrence at surgical anastomosis.
Stage based upon American Joint Committee on Cancer, Sixth Edition.
Microsatellite Instability Status
MSI status had been evaluated by fluorescence-labeled microsatellite marker PCR followed by capillary electrophoresis fragment size analysis using an ABI 3130 sequencer and Genescan software (Applied Biosystems, Foster City, CA), and/or by immunohistochemistry for MSH2, MLH1, and in some cases MSH6 and PMS2 mismatch repair gene products. Forty-five cases were evaluated for MSI by both PCR and immunohistochemistry and had concordance of MSI status with immunohistochemical status of proteins in 43, 95.6%; nine by immunohistochemistry alone; and one by PCR alone. For microsatellite marker analysis by PCR, five to seven markers (BAT25, BAT26, D2S123, D5S346, D17S250 of the National Cancer Institute panel with or without BAT40 and/or TGFBR2) were analyzed. The carcinomas were classified as MSI-H if two or more markers showed altered allelic size in tumor DNA, MSI-L if one marker showed allelic shift and MSS if none showed allelic shift. For immunohistochemistry, tumors were considered MSI-H if there was loss of nuclear staining for MSH2 or MLH1. MSI-H cases were considered due to HNPCC (n = 6) if Amsterdam criteria were met and/or germline mutational analysis was positive for MSH2 or MLH1 mismatch repair gene (See Supplemental Table S1 at http://jmd.amjpathol.org).
RNA Isolation
Manual microdissection of paired tumor and non-neoplastic FFPE tissue from unstained glass slides was done with a corresponding H&E slide as a guide. Total RNA was extracted and treated with DNAase using the Ambion RecoverAll kit (ABI) according to the manufacturer's instructions. RNA concentrations were evaluated by measuring absorbance (A260) with a NanoDrop Spectrophotometer ND-1000 (Thermo Scientific, Wilmington, Delaware). Extracted total RNA was diluted with nuclease-free water to ensure a constant starting concentration of 5 ng/μL for each reverse transcription (RT) reaction.
Selection of microRNAs for Analysis
microRNA were chosen on the basis of: i) prior pilot studies of CRC performed in our laboratory (miR-135a, -155, -31, -196a, -16, and let-7a); ii) the work of Lanza et al16 on miRNA array expression from fresh frozen tissue of CRC, which showed differential expression of miR-223, -215, -192, -191, -203, -32, -17, -25, -92, -93-1, and -20 between MSI and MSS CRC; and iii) miRNA reported previously to be abnormally expressed in CRC, including miR-143 and -145,32 -96, -133b, -135b, and -183.15 As there is no well accepted miRNA normalizer, we incorporated miR-26b, which was suggested as a potential endogenous control by Applied Biosystems in an application note based on examination of the expression levels of 247 miRNAs using TaqMan MicroRNA assays across 38 normal human tissues and 59 NCI-60 cell lines. miR-106a that was shown by Lanza et al16 to be differentially expressed in MSI CRC was not included because the stem-loop reverse transcriptase was not available in the RT pools available from ABI. The miRNA characteristics, including chromosomal locations, sequences, overlapping transcripts and colocalized miRNA (within 10kb) of studied miRNA, are shown in Supplemental Table S2 (see http://jmd.amjpathol.org).
RT and Real-Time PCR
Multiplex RT reactions were performed using pre-defined pools of stem-loop RT-primers (ABI). Samples were stored at −20°C until used for real-time PCR. Immediately before real-time PCR, cDNA was diluted 10-fold by adding water. Real-time PCR reactions using 2.0 μl of diluted cDNA were performed on a 7900HT analyzer (ABI) with 384-well PCR plates preloaded with specific TaqMan primer/probe mixes custom formatted according to our specific miRNA targets. The first cycle was at 95°C for 10 minutes, then 95°C for 15 seconds and 60°C for 60 seconds × 50 cycles. With the exception of final volumes of 10 μl for RT and 20 μl for qPCR, reactions were performed according to the manufacturer's recommendations (all reagents supplied by ABI).
RT-qPCR Data Analysis
To allow for unbiased comparison across all plates and target miRNAs, a single threshold of 0.05 was set, with a baseline from 3 to 15 Cts. The RT-qPCR results were imported into Microsoft Excel, and the average values of duplicate Ct values were analyzed with the mean Ct values of all miRNA analyzed for a particular specimen as normalizer, using the ΔCt method. For the few instances with successful amplification of only one duplicate, this single Ct value was used to calculate ΔCt. For comparisons of CRC to paired mucosa, relative expression was calculated using the 2−ΔCt method.34
Statistical Analysis
The 2−ΔCt values for tumor and non-neoplastic mucosa samples were compared using a Wilcoxon signed rank test. Within the set of tumor values, the effect of the three categories of MSI on ΔCt values for each miRNA were compared using a one-way analysis of variance with pairwise differences investigated by the Ryan-Einot-Gabriel-Welsch multiple range test. The Kruskal-Wallis test was used to perform a non-parametric analysis of the 2−ΔCt values with pairwise differences investigated by applying the Ryan-Einot-Gabriel-Welsch multiple range test to the rank transformed 2−ΔCt values. P values of <0.05 were considered significant after using a false discovery rate (FDR) to correct for multiple simultaneous comparisons.35 The rough FDR is an estimate of the proportion of errors among the identified differentially expressed miRNA and is defined as (m + 1)/(2m) where m is the number of potential markers being investigated. Figure 1 presents the results of our analysis of the 2−ΔCt values. The P values presented in Figure 1 represent FDR adjusted Kruskal-Wallis P values and the results of the multiple comparisons are indicated through color coding. Within the set of tumor values, regression analyses were performed to determine whether ΔCt values or the rank transformed 2−ΔCt values had significant relationships with MSI status, age, HNPCC, gender, clinical stage, and anatomical site. A forward-stepwise selection method was used with the significance level for a variable to enter/stay in the model of 0.026. This significance level represents the nominal level of 0.05 adjusted for the FDR. The regression analysis is based on the rank transformed 2−ΔCt values. In general, there was good agreement between the regressions of the ΔCt values and the rank transformed 2−ΔCt values (data not shown).
Figure 1.
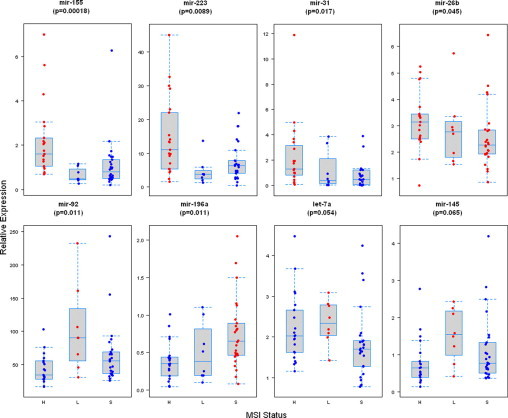
miRNAs differentially expressed among MSI-H, MSI-L, and MSS colorectal adenocarcinomas. Box plots of the log transformed relative levels of differentially expressed miRs in MSI-H, MSI-L, and MSS. Each box shows the variation of relative values of miRs, and the black horizontal bar shows the median value in each box. Increased relative expression of miR-155, −31, −223, and −26b was significantly associated with MSI-H status (top panel). Increased relative expression of miR-92, let-7a, and miR-145 was associated with MSI-L status, whereas increased relative expression of miR-196a was associated with MSS status (bottom panel). The P values presented represent FDR adjusted Kruskal-Wallis P values. Significant differences for a given pairwise comparison among three subgroups are indicated by red versus blue.
Results
Technical Performance of miRNA Assays
The no-template controls run for each target miRNA on each PCR plate showed no amplification. For miR-135b, all results were “undetermined” and for miR-96, results were either “undetermined” or had Ct values ranging from 33.7 to 43.9, suggesting a problem with the primer/probes for these miRNAs on the pre-formatted plate. After exclusion of miR-135b and -96 from the analysis, results for the 22 remaining miRNAs were based on successful duplicate amplification in 2321 of 2420 experiments (95.9%) and successful single amplification in 71 (2.9%). Duplicate Ct values generally had excellent agreement, with a mean difference of <0.5 Cts.
Comparison of Tumors and Non-Neoplastic Mucosa
Twenty of the 22 miRNA selected for evaluation were differentially expressed between CRC and mucosa. Eleven miRNAs were over-expressed in CRC (Table 2). This group was comprised of miR-183, -31, -20, -25, -92, -93, -17, -135a, -203, -133b, and -223. By contrast, nine, consisting of miR-192, -215, -26b, -143, -145, -191, -196a, -16, and let-7a, were underexpressed in CRC (Table 3). Overexpression of miR-183, -31 and -135a, and underexpression of miR-215, -143, -145, and -192 were the most apparent differences. Two miRNAs were not statistically differentially expressed (miR-155 and -32).
Table 2.
miRNAs Significantly Overexpressed in Tumors Relative to Non-Neoplastic Mucosa
| miRNA | Relative expression* in tumors (Mean ± SD) | Relative expression* in NM† (Mean ± SD) | Fold increase in tumors‡ | p-Value§ |
|---|---|---|---|---|
| miR-183 | 0.04 ± 0.03 | 0.007 ± 0.004 | 5.40 | 2.47E-16 |
| miR-31 | 1.4 ± 1.9 | 0.15 ± 0.87 | 8.94 | 6.02E-13 |
| miR-20a | 3.5 ± 2.3 | 2.1 ± 2.5 | 1.66 | 1.52E-10 |
| miR-25 | 1.6 ± 0.70 | 1.1 ± 0.54 | 1.49 | 3.92E-07 |
| miR-92 | 61.8 ± 45.8 | 35.4 ± 11.4 | 1.75 | 1.96E-06 |
| miR-93 | 3.9 ± 1.9 | 2.8 ± 0.74 | 1.41 | 3.33E-05 |
| miR-17 | 0.44 ± 0.32 | 0.28 ± 0.10 | 1.55 | 4.38E-04 |
| miR-135a | 0.010 ± 0.009 | 0.004 ± 0.005 | 2.53 | 1.12E-03 |
| miR-203 | 1.6 ± 1.0 | 1.1 ± 0.49 | 1.44 | 3.14E-03 |
| miR-133b | 0.21 ± 0.59 | 0.17 ± 0.12 | 1.20 | 1.36E-02 |
| miR-223 | 9.8 ± 9.0 | 6.6 ± 3.8 | 1.49 | 3.69E-02 |
FDR is defined as (m + 1)/(2m) where m is the number of potential markers being investigated.
2-ΔCt.
NM = Non-neoplastic mucosa.
Relative expression in T/NM.
Wilcoxon Signed Rank test, adjusted for FDR.
Table 3.
miRNAs Significantly Underexpressed in Tumors Relative to Non-Neoplastic Mucosa
| miRNA | Relative expression* in tumors (Mean ± SD) | Relative expression* in NM† (Mean ± SD) | Fold decrease in tumors‡ | p-Value§ |
|---|---|---|---|---|
| miR-192 | 17.7 ± 9.0 | 39.1 ± 14.9 | 2.21 | 3.51E-16 |
| miR-215 | 0.28 ± 0.40 | 1.3 ± 0.88 | 4.68 | 2.37E-13 |
| miR-26b | 2.8 ± 1.2 | 4.8 ± 2.2 | 1.69 | 8.91E-09 |
| miR-143 | 0.09 ± 0.07 | 0.20 ± 0.11 | 2.29 | 2.47E-08 |
| miR-145 | 1.0 ± 0.81 | 2.2 ± 1.6 | 2.13 | 9.39E-08 |
| miR-191 | 4.3 ± 1.7 | 5.7 ± 1.5 | 1.33 | 3.71E-06 |
| let-7a | 2.0 ± 0.83 | 2.7 ± 1.2 | 1.30 | 4.93E-04 |
| miR-196a | 0.57 ± 0.42 | 0.83 ± 0.40 | 1.46 | 7.72E-04 |
| miR-16 | 14.5 ± 6.5 | 18.4 ± 7.7 | 1.27 | 1.78E-03 |
FDR is defined as (m + 1)/(2m) where m is the number of potential markers being investigated.
2-ΔCt.
NM = Non-neoplastic mucosa.
Relative expression in NM/T.
Wilcoxon Signed Rank test, after correction for FDR.
Comparison of MSI-H, MSI-L and MSS CRC
The MSI categories of CRC differed in expression of miRNA in the tumors (Figure 1). Statistically significant differences in relative expression (2−ΔCt) with Kruskal-Wallis test were observed for miR-155 (P = 0.00017), miR-223 (P = 0.0086), miR-92 (P = 0.011), miR-196a (P = 0.011), miR-31 (P = 0.017), and miR-26b (P = 0.044). Differences in relative expression trended toward statistical significance for let-7a (P = 0.052) and miR-145 (P = 0.064). Increased relative expression of miR-92, let-7a and miR-145 were associated with MSI-L status, while increased relative expression of miR-155, −223, −31, and −26b were significantly associated with MSI-H status. Increased relative expression of miR-196a was associated with microsatellite-stable CRC. Excluding HNPCC-associated CRCs from analysis did not alter the results observed (data not shown).
Associations of Relative miRNA Expression with Clinicopathologic Characteristics
In CRC, statistically significant relationships on multiple regression analysis were found between multiple miRNA and several clinicopathologic characteristics (Table 4). The results presented in the table represent the regressions based on the rank transformed 2−ΔCt values. Significant associations between miRNA expression and MSI status identified in the multivariate analysis were concordant with those identified in the univariate (Kruskall-Wallis) comparison. Additional observations from these comparisons were over-expression of miR-223 and -31 in CRC from HNPCC patients; increasing relative expression of miR-155, -223, and -135a associated with increasing age; and differential expression of miR-31 (increased) and -135a (decreased) associated with right-sided location of tumors.
Table 4.
Statistically Significant Associations between miRNAs and Clinicopathologic Parameters (Multivariate Regression Analysis)
| miRNA | Clinicopathologic parameter | p-Value | Relationship |
|---|---|---|---|
| miR-155 | Age | 1.14E-04 | Higher expression levels are associated with increasing age |
| MSI-L | 7.19E-03 | Lower expression levels are associated with MSI-L status | |
| MSS | 2.03E-03 | Lower expression levels are associated with MSS status | |
| miR-223 | Age | 5.50E-04 | Higher expression levels are associated with increasing age |
| HNPCC | 2.08E-02 | Higher expression levels are associated with HNPCC | |
| miR-31 | Side | 6.61E-04 | Higher expression levels are associated with tumors on the right side |
| HNPCC | 1.63E-02 | Higher expression levels are associated with HNPCC | |
| miR-196a | MSS | 1.24E-03 | Higher expression levels are associated with MSS status |
| miR-26b | MSS | 1.24E-03 | Lower expression levels are associated with stability. |
| let-7a | MSS | 1.12E-02 | Lower expression levels are associated with MSS status |
| miR-92 | MSI-L | 1.35E-02 | Higher expression levels are associated with MSI-L status |
| MSS | 2.50E-02 | Lower expression levels are associated with MSS status | |
| miR-192 | Side | 1.48E-02 | Lower expression levels are associated with tumors on the right side |
| miR-135a | Age | 1.42E-02 | Lower expression levels are associated with increasing age |
Discussion
Our study provides new information regarding miRNA expression in relation to MSI subgroups including MSI-L CRC and associations with HNPCC, patient age, and anatomical site of the cancer. Further, using RT-qPCR and RNA extracted from FFPE tissue of the type that is widely available in pathology specimen repositories, we corroborated findings from previous publications on miRNA expression in CRC using different detection methods and fresh/frozen tissues.
We found overexpression of miR-20a -203, -183, -31, and -135b and underexpression of miR-143 and -145 in CRC compared with mucosa, in agreement with previously published data in CRC.15,28,32 Several miRNA investigated for association with MSI status of CRC were also shown to be differentially expressed in our comparisons between CRC and mucosa. Additionally, the miRNA previously described in multiple literature sources as differentially expressed in CRC tended to show high levels of differential expression (ratio of miRNA expression in tumor compared with mucosa) (Tables 2and 3). These large relative differences may imply a greater biological significance in tumor development, whether causative or consequential.
Of the eight miRNAs differentially expressed or trending toward differential expression in the three MSI groups we studied, three (miR-223, -155, and -92) had been described by Lanza et al16 to be differentially expressed between MSI-H and MSS CRC in a study involving 39 frozen specimens (23 MSS and 16 MSI-H) evaluated with miRNA-chip technology. Nine other differentially expressed miRNAs (miR-17, -215, -191, -192, -203, -32, -25, -93, and -20) from their work that were included in our panel were not significantly differentially expressed by our methods. However, evaluation of our results with the Ryan-Einot-Gabriel-Welsch multiple range test revealed trends toward under- and overexpression in the same direction as in the work of Lanza et al,16 with the exception of miR-32 and -203 that trended in the opposite direction (data not shown).
There are many possible explanations for these differences between the two studies. First, we included the third category of tumors, the MSI-L group, in our analysis. In addition, the sample sizes differed, producing differences between the statistical power of the two studies. Thirdly, there are differences in the patient populations, such as geographic region, tissue procurement, and relative amount of tumor and “contaminating” non-neoplastic tissue, and in laboratory methodologies, such as hybridization efficiency, nucleic acid amplification versus signal amplification, normalization technique, etc Another important consideration is that, although differential expression between MSI-H and MSS groups was significantly different in the study of Lanza et al,16 the ratios of expression levels between the MSI-H and MSS groups were close to unity, indicating small differences between the groups. Expression in malignant epithelium may be disguised by the presence of a miRNA signature derived from “contaminating” non-neoplastic stroma or inflammatory cells, which are often plentiful in MSI-H CRC, but can be seen in varying degrees in non-neoplastic mucosa as well. Importantly, the expression ratios for MSI-H to MSS for miR-203 and −32, for which our results were discordant with those of the previously reported study, are among the closest to unity that the authors reported. In our analysis, some miRNAs had significant differential expression associated with HNPCC syndrome, age, and site in the large bowel where the tumor was located (Table 4). All of these factors may confound the concordance of results because associations between all of these variables and MSI status are well known.
Most clinical and pathological associations with MSI tumors have been based on MSI-H CRC compared with MSS and MSI-L CRC grouped together, or comparisons of MSI-H CRC to MSS CRC without inclusion of MSI-L tumors. However, recent studies have shown morphological and molecular differences among non-MSI-H CRC.20,36 In our study, we found in both the univariate Kruskall-Wallis and multivariate regression analyses that MSI-L tumors have significant differences in miRNA expression from both MSI-H and MSS CRC. Our findings substantiate the previous findings and provide additional evidence suggesting that MSI-L tumors have specific clinical and molecular characteristics.37 The grouping of MSI-L tumors with MSS for research purposes may need to be re-evaluated as more is learned about the relationships among gene regulatory mechanisms and the categories of MSI CRC.
Current challenges in the field of miRNA include understanding the biological roles, identifying mRNA targets, and demonstrating the effects of specific miRNA in specific tumor types. These areas have been the focus of numerous studies that have demonstrated potential oncogenic or tumor suppressor roles for many of the miRNAs evaluated in CRC in our study. Targets for the miRNA we studied include: for miR-143, ERK532,38; for miR-145, MYCN, FOS, YES, FLI, cyclin D2, cyclin L1, MAP3K3, MAPK4K415 and HOXA9;39 for miR-215, SIP140; for miR-31, AXIN1 (Wnt pathway), FOXC2 and FOXP3;15 and for miR-183, FOXF2, FOXK2, FOXO1A, FOXO3A, and FOXQ1.15 These relationships highlight the complexity of the molecular pathogenesis of the subtypes of CRC and potential influences on biological behavior.41
An additional issue regarding the understanding of miRNA expression is the upstream regulatory control of miRNA expression. In our study, miR-17, -20, and -92 were all significantly overexpressed in CRC at similar levels (ratio of expression in tumor to mucosa of 1.55, 1.66, and 1.75, respectively). These miRNA are located on the well-known miR-17–92 polycistron on chromosome 13 (see Supplemental Table S2 at http://jmd.amjpathol.org), and their transcription is induced by c-myc.42,43 Likewise, miR-25 and −93 are co-localized within 10kb on chromosome 7, and we observed similar levels of over-expression of these miRNAs in CRC (ratio of expression in tumor to mucosa of 1.49 and 1.41, respectively). Similar observations were made for the underexpressed miR-143 and −145 that are colocalized within 10kb on chromosome 5 (ratio of expression of 2.29 and 2.13, respectively). The finding of evidence of co-regulation suggests important roles in CRC for these types of alterations.
In conclusion, the findings of our study reinforce those of previous publications on miRNA expression in CRC and provide proof of concept for quantitative evaluation of miRNA by RT-qPCR following nucleic acid extraction from FFPE specimens. If miRNA are to be used as biomarkers, therapeutic targets or therapeutic agents, as is predicted,44 consistency of results and methodological considerations will become increasingly important because of the ubiquity of FFPE specimens in the clinical practice of pathology and the familiarity with RT-qPCR platforms in clinical laboratories. Our work also highlights a small panel of miRNA that may be of future use as prognostic or therapy-response indicators in CRC, identifies differences in miRNA expression in relation to MSI-H, and MSI-L status and HNPCC-associated CRC. The significance of these findings and potential roles as molecular classifiers and/or clinical biomarkers will require vigorous validation in larger cohort studies.
Acknowledgements
We thank James M. Gilbert and Gloria Rice for assistance with preparation of the manuscript and illustrations and Kerry Sieger for assistance with acquisition of case material.
Footnotes
Supported by a Fellow Research Fund award from the Division of Pathology and Laboratory Medicine at The University of Texas M.D. Anderson Cancer Center (to J.S.L.E.) and by Cancer Center Support grant CA16672 from the National Cancer Institute, National Institutes of Health.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Web Extra Material
Patient and Tumor Characteristics
miRNA Information
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer Statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–69. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V, Abbas A, Fausto N: Robbins and Cotran Pathologic Basis of Disease, 7th Edition, 2005 306–307
- 4.Desai TK, Barkel D. Syndromic colon cancer: Lynch syndrome and familial adenomatous polyposis. Gastroenterol Clin North Am. 2008;37:47–72. doi: 10.1016/j.gtc.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samowitz WS. The CpG island methylator phenotype in colorectal cancer. J Mol Diagn. 2007;9:281–283. doi: 10.2353/jmoldx.2007.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 9.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 10.Lanza G, Gafa R, Santini A, Maestri I, Guerzoni L, Cavazzini L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol. 2006;24:2359–2367. doi: 10.1200/JCO.2005.03.2433. [DOI] [PubMed] [Google Scholar]
- 11.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 12.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, Warren RS, Redston M. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 15.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM, Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 18.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 19.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 20.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 21.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 26.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattes J, Yang M, Foster PS. Regulation of microRNA by antagomirs: a new class of pharmacological antagonists for the specific regulation of gene function? Am J Respir Cell Mol Biol. 2007;36:8–12. doi: 10.1165/rcmb.2006-0227TR. [DOI] [PubMed] [Google Scholar]
- 30.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 31.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 32.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 33.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 36.Ogino S, Kawasaki T, Kirkner GJ, Suemoto Y, Meyerhardt JA, Fuchs CS. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56:1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohonen-Corish MR, Daniel JJ, Chan C, Lin BP, Kwun SY, Dent OF, Dhillon VS, Trent RJ, Chapuis PH, Bokey EL. Low microsatellite instability is associated with poor prognosis in stage C colon cancer. J Clin Oncol. 2005;23:2318–2324. doi: 10.1200/JCO.2005.00.109. [DOI] [PubMed] [Google Scholar]
- 38.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 39.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JP, Yuen ST, Leung SY, Hamilton SR. Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res. 2008;14:2560–2569. doi: 10.1158/1078-0432.CCR-07-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coller HA, Forman JJ, Legesse-Miller A. “Myc'ed messages”: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 44.Mack GS. MicroRNA gets down to business. Nature Biotechnol. 2007;25:631–638. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient and Tumor Characteristics
miRNA Information


