Abstract
Chromosomal rearrangements involving the mixed lineage leukemia (MLL) gene at 11q23 are frequent in adult and childhood acute leukemia and have been associated with an unfavorable prognosis. Recent evidence suggests that MLL gene partners may influence prognosis. Five translocations account for ∼80% of MLL rearrangements: t(4;11)(q21;q23), AFF1/MLL; t(6;11)(q27;q23), MLLT4/MLL; t(9;11)(p22;q23), MLLT3/MLL; t(11;19)(q23;p13.1), MLL/ELL; and t(11;19)(q23;p13.3), MLL/MLLT1. We have designed dual-color, double-fusion fluorescence in situ hybridization (D-FISH) probe sets to identify these translocations. A blinded study was performed for each probe set using 25 normal bone marrow samples, 25 t(4;11), 20 t(6;11), 20 t(9;11), 18 t(11;19p13.1), and 20 t(11;19p13.3) leukemia specimens as defined by chromosome analysis. The findings demonstrated abnormal D-FISH results for 24 of 25 AFF1/MLL, 19 of 20 MLLT4/MLL, all 20 MLLT3/MLL, all 18 MLL/ELL, and all 20 MLL/MLLT1 samples, confirming the efficacy of these D-FISH assays in detecting these common MLL/partner translocations. Our D-FISH assays were more accurate than chromosome analysis at distinguishing disruption of 19p13.1/ELL from that of 19p13.3/MLLT1. We also demonstrated a statistically significant increase in complex/unbalanced MLL/partner translocations occurring in pediatric patients versus adult patients (P = 0.02). A normal cutoff of 0.6% was established, suggesting an application for these assays in minimal residual disease detection and disease monitoring.
Reciprocal chromosomal rearrangements involving the mixed lineage leukemia (MLL) gene have been associated with the development of a heterogeneous group of leukemias including acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), mixed phenotype leukemia, and acute leukemias associated with topoisomerase II chemotherapy.1,2,3,4,5,6,7,8,9,10 Overall, rearrangements of MLL occur in ∼6% of acute leukemia cases in adults and children. However, MLL rearrangements are significantly more frequent in specific patient cohorts, particularly ALL occurring in children < 12 months of age, where MLL disruption accounts for 80% of cases.7,10 In AML, rearrangements of MLL are typically associated with a myelomonoblastic or monoblastic phenotype (M4 and M5 subtypes) and are more frequent in children, representing 9 to 12% of pediatric cases and ∼65% of infant cases.2,11 Similarly, MLL rearrangement is frequent in mixed phenotype acute leukemia in children.8,9 Therapy-related AML, occurring in patients who have been previously treated with chemotherapeutic drugs that inhibit DNA topoisomerase II, also show a high frequency of rearrangements involving MLL.4,5,6
The MLL gene, found on the long arm of chromosome 11 at band q23, contains 37 exons and spans >100 kb.11,12,13 The majority of recurrent MLL translocation breakpoints occur within an 8.3-kb breakpoint cluster region located between exons 8 and 14.11,12,13,14,15,16 A remarkable feature of MLL gene rearrangements in leukemias is the large number and assortment of MLL fusion partners. To date, >80 recurrent chromosomal translocations disrupting MLL have been identified, and >50 MLL partner genes have been defined.1,2,17,18 Despite the diversity of MLL pairing partners, five translocations account for ∼80% of all MLL-associated leukemia.1,2,16,17,18 The five most frequent MLL rearrangements include t(4;11)(q21;q23), AFF1(AF4)/MLL; t(6;11)(q27;q23), MLLT4(AF6)/MLL; t(9;11)(p22;q23), MLLT3(AF9)/MLL; t(11;19)(q23;p13.1), MLL/ELL; and t(11;19)(q23;p13.3), MLL/MLLT1(ENL).
The presence of MLL rearrangements in patients with acute leukemia generally portends a less favorable prognosis and response to chemotherapy.7,19,20,21,22,23 However, recent studies have suggested that the specific MLL rearrangement partner may also influence response to therapy and overall prognosis depending on the clinical context.21,24,25,26,27,28 This association seems to be especially pronounced in AML, leading the most recent 2008 World Health Organization consensus to separate the t(9;11)(p22;q23) as a distinct clinical entity with a more favorable prognosis. The World Health Organization also recommended that the diagnosis of AML include the presence of any MLL rearrangement and the pairing partner gene (if known).29
Typically, conventional cytogenetics has been used to detect rearrangements involving the MLL gene. However, conventional cytogenetics may fail to detect nearly one-third of MLL rearrangements; therefore, fluorescence in situ hybridization (FISH) has emerged as the modality of choice for detection of such rearrangements. Commercially available FISH probes are only available for the MLL gene region and are able to detect a simple disruption of the gene but do not identify specific gene fusion partners. Additionally, this strategy is unable to detect with confidence the presence of affected cells below a level of 5%, similar to a conventional chromosome analysis, thus limiting the utility in its application for minimal residual disease (MRD) detection. Herein, we describe our development and validation of dual color, double-fusion FISH (D-FISH) probe sets for each of the five most frequent MLL translocation partners.
Materials and Methods
Study Design
Following Institutional Review Board approval, we conducted a retrospective review of the Mayo Clinic Cytogenetics database to identify blood and bone marrow samples in which a t(4;11)(q21;q23), t(6;11)(q27;q23), t(9;11)(p22;q23), t(11;19)(q23;p13.1), or t(11;19)(q23;p13.3) was identified by conventional cytogenetics.30 We selected 25 samples with a t(4;11), 20 samples with a t(6;11), 20 samples with a t(9;11), 18 samples with a t(11;19)(q23;p13.1), and 20 samples with a t(11;19)(q23;p13.3) for inclusion in the study. Although the majority of selected specimens represented the diagnostic sample, we also included a few posttherapy and relapse specimens to challenge the probe sets. In addition, as a large cytogenetics reference laboratory, we do not always receive the diagnostic pathology reports with the bone marrow specimens submitted for chromosome studies. Thus, the specific subtype of acute leukemia associated with our chromosome results is not always known. However, we included both pediatric and adult patients with B cell ALL, T cell ALL, de novo AML, and therapy-related AML in these evaluations.
For each probe set, 25 normal bone marrow samples (obtained from patients undergoing hip replacement surgery) were included to establish normal cutoff ranges. The patient specimens for each MLL translocation partner and corresponding 25 negative controls were blinded for each study, and 250 representative interphase nuclei were independently scored by two technologists (total of 500 interphase nuclei scored per sample). A total of 228 specimens were evaluated and included 50 total samples for the AFF1(AF4)/MLL probe set, 45 total samples for the MLLT4(AF6)/MLL probe set, 45 total samples for the MLLT3(AF9)/MLL probe set, 43 total samples for the MLL/ELL probe set, and 45 total samples for the MLL/MLLT1(ENL) probe set.
Probe Design
Direct-labeled FISH probes were designed from bacterial artificial chromosomes (BACs) and validated according to standard methods.31 Bacterial clones were chosen from the University of California Santa Cruz website (http://genome.ucsc.edu; May 2004 Assembly (hg17)) based on their coverage of the gene region of interest and were then ordered from Invitrogen (Carlsbad, CA). The BACs were cultured, and plasmid DNA was extracted using a Qiagen Plasmid Maxi Kit (Qiagen, Valencia, CA). The isolated DNA was fluorescently labeled via nick translation (Abbott Molecular, Abbott Park, IL), and clone specificity was verified on metaphases from normal XY blood specimens. PCR was also performed to ensure that the DNA clone corresponded to the region of interest. Once individually validated, all of the clones were mixed to create a complete probe mixture. For the MLL gene probe, the five clones used were RP11-832A4, RP11-215H18, RP11-770J1, RP11-861M13, and CTC-89C10 (Figure 1A). For the AFF1(AF4) gene probe, the four clones used were RP11-397E7, RP11-168E22, RP11-476C8, and RP11-529H2 (Figure 1B). For the MLLT4(AF6) gene probe, the five clones used were RP11-931J21, RP3-431P23, RP11-471L1, RP3-470B24, and RP11-164L23 (Figure 1C). For the MLLT3(AF9) gene probe, the four clones used were RP11-15P13, RP11-336O12, RP11-73E6, and RP11-4E23 (Figure 1D). For the ELL gene probe, the four clones used were RP11-282P1, CTD-3137H5, CTD-2516F17, and CTD-2643B8 (Figure 1E). For the MLLT1(ENL) gene probe, the five clones used were CTC-232P5, CTB-66B24, CTC-503J8, RP11-114A7, and RP11-30F17 (Figure 1F). The AFF1, MLLT4, MLLT3, ELL, and MLLT1 BACs were labeled with Spectrum Orange-dUTP (Abbott Molecular), herein referred to as Red. The MLL BACs were labeled with Spectrum Green-dUTP (Abbott Molecular). A commercially available break-apart MLL FISH probe (Abbott Molecular) was used according to the manufacturer's instructions.
Figure 1.
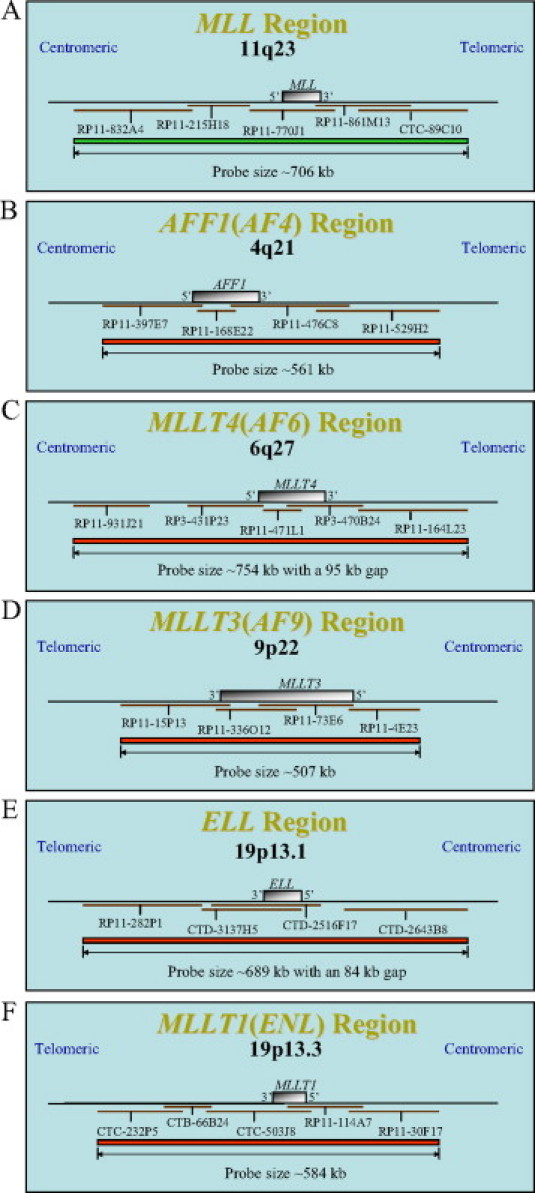
Probe design schematics for MLL, AFF1, MLLT4, MLLT3, ELL, and MLLT1 as used in the dual-color D-FISH probe assays. A: Probe schematic for the five BACs labeled in Spectrum Green, RP11-832A4, RP11-215H18, RP11-770J1, RP11-861M13, and CTC-89C10, spanning the 11q23 MLL region. B: Probe schematic for the four BACs labeled in Spectrum Orange, RP11-397E7, RP11-168E22, RP11-476C8, and RP11-529H2, spanning the 4q21 AFF1(AF4) region. C: Probe schematic for the five BACs labeled in Spectrum Orange, RP11-931J21, RP3-431P23, RP11-471L1, RP3-470B24, and RP11-164L23, spanning the 6q27 MLLT4(AF6) region. D: Probe schematic for the four BACs labeled in Spectrum Orange, RP11-15P13, RP11-336O12, RP11-73E6, and RP11-4E23, spanning the 9p22 MLLT3(AF9) region. E: Probe schematic for the four BACs labeled in Spectrum Orange, RP11-282P1, CTD-3137H5, CTD-2516F17, and CTD-2643B8, spanning the 19p13.1 ELL region. F: Probe schematic for the five BACs labeled in Spectrum Orange, CTC-232P5, CTB-66B24, CTC-503J8, RP11-114A7, and RP11-30F17, spanning the 19p13.3 MLLT1(ENL) region. Note: figures are not drawn to scale.
FISH Protocol
Archived blood and bone marrow pellets were stored at −70°C in methanol:glacial acetic acid (2:1) fixative. After a change of fixative, the fixed-cell pellet suspensions were manually dropped onto microscope slides and checked by phase contrast microscopy to verify appropriate cellularity. Samples were then subjected to standard FISH pretreatment, hybridization, and fluorescence microscopy.32,33
FISH Signal Patterns
The same FISH probe strategy is used for the detection of the five specific MLL/partner fusions, which include t(4;11)(q21;q23), AFF1(AF4)/MLL; t(6;11)(q27;q23), MLLT4(AF6)/MLL; t(9;11)(p22;q23), MLLT3(AF9)/MLL; t(11;19)(q23;p13.1), MLL/ELL; and t(11;19)(q23;p13.3), MLL/MLLT1(ENL) (Figure 2A). Representative ideograms of chromosomes 4 and 11 illustrate the D-FISH signal pattern observed on normal metaphase chromosomes (Figure 2B). A signal pattern of two red (2R) and two green (2G) corresponds to two red partner probe signals on the normal chromosome 4 homologues and two green MLL signals on the normal chromosome 11 homologues. Representative ideograms of abnormal metaphase chromosomes with a 4;11 translocation (Figure 2B) exhibit a 1R1G2F (F, fusion) signal pattern corresponding to one R signal from AFF1 on the normal chromosome 4, one G signal for MLL on the normal chromosome 11, and two yellow F signals from the AFF1/MLL fusions on the derivative chromosomes 4 and 11.
Figure 2.
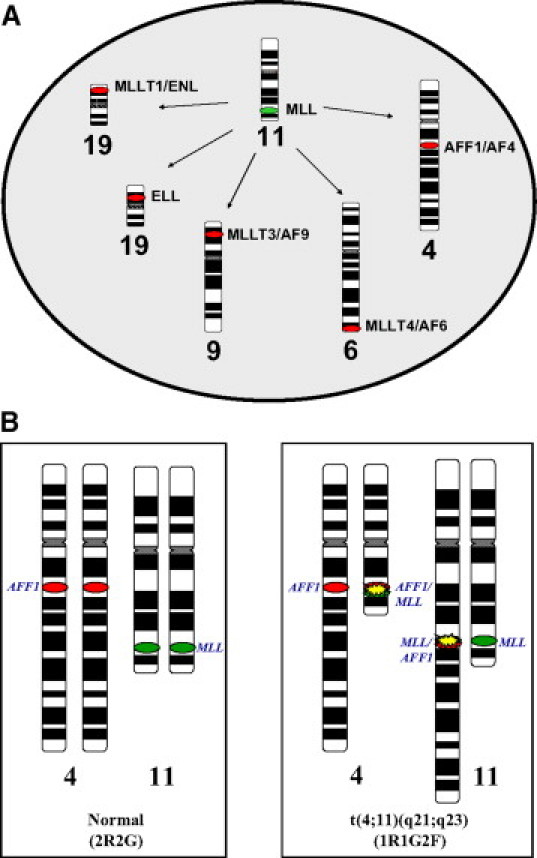
General strategy for the MLL pairing partner D-FISH assay. A: Each of the five assays uses the MLL probe labeled in green and a second probe labeled in red for one of the translocation partner genes (AFF1, MLLT4, MLLT3, ELL, or MLLT1). B: Ideograms of chromosome 4 and 11 with red (R) and green (G) ovals representing the 4q21 AFF1 and 11q23 MLL probe regions, respectively. Normal chromosome four homologues display a 2R signal pattern and normal chromosome 11 homologues display a 2G signal pattern. Reciprocal translocation of chromosomes 4 and 11 resulting in disruption of AFF1 and MLL causes splitting of one of the R and G signals, respectively. Subsequent juxtaposition of the split AFF1 and MLL signals results in two sets of paired R and G signals, observed as two yellow F signals. The normal chromosomes 4 and 11 remain as separate R and G signals. An overall 1R1G2F FISH signal pattern is observed.
Statistics
Two statistical evaluations were performed for this data set. The first included an evaluation of the percentage of complex D-FISH signal patterns in pediatric patients (8 of 24) versus adult patients (6 of 76). To answer this question, we used Fisher's exact test for equal proportions with small expected counts, yielding a two-sided P value of 0.02. The second included an evaluation of the percentage of complex karyotypes in pediatric patients (15 of 24) versus adult patients (29 of 76). To address this question, we used a χ2 test for equal proportions, which produced a two-sided P value of 0.06.
Results
FISH Probe Validation
The study design included the analysis of 500 interphase nuclei from 103 patient specimens and 125 normal control specimens for a total of 114,000 interphase nuclei scored for this study. On completion, the data for each probe set were unblinded. Results obtained from the 25 normal bone marrow specimens were used to establish a normal cutoff for each signal pattern observed with each specific probe set. All normal controls were found to be normal for each of the five probe sets, and a normal cutoff of 0.6% was established for the expected 1R1G2F signal pattern using a one-sided binomial distribution with a 95% confidence interval. This same statistical strategy was used to generate cutoffs for atypical and unexpected signal patterns. Normal cutoffs for both typical (2R2G1F, 1R2G1F, and 2R1G1F) and atypical (1R1G3F, 1R3G1F, and 2R1G2F) D-FISH patterns found in this experiment were determined to be 0.6%. The cutoff range for 1R1G1F was 5.8 to 10%, depending on which MLL/partner probe set was being analyzed. The cutoffs for the signal patterns without fusion signals, 3R1G and 2R3G, were 0.6 and 1.0 to 1.6%, respectively.
Representative FISH signal patterns observed in interphase nuclei from the five D-FISH probe sets are illustrated in Figure 3. The MLL probe on chromosome 11q23 is labeled in green (G), and the various translocation partners are labeled in red (R). Normal interphase nuclei have a 2R2G signal pattern (Figure 3A). A coincidental overlap of R and G signals in normal interphase nuclei can yield a signal pattern of 1R1G1F (Figure 3B). For the majority of patients with a balanced reciprocal MLL translocation, abnormal interphase nuclei will display a D-FISH signal pattern of 1R1G2F (Figure 3C). For some patients, variable signal patterns are observed as a result of complex translocations, 2R2G1F (Figure 3D), or loss of DNA at the breakpoint junctions, 1R2G1F and 2R1G1F (Figures 3, E and F, respectively). Alternate signal patterns are also observed due to gain of an additional derivative chromosome, 1R1G3F (Figure 3G), and duplication/trisomy of the chromosomes covered by the probe set, 1R3G1F and 2R1G2F (Figure 3H and 3I). Abnormal signal patterns lacking a fusion are observed as a result of translocations that disrupt only one of the two genes covered by the probe set, 3R1G and 2R3G (Figure 3, J and K, respectively) (see Discussion). The 3R1G signal pattern also denotes the deletion of 1 MLL signal.
Figure 3.
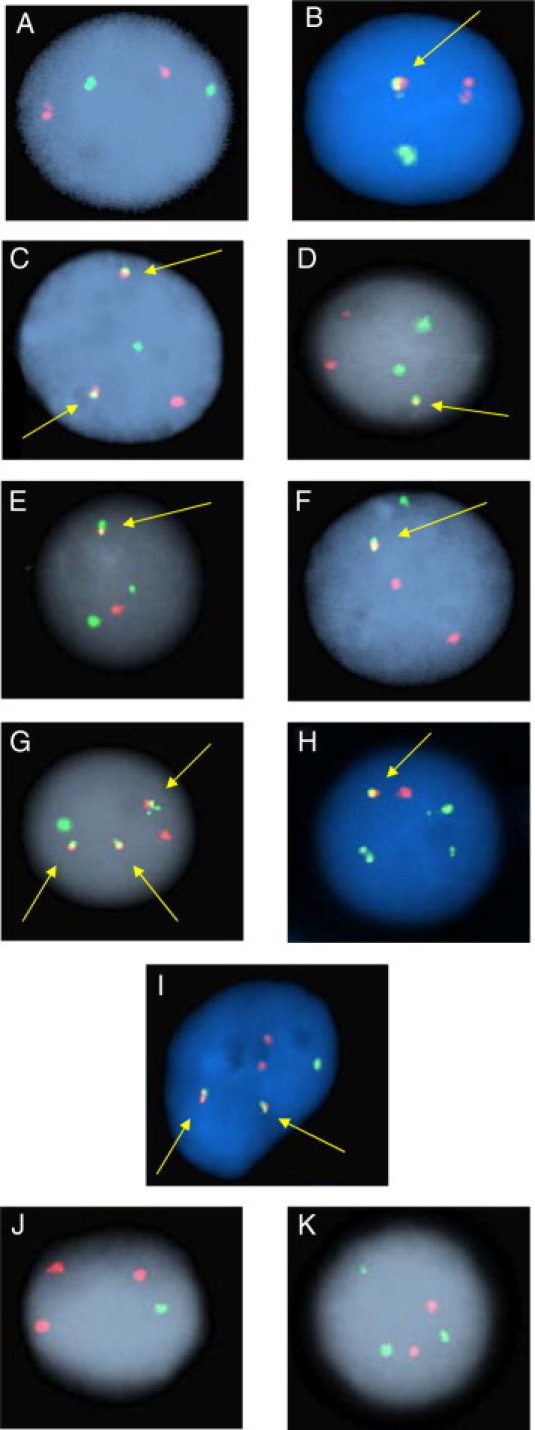
Representative interphase FISH signal patterns observed with the MLL/partner D-FISH assays. A: Nonneoplastic nucleus with a normal 2R2G signal pattern. B: Nonneoplastic nucleus with coincidental R and G signal overlap resulting in a 1R1G1F signal pattern. C: Neoplastic nucleus with a 1R1G2F signal pattern representing a balanced translocation resulting in two MLL/partner fusion signals. D: Neoplastic nucleus demonstrating a 2R2G1F signal pattern representing a complex translocation. E: Neoplastic nucleus demonstrating a 1R2G1F signal pattern indicating an unbalanced MLL/partner translocation with an associated deletion of the partner DNA at one of the translocation junctions. F: Neoplastic nucleus demonstrating a 2R1G1F signal pattern indicating an unbalanced MLL/partner translocation with an associated deletion of the MLL DNA at one of the translocation junctions. G: Neoplastic nucleus with a 1R1G3F signal pattern due to gain of an additional copy of one of the derivative chromosomes. H: Neoplastic nucleus with a 1R3G1F signal pattern indicating an unbalanced translocation and MLL duplication/trisomy of chromosome 11. I: Neoplastic nucleus with a 2R1G2F signal pattern indicating a balanced translocation and an additional copy of the partner chromosome. J and K: Abnormal signal patterns lacking a fusion (3R1G and 2R3G) are observed as a result of translocations that disrupt only one of the two genes covered by the probe set. The 3R1G signal pattern also denotes a deletion of 1 MLL signal. Arrows denote yellow fusion signals.
AFF1(AF4)/MLL t(4;11)(q21;q23) D-FISH Assay
The conventional chromosome results for the 25 leukemic bone marrow samples with corresponding results by AFF1/MLL D-FISH assay are summarized in Table 1. Patient ages ranged from 4 months to 80 years (median age, 40 years) and included 6 pediatric patients and 19 adult patients. Of the 25 samples, 23 specimens showed a balanced t(4;11)(q21;q23) by chromosome analysis, one specimen demonstrated a der(11)t(4,11) only, and one patient showed a t(4;11)(q21;q23) plus a der(4)t(4,11) in the same clone.
Table 1.
Conventional Cytogenetic and D-FISH Results Using a Homebrew MLL/AFF1 Probe for 25 Patients with t (4;11) (q21;q23)
| Conventional cytogenetic analysis |
FISH analysis* |
|||
|---|---|---|---|---|
| Patient no. | Age (years) | Karyotype | Abnormal nuclei (%) | Interphase signal pattern |
| 1 | 4 M | 46,XY,t (4;11) (q21;q23) [15]/46,XY[5] | 95 | 1R1G2F |
| 2 | 6 M | 46,XY,t (4;11) (q21;q23) [30] | 92.2 | 1R1G2F |
| 3 | 21 M | 49-53,XX,+X,+add(1) (p13),t (4;11) (q21;q23),+der(4)t (4;11) (q21;q23),+5,del(6) (q21q25),+8,+8,+13[cp20] | 91.6 | 1R1G3F |
| 4 | 4 | 46,XX,t (4;11) (q21;q23) [4]/46,XX[16] | 85.2 | 1R1G2F |
| 5 | 17 | 46,XY,t (4;11) (q21;q23) [3]/46,XY[3] | 88.8 | 1R1G2F |
| 6 | 18 | 46,XY,t (4;11) (q21;q23) [3]/46,XY[3] | 92.4 | 1R1G2F |
| 7 | 24 | 48,XX,+X,add(1) (p36.3),+4,add(4) (q12),t (4;11) (q21;q23) [cp4]/ 48,idem,t (1;19) (q23;p13.3) [1] | 95.8 | 1R1G3F |
| 8 | 30 | 47,XX,+X,t (4;11) (q21;q23) [cp10]/48,idem,+8,i(17) (q10) [3]/46,XX,t (4;11) (q21;q23),add(21) (q22) [7] | 98.6 | 1R1G2F |
| 9 | 31 | 46,XY,t (4;11) (q21;q23) [20] | 97.6 | 1R1G2F |
| 10 | 31 | 48,XX,t (4;11) (q21;q23),+6,+13[9]/46,XX[11] | 95.6 | 1R1G2F |
| 11 | 38 | 46,XX,t (4;11) (q21;q23),i(7) (q10) [9]/46,XX[11] | 92.4 | 1R1G2F |
| 12 | 39 | 46,XY,t (4;11) (q21;q23) [4]/46,XY[8] | 94.8 | 1R1G2F |
| 13 | 40 | 46,XY,t (4;11) (q21;q23) [20] | 97 | 2R2G1F |
| 14 | 41 | 46,XY,t (4;11) (q21;q23) [5]/46,XY[8] | 94.2 | 1R1G2F |
| 15 | 42 | 46,XX,t (4;11) (q21;q23) [19]/46,XX[1] | 98.2 | 1R1G2F |
| 16 | 45 | 46,XX,t (4;11) (q21;q23) [20] | 91 | 1R1G2F |
| 17 | 51 | 46,XX,t (4;11) (q21;q23),add(12) (p11.2) [11]/46,XX[9] | 97.8 | 1R1G2F |
| 18 | 54 | 46,XY,t (4;11) (q21;q23) [14] | 95.4 | 1R1G2F |
| 19 | 58 | 46,XX,t (4;11) (q21;q23) [14]/46,XX[6] | 94.4 | 1R1G2F |
| 20 | 58 | 47,XX,+X,t (4;11) (q21;q23) [14]/46,XX[6] | 85 | 1R1G2F |
| 21 | 58 | 46,XX,t (4;11) (q21;q23) [6]/46,XX[14] | 50.4 | 1R1G2F |
| 22 | 65 | 46,XX,t (4;11) (q21;q23) [6]/46,XX[3] | 96.2 | 1R1G2F |
| 23 | 65 | 46,XX,t (4;11) (q21;q23) [15]/47,idem,+6[1]/46,XX[4] | 94.4 | 1R1G2F |
| 24 | 74 | 47,XX,+X,dup(1) (q21q25),add(2) (q11.2),add(4) (q35),del(9) (q13q22), der(11)t (4;11) (q21;q23),−18,+19[cp4]/46,XX[16] | 0 | 2R2G |
| 25 | 80 | 41-43,XX,add(1) (q21),t (4;11) (q21;q23),−5,del(9) (p13),−10,i(14) (q10),−15,del(16) (q12.1),−17,−19,−20,+4mar[cp18]/46,XX[3] | 27.6 | 3R1G |
F, fusion (yellow); G, green; M, months; R, red.
A total of 500 interphase nuclei were analyzed by two technologists (250 nuclei each).
Abnormal FISH results were obtained for 24 (96%) of the 25 AFF1/MLL samples. Twenty samples yielded the expected abnormal signal pattern of 1R1G2F with an abnormal reference range from 50.4 to 98.6% (mean, 96.3%). Samples from patients 3 and 7 both showed an abnormal FISH signal pattern of 1R1G3F. The sample from patient 13 showed a 2R2G1F signal pattern. Patient 24 showed a normal FISH signal pattern. The sample from patient 25 showed only a 3R1G signal pattern. Interrogation of the specimens from patients 24 and 25 using the commercially available break-apart MLL FISH probes showed a signal pattern of 2F confirming an intact MLL locus in both cases.
MLLT4(AF6)/MLL t(6;11)(q27;q23) D-FISH Assay
The conventional chromosome study findings for 20 leukemic bone marrow samples with corresponding results by MLLT4/MLL D-FISH assay are summarized in Table 2. The patient age ranged from 5 months to 86 years (median, 40 years) and included 4 pediatric and 16 adult patients. For the 20 diagnostic samples, conventional cytogenetics defined 17 samples with a t(6;11)(q27;q23), two samples with a t(6;11)(q27;q23) and a der(6)t(6;11) present in the same clone and one sample with a cryptic, insertional t(6;11), patient 28.
Table 2.
Conventional Cytogenetic and D-FISH Results Using a Homebrew MLL/MLLT4 Probe for 20 Patients with t (6;11) (q27;q23)
| Patient no. | Age (years) | Conventional cytogenetic analysis Karyotype | FISH analysis* Abnormal nuclei (%) | Interphase signal pattern(s) |
|---|---|---|---|---|
| 26 | 5 M | 50,XX,+5,t (6;11) (q27;q23),+12,+15,+18[8]/51-52,idem,+4,+15[cp9]/46,XX[3] | 35.8 | 1R1G2F |
| 27 | 5 | 45,XX,t (6;11) (q27;q23),+der(6)t (6;11),−9,−20[1]/47,XX, t (6;11) (q27;q23),+der(6)t (6;11),add(10) (p15) [19] | 88.4 | 1R1G3F |
| 28 | 10 | 46,XY,der(11)del(11) (p13)add(11) (q21),add(12) (p11.2) [5]/46,sl, t (8;18)(q13;q21) [15].ish t (6;11) (q27;q23) (3′MLL+;5′MLL+) | 11.6, 85.2 | 1R2G1F, 1R3G1F |
| 29 | 11 | 46,XX,t (6;11) (q27;q23),t (15;19) (q24;p13.3)?c[14]/92,XXXX, add(1) (p32),add(5) (q13),t (6;11) (q27;q23)x2, t (15;19) (q24;p13.3)?cx2[6] | 80, 13.6 | 1R1G2F, 2R2G4F |
| 30 | 25 | 47,XY,der(6)t (6;11) (q27;q23),der(11)add(11) (p11.2)t (6;11),+21[19]//46,XX[1] | 74.8 | 1R1G2F |
| 31 | 26 | 46,XX,t (6;11) (q27;q23) [20] | 98 | 1R1G2F |
| 32 | 29 | 46,XY,t (6;11) (q27;q23) [10]/47,XY,idem,+9[10] | 96.6 | 1R1G2F |
| 33 | 33 | 46,XY,t (6;11) (q27;q23) [20] | 87.6 | 1R1G2F |
| 34 | 38 | 46,XX,t (6;11) (q27;q23) [20] | 97.6 | 1R1G2F |
| 35 | 38 | 46,XY,t (6;11) (q27;q23) [2]/46,XY[18] | 6.8 | 1R1G2F |
| 36 | 41 | 46,XY,t (6;11) (q27;q23) [15]/54,XY,+4,t (6;11)(q27;q23),+der(6)t (6;11),+8,+8,+12,+15,+16,+18[8] | 4.6, 6.6, 7, 65.6 | 1R1G3F, 2R1G2F, 2R1G1F, 1R1G2F |
| 37 | 42 | 46,XX,t (6;11) (q27;q23) [cp10]/47,idem,add(4) (q31.1),+8[cp3]/51,idem,+X,+6,+8,+15,+21[3]/46,XX[4] | 15.8, 65.6 | 2R1G2F, 1R1G2F |
| 38 | 44 | 46,XX,t (6;11) (q27;q23) [13]/48,sl,+19,+21[2]/46,XX[5] | 11.4 | 1R1G2F |
| 39 | 55 | 46,XX,t (6;11) (q27;q23) [1]/50,idem,+19,+21,+21[18]/46,XX[1] | 1.2, 5.8, 44.2 | 2R1G1F, 1R1G2F, 2R1G2F |
| 40 | 55 | 46,XY,t (6;11) (q27;q23) [20] | 88 | 1R1G2F |
| 41 | 58 | 46,XX,t (6;11) (q27;q23) [14]/46,XX[6] | 94 | 1R1G2F |
| 42 | 60 | 46,XY,t (1;5) (p22;q11.2)?c,t (6;11) (q27;q23) [20] | 95.8 | 1R1G2F |
| 43 | 65 | 47,XY,t (6;11) (q27;q23),+8[20] | 95.6 | 1R1G2F |
| 44 | 81 | 45,X,−Y,t (6;11) (q27;q23) [20] | 98.4 | 1R1G2F |
| 45 | 86 | 47,XX,t (6;11) (q27;q23),+19[8]/46,XX,del(11) (q13q23) [4]/46,XX[8] | 0 | 2R2G |
F, fusion (yellow); G, green; M, months; R, red.
A total of 500 interphase nuclei were analyzed by two technologists (250 nuclei each).
Abnormal FISH results were obtained for 19 of the 20 MLLT4/MLL samples. Seventeen of the 20 samples yielded the expected 1R1G2F signal pattern with abnormal reference range from 5.8 to 98.4% (mean, 69.4%). The sample from patient 27 had a 1R1G3F signal pattern. The sample from patient 28 showed 1R2G1F and 1R3G1F abnormal signal patterns and both a 2R1G2F and a 1R1G2F pattern were seen in the sample from patient 37. Patient 39 showed the expected 1R1G2F abnormal signal pattern as a secondary pattern with 2R1G2F as the dominant pattern. The sample from patient 45 had a normal 2R2G signal pattern with the MLLT4/MLL probe set. Interrogation of the specimen from patient 45 using the commercially available break-apart MLL FISH probes showed a signal pattern of 2F confirming an intact MLL locus.
MLLT3(AF9)/MLL t(9;11)(p22;q23) D-FISH Assay
Conventional cytogenetic analysis findings for 20 leukemic bone marrow samples with corresponding results by MLLT3/MLL D-FISH assay are summarized in Table 3. The patient ages were between 7 months and 77 years with a median age of 44 years. Five patients were pediatric and 15 were adults. Of the 20 samples, conventional cytogenetics defined 19 specimens with t(9;11)(p22;q23) and 1 specimen with a t(9;11)(p22;q23) and a der(11)t(9,11) in the same clone.
Table 3.
Conventional Cytogenetic and D-FISH Results Using a Homebrew MLL/MLLT3 Probe for 20 Patients with t (9;11) (p22;q23)
| Conventional cytogenetic analysis |
FISH analysis* |
|||
|---|---|---|---|---|
| Patient no. | Age (years) | Karyotype | Abnormal nuclei (%) | Interphase signal pattern |
| 46 | 7 M | 43-46,XX,t (9;11) (p22;q23),dic(9;13) (p22;q12),der(11)t (9;11)(p22;q23), del(12) (p11.2p13),del(13) (q12q14) [cp18]/46,XX[2] | 87.4 | 1R2G1F |
| 47 | 23 M | 48,XX,der(9)t (9;11) (p24;p11.2),der(9)t (9;11) (p22;q23),+10, der(11)t (9;11) (p24;p11.2),t (9;11) (p22;q23),+19[7]/49,idem,+8[2]/46,XX[11] | 10.6 | 1R1G2F |
| 48 | 11 | 46,X,t (X;Y) (q28;q11.2)?c[3]/46,idem,t (9;11) (p22;q23) [17] | 85.8 | 1R1G2F |
| 49 | 15 | 46,XY,t (9;11) (p22;q23) [11]/46,idem,der(?)t (1;?) (q23;?) [2]/ 47,idem,+8[2]/48,idem,+8,+19[5] | 93.4 | 1R1G2F |
| 50 | 18 | 46,XX,t (9;11) (p22;q23) [19]/47,XX,+9,t (9;11) (p22;q23) [1] | 82.2 | 1R1G2F |
| 51 | 30 | 46,XY,t (9;11) (p22;q23) [20] | 93.6 | 1R1G2F |
| 52 | 31 | 48,XX,+4,t (9;11) (p22;q23),+12,t (17;18) (p10;q10)?c[cp20] | 88.4 | 1R1G2F |
| 53 | 39 | 46,XY,t (9;11) (p22;q23) [20] | 90.4 | 1R1G2F |
| 54 | 41 | 48,XX,+8,t (9;11) (p22;q23),+19[20] | 97 | 1R1G2F |
| 55 | 42 | 47,XY,t (9;11) (p22;q23),+21[3]/46,X,-Y,t (9;11),+21[16]/46,XY[1] | 95 | 1R1G2F |
| 56 | 46 | 46,XY,t (9;11) (p22;q23) [20] | 74 | 1R1G2F |
| 57 | 48 | 46,XX,t (9;11) (p22;q23) [20] | 89 | 1R1G2F |
| 58 | 54 | 46,XX,t (9;11) (p22;q23) [20] | 91.6 | 1R1G2F |
| 59 | 58 | 50,XX,+4,+8,t (9;11) (p22;q23),+12,+20[20] | 93.2 | 1R1G2F |
| 60 | 60 | 46,XX,t (9;11) (p22;q23) [20] | 92.4 | 1R1G2F |
| 61 | 61 | 46,XY,t (9;11) (p22;q23) [20] | 92.6 | 1R1G2F |
| 62 | 62 | 46,XY,t (9;11) (p22;q23) [20] | 95.2 | 1R1G2F |
| 63 | 70 | 46,XX,t (9;11) (p22;q23) [19]/46,XX[1] | 85.2 | 1R1G2F |
| 64 | 76 | 46,XX,t (9;11) (p22;q23) [23] | 92.6 | 1R1G2F |
| 65 | 77 | 46,XX,t (9;11) (p22;q23) [20] | 85.6 | 1R1G2F |
F, fusion (yellow); G, green; M, months; R, red.
A total of 500 interphase nuclei were analyzed by two technologists (250 nuclei each).
Abnormal FISH results were obtained for all 20 samples. Nineteen of the 20 t(9;11)(p22;q23) samples yielded the expected abnormal 1R1G2F signal pattern with an abnormal reference range from 10.6 to 95.2% (median, 85.7%). Despite patient sample 46 demonstrating both an apparently balanced 9;11 and an extra derivative chromosome 11, the sample demonstrated a single FISH signal pattern of 1R2G1F.
MLL/ELL t(11;19)(q23;p13.1) D-FISH Assay
The conventional cytogenetic analysis findings for 18 leukemic bone marrow samples with corresponding results by MLL/ELL D-FISH assay are summarized in Table 4. Patient ages ranged from 7 weeks to 78 years with a median age of 52 years and included 3 pediatric patients and 15 adult patients. These 18 patients include five samples (66, 69, 71, 77, and 78) that were originally defined cytogenetically as t(11;19)(q23;p13.3). However, the FISH results for the MLL/MLLT1 D-FISH assay indicated a split MLL signal without fusion to MLLT1. Subsequent testing with the MLL/ELL D-FISH assay revealed a double-fusion signal pattern in each patient, indicating the original 19p13.3 breakpoint was incorrectly designated. Thus, these five patients had the chromosome nomenclature correctly reassigned to the 19p13.1 breakpoint and are included in Table 4. For the 18 specimens demonstrating MLL/ELL fusion, 17 specimens had an apparently balanced t(11;19)(q23;p13.1), and one sample had a three-way translocation involving chromosomes 2, 11, and 19.
Table 4.
Conventional Cytogenetics and D-FISH Results Using a Homebrew MLL/ELL Probe for 18 Patients with t (11;19) (q23;p13.1)
| Conventional cytogenetic analysis |
FISH analysis* |
|||
|---|---|---|---|---|
| Patient no. | Age (years) | Karyotype | Abnormal nuclei (%) | Interphase signal pattern |
| 66† | 7 W | 47,XY,+8,t (11;19) (q23;p13.1) [20] | 82.6 | 1R1G2F |
| 67 | 5 M | 46,XY,t (2;19;11) (p11.2;p13.1;q23) [2] | 2.8 | 2R2G1F |
| 68 | 13 | 46,XX,t (2;12) (p11.2;q13),add(7) (p15),t (11;19) (q23;p13.1),add(17) (q25) [20] | 91.2 | 1R1G2F |
| 69† | 20 | 46,XX,t (11;19) (q23;p13.1) [18]/46,XX[12] | 71.4 | 1R1G2F |
| 70 | 30 | 46,XX,t (11;19) (q23;p13.1) [7]/46,idem,add(13) (p12) [13] | 56.4 | 1R1G2F |
| 71† | 34 | 47,XX,+8,t (11;19) (q23;p13.1),del(13) (q12q14) [20] | 88.8 | 1R1G2F |
| 72 | 44 | 46,XY,t (11;19) (q23;p13.1),−14,t (15;21) (q22;q22) [1]/46,XY[29] | 12.8 | 1R1G2F |
| 73 | 49 | 46,XX,t (11;19) (q23;p13.1) [19]/46,XX[1] | 81.8 | 1R1G2F |
| 74 | 51 | 46,XX,t (11;19) (q23;p13.1) [20] | 96.8 | 1R1G2F |
| 75 | 52 | 46,XX,t (11;19) (q23;p13.1) [21] | 81.6 | 1R1G2F |
| 76 | 55 | 46,XX,t (11;19) (q23;p13.1) [20] | 80.6 | 1R1G2F |
| 77† | 55 | 46,XX,t (11;19) (q23;p13.1) [12]/46,XX[10] | 50.4 | 1R1G2F |
| 78† | 58 | 45-48,XY,t (11;19) (q23;p13.1),+i(13) (q10) [cp25]/46,XY,i(17) (q10) [2]/ 46,XY[13] | 96.8 | 1R1G2F |
| 79 | 58 | 46,XX,t (11;19) (q23;p13.1) [20] | 85 | 1R1G2F |
| 80 | 60 | 46,XX,del(7) (q22),t (11;19) (q23;p13.1) [16]/46,XY[4] | 47.6 | 1R1G2F |
| 81 | 60 | 46,XY,t (11;19) (q23;p13.1) [20] | 85.4 | 1R1G2F |
| 82 | 68 | 46,XX,t (11;19) (q23;p13.1) [30] | 94.8 | 1R1G2F |
| 83 | 78 | 46,XX,t (11;19) (q23;p13.1) [20] | 96.8 | 1R1G2F |
F, fusion (yellow); G, green; M, months; R, red; W, weeks.
A total of 500 interphase nuclei were analyzed by two technologists (250 nuclei each).
The breakpoint for these patients was initially evaluated as 19p13.3.
Following reassignment of the five patients listed above, abnormal FISH results were obtained for all of the ELL samples. Seventeen of the 18 samples yielded the 1R1G2F signal pattern with an abnormal reference range from 12.8 to 96.8% (mean, 76.5%). The sample from patient 67 yielded a predominant abnormal FISH signal pattern of 2R2G1F.
MLL/MLLT1(ENL) t(11;19)(q23;p13.3) D-FISH Assay
The conventional cytogenetic analysis findings for 20 leukemic bone marrow samples with corresponding results by MLL/MLLT1 D-FISH assay are summarized in Table 5. Six patients were pediatric and 14 were adults, with an age range from 4 weeks of age to 75 years and a median age of 27 years. Three of the 20 patients (96, 97, and 103; Table 5) were originally defined cytogenetically with a t(11;19)(q23;p13.1). However, the FISH results for the MLL/ELL D-FISH assay indicated a split MLL signal without fusion to ELL. Subsequent testing with the MLL/MLLT1 D-FISH assay revealed a double-fusion signal pattern in each patient, indicating the original 19p13.1 breakpoint was incorrectly designated. Thus, these three patients had the chromosome nomenclature correctly reassigned to the 19p13.3 breakpoint and are included in Table 5. Each of the 20 specimens had an apparently balanced t(11;19)(q23;p13.3).
Table 5.
Conventional Cytogenetic and D-FISH Results Using a Homebrew MLL/MLLT1 Probe for 20 Patients with t (11;19) (q23;p13.3)
| Conventional cytogenetic analysis |
FISH analysis* |
|||
|---|---|---|---|---|
| Patient no. | Age (years) | Karyotype | Abnormal nuclei (%) | Interphase signal pattern(s) |
| 84 | 4 W | 46,XY,t (11;19) (q23;p13.3) [7]/46,XY[13] | 92 | 1R2G1F |
| 85 | 12 M | 46,XY,t (11;19) (q23;p13.3) [3]/46,XY,del(6) (q21q23),t (11;19) (q23;p13.3) [7]/46,XY[10] | 91 | 2R2G1F |
| 86 | 22 M | 46,XY,t (11;19) (q23;p13.3) [2]/46,XX[18] | 52.2 | 1R1G2F |
| 87 | 12 | 46,XX,t (9;17) (q34;q25),t (11;19) (q23;p13.3) [17]/46,XX[3] | 72.4 | 1R1G2F |
| 88 | 15 | 46,XX,t (11;19) (q23;p13.3) [30] | 87.4 | 1R1G2F |
| 89 | 16 | 52,XX,+X,+5,+6,+8,t (11;19) (q23;p13.3),+19,+21[19]/46,XX[1] | 5.8, 52.4 | 1R1G2F, 2R1G2F |
| 90 | 20 | 46,XX,t (11;19) (q23;p13.3) [15]/46,XX[5] | 90.8 | 1R1G2F |
| 91 | 20 | 46,XX,t (11;19) (q23;p13.3) [13]/46,XX[7] | 91 | 1R1G2F |
| 92† | 20 | 45-47,XY,i(9) (q10),t (11;19) (q23;p13.3),+2-3mar[cp3]/46,XY[27] | 0.4 | 1R1G2F |
| 93 | 23 | 48,XY,+8,t (11;19) (q23;p13.3),+20[2]/48,idem,del(5) (q22q33) [16]/ 48,idem,t (2;12;10) (q37;q13;q22) [2] | 89.6 | 1R1G2F |
| 94 | 30 | 46,XX,t (11;19) (q23;p13.3) [16]/46,XX[4] | 86.2 | 1R1G2F |
| 95 | 36 | 47,XXY?c,t (11;19) (q23;p13.3) [20] | 98.6 | 1R1G2F |
| 96‡ | 37 | 46,XX,t (11;19) (q23;p13.3) [20] | 75 | 1R1G2F |
| 97‡ | 39 | 46,XY,t (11;19) (q23;p13.3),der(20)t (17;20) (q11.2;q13.3) [20] | 96.6 | 1R1G2F |
| 98 | 43 | 46,XY,t (11;19) (q23;p13.3) [18]/47,idem,+8[2] | 1.6, 92.4 | 1R1G3F, 1R1G2F |
| 99 | 45 | 47,XX,+X,t (11;19) (q23;p13.3) [19]/46,XX[1] | 95 | 1R1G2F |
| 100 | 47 | 46,XY,t (11;19) (q23;p13.3) [20] | 96.4 | 1R1G2F |
| 101 | 53 | 46,XX,t (11;19) (q23;p13.3) [9]/46,XX[1] | 94.8 | 1R1G2F |
| 102 | 71 | 47,XX,+X,t (11;19) (q23;p13.3) [4]/46,XX[26] | 92.2 | 1R1G2F |
| 103‡ | 75 | 46,XX,t (11;19) (q23;p13.3) [20] | 96.4 | 1R1G2F |
F, fusion (yellow); G, green; M, months; R, red; W, weeks.
A total of 500 interphase nuclei were analyzed by two technologists (250 nuclei each).
This sample represents a posttherapy sample.
The breakpoint for these patients was initially evaluated as 19p13.1.
Following reassignment of the three patients listed above, abnormal FISH results were obtained for all of the MLL/MLLT1 samples. Nineteen of the 20 samples yielded the signal pattern of 1R1G2F with an abnormal reference range from 0.4 to 98.6% (mean, 78.5%). The sample from patient 85 showed a 2R2G1F D-FISH pattern. The sample from patient 89 demonstrated a predominant 2R1G2F D-FISH signal pattern with a secondary 1R1G2F pattern. Patient 98 showed a predominant 1R1G2F signal pattern with a minor pattern of 1R1G3F.
Discussion
Numerous MLL pairing partners have been identified in acute leukemias; however, gene pairing with MLL most commonly involves AFF1(AF4) at 4q21, MLLT4(AF6) at 6q27, MLLT3(AF9) at 9p22, ELL at 19p13.1, and MLLT1(ENL) at 19p13.3.1,17,18,24,34 Recent findings suggest that the specific pairing partners involved in MLL-rearranged leukemias may significantly impact prognosis and/or have therapeutic implications depending on acute leukemia subtype and clinical context.10,28,35,36 Therefore, providing diagnostic information with regard to the presence of MLL rearrangement as well as the identity of the MLL pairing partner may have relevance for acute leukemia prognostication.
Herein, we evaluated 103 bone marrow and blood specimens from patients with acute leukemia. The patients ranged in age from only 4 weeks of age to 86 years old. Overall, we evaluated 24 pediatric leukemia specimens, including 8 under the age of 1 year (infant leukemia). A total of 79 adult leukemia specimens were interrogated with these novel probe assays, with 42 below the age of 50 years and 37 between the ages of 51 and 86 years of age. The average patient age for four of the five translocation patient cohorts was similar (median age range, 40 to 52 years). The exception was the t(11;19p13.3) patient cohort for who the median patient age was only 27 years, suggesting this particular translocation may occur more frequently in children and younger adults. However, as each D-FISH probe set was successful in generating the expected MLL/partner double-fusion signal patterns in most patients, our results indicate the five probe assays can be used for acute leukemias occurring across all age groups.
The patient samples evaluated in this study were chosen because of a previously defined, chromosomally visible 11q23/partner translocation. Approximately half of the patients had the 11q23/partner translocation as the sole chromosome abnormality (56 of 103; 54%). The remaining 47 patients had additional clonal chromosome abnormalities, from single trisomies to very complex structural and numeric abnormalities. The frequency of additional chromosome abnormalities demonstrated a statistical trend toward more complex clones in the 24 pediatric samples (15 of 24, 63%) versus the 79 adult samples (32 of 79, 40%; P = 0.06).
The current commercially available break-apart FISH probe set for interrogation of MLL offers a sensitive means of detecting the presence of MLL rearrangements but provides no information regarding the identity of the gene with which MLL is paired. The development of the current set of homebrew D-FISH probes allows for detection of MLL rearrangements and the identification of the five most common MLL gene fusion partners, t(4;11)(q21;q23), t(6;11)(q27;q23), t(9;11)(p22;q23), t(11;19)(q23;p13.1), and t(11;19)(q23;p13.3). Overall, the homebrew MLL D-FISH assays performed well on specimens demonstrating these five translocations by conventional banding. As detailed in Results, the expected 1R1G2F signal pattern indicative of balanced translocations involving MLL, and one of these five partner genes was observed in 89 (86%) of the 103 leukemic bone marrow specimens.
A minor degree of variation from the expected abnormal FISH signal pattern was observed in the assays for all five MLL partners. Of the 100 patient samples with a documented MLL/partner fusion by FISH, only 14 patients demonstrated an atypical FISH signal pattern other than 1R1G2F. Virtually no signal pattern variation was observed with the MLLT3/MLL and the MLL/ELL probe sets, with a single patient in each having an atypical pattern. The AFF1/MLL, MLL/MLLT1, and MLLT4/MLL probe sets had three, four, and five patients, respectively, with atypical signal patterns detected, of which six cases represented an additional signal pattern in subclones. FISH signal pattern variation was significantly more prevalent in the pediatric patient cohort (8 of 24, 33%) than in the adult patient cohort (6 of 76, 8%) (P = 0.02; Fisher's exact test). These results suggest the pediatric leukemia specimens may be more prone to atypical chromosomal mechanisms generating MLL/partner fusion, instability of the translocation event, or the presence of additional abnormalities involving chromosomes 11 and the MLL partner chromosome (see subsequent Discussion). Regardless of the etiology for the atypical signal patterns observed with the MLL/partner D-FISH assays, it is clear that the atypical patterns are much more likely to be encountered in pediatric samples, and a laboratory using these MLL/partner D-FISH assays should anticipate the identification of atypical signal patterns when evaluating pediatric leukemia specimens.
The atypical abnormal FISH signal patterns made up four subgroups. The first atypical abnormal signal pattern involved the absence of one of the two expected fusion signals. Eight patients were observed with this atypical signal pattern (patients 13, 28, 36, 39, 46, 67, 84, and 85). Three of these patients (13, 67, and 85) demonstrated a signal pattern indicative of a complex, three-way translocation (2R2G1F), as illustrated in Figure 3D. As only one of these patients had a demonstrable three-way translocation, it is likely the other two patients had a third partner chromosome that was not appreciated by the original G-banded chromosome study. Unfortunately, metaphase FISH was not possible on these two cases to identify the third partner chromosome. The most likely explanation for the atypical single fusion results observed in the other five patients is loss of DNA at the fusion junction (Figure 3E and 3F). Thus, despite the apparently balanced translocation observed by conventional chromosome analysis in these samples, DNA has been deleted at the molecular level. Alternatively, it may be that the original conventional analysis failed to detect subtle deletions at these translocation junctions. However, because these five patients still generated a MLL/partner fusion signal, these leukemia samples can still be consistently identified by these D-FISH assays.
The second FISH pattern variation involved the presence of a third fusion signal in addition to the two expected fusion signals (Figure 3G), yielding an overall pattern of 1R1G3F (patients 3, 7, 27, 36, and 98). This variation reflects the presence of an extra copy of the derivative chromosome containing the MLL/partner fusion in addition to the expected balanced translocation fusion signals. Such derivative chromosomes were identified by conventional analysis in three of the five patient samples that produced the 1R1G3F signal pattern (patients 3, 27, and 36). The sample from patient 7 did not have an additional derivative chromosome identified by conventional cytogenetic analysis. However, it is likely that the chromosomal abnormality identified as an add(4)(q12) represents a der(4)t(4,11)(q21;q23), which was not obvious by conventional banded analysis. The sample from patient 98 predominantly showed the expected abnormal 1R1G2F pattern but also had a small percentage of cells (1.6%) with a 1R1G3F pattern not explained by the karyotype. Given the small percentage, it is not surprising that this second clonal population was not detected by conventional analysis. This small population likely represents a subclone with an additional copy of a derivative chromosome from the translocation.
The third atypical FISH signal pattern was associated with extra copies of the green or red signals in addition to the fusion signals (Figure 3H and 3I). These FISH signal patterns were observed in five patients (patients 28, 36, 37, 39, and 89). In each leukemic sample, this signal pattern was observed as a second subclonal population because of the presence of additional copies of the chromosomes (trisomy 6, 11, or 19) that corresponded to the chromosomal location of the FISH probe. Although these additional signals do not denote any disruption of the actual genes covered by the probes, these patterns need to be recognized as possible variant FISH signal patterns despite not representing true translocation variants.
The fourth atypical FISH signal pattern lacked the generation of any fusion signals (Figure 3A and 3J), as seen in three patient samples. Two patient samples showed only a normal 2R2G signal pattern (patients 24 and 45), and the sample from patient 25 only showed a 3R1G signal pattern. In this scenario, the most likely explanation for patterns lacking a fusion signal is that, despite a chromosomal translocation involving 11q23 and a classic partner chromosome, the translocations in these tumor cells did not disrupt MLL or the partner gene but instead involved adjacent gene regions. Subsequent evaluation with a commercially available MLL split-signal probe indicated the MLL gene region was not disrupted in any of these three patients. Given the D-FISH pattern, the original metaphase chromosomes from each of these three patients were re-evaluated to confirm the original karyotype descriptions. On re-examination, chromosomes from patient 24 verified a 4;11 translocation, but the breakpoints were revised to 4q13 and 11q21. These atypical breakpoints were confirmed with subsequent metaphase FISH, which showed intact AFF1 and MLL FISH signals localizing to 11q and 4q, respectively. For patient 25, a review of the conventional chromosome analysis also verified the presence of a t(4;11)(q21;q23). However, application of the AFF1/MLL D-FISH probe set on the abnormal metaphase chromosomes indicated a rearrangement of the AFF1 probe and a deletion of the MLL gene region, verifying an atypical 4;11 translocation in this patient. Re-evaluation of the original karyotypes in patient 45 also suggested the breakpoints for the 6;11 translocation were proximal to 6q27 and 11q23; however, insufficient sample was available for metaphase FISH to verify the revised chromosomal breakpoints.
Because of the close proximity of the ELL (19p13.1) and MLLT1 (19p13.3) genes, conventional cytogenetic analysis may at times be unable to predict the correct 19p13 breakpoint involved in the 11q23 translocation. This difficulty is exemplified by findings in the present study where eight patients were shown to have either the 19p13.1 or the 19p13.3 breakpoints incorrectly assigned (Tables 4and 5). During evaluation of the MLL/MLLT1 (19p13.3) D-FISH probe set, five samples that were originally classified as carrying a t(11;19)(q23;p13.3) by conventional chromosome analysis (patients 66, 69, 71, 77, and 78) failed to demonstrate the expected 1R1G2F signal pattern. In these samples, the predominant signal pattern was 2R3G (Figure 3K), which was present in 53 to 80% of cells, whereas a second signal pattern, 1R2G1F (Figure 3E), was present in a minority (10 to 12%) of cells. Subsequent interrogation of these five samples using the MLL/ELL D-FISH probe set produced a predominant FISH pattern of 1R1G2F present in 50 to 97% of cells. These results indicate that these five samples carry a t(11;19)(q23;p13.1), contrary to the original designation of a t(11;19)(q23;p13.3) by conventional cytogenetic analysis.
Similarly, during evaluation of the MLL/ELL probe set, three patient samples (patients 96, 97, and 103) originally classified by conventional chromosome analysis as carrying a t(11;19)(q23;p13.1) showed only a small proportion (12.8 to 14.8%) of cells with an atypical 1R2G1F signal pattern, whereas the majority of cells had a 2R3G signal pattern (70.4 to 80%). Subsequent evaluation using the MLL/MLLT1 probe set showed all three cases to have a 1R1G2F signal pattern in 75 to 96.6% of cells.
The primary 2R3G signal pattern of the eight samples with misidentified 11;19 translocations indicates that the MLL gene is disrupted without disruption of MLLT1 (five samples) or ELL (three samples). The cells with the atypical 1R2G1F signal pattern simply correspond to random overlap of 1R and 1G signal in the abnormal interphase nuclei, based on the close proximity of these signals on the derivative chromosome 19.
These results indicate D-FISH is more accurate than conventional chromosome studies in the correct identification of the 19p13.1 and 19p13.3 breakpoints and thereby distinguishing the involvement of ELL from MLLT1. In addition, these results demonstrate that a true MLL/partner fusion signal should be part of the predominant abnormal FISH pattern observed with a D-FISH probe set. If an additional red or green signal is identified in most nuclei and a small population of cells with a single apparent fusion is also observed, it is very likely that the subset of cells with fusion represent random signal overlap rather than true fusion. This predisposition to signal overlap is particularly pertinent when multiple gene targets are in close proximity to the true chromosomal breakpoint (such as ELL at 19p13.1, which is physically close to MLLT1 at 19p13.3). The proximity of these genes can result in the misinterpretation of an atypical fusion signal pattern and the incorrect assignment of the translocation partner.
In addition to the advantage of identifying the specific translocation partner, these D-FISH assays also substantially increase the ability to detect leukemic cells present at very low levels. This application in the detection of FISH MRD enables a more informative means of monitoring response to therapy in posttreatment samples. The current method of monitoring disease characterized by MLL rearrangements relies on either chromosome studies or the commercially available MLL disruption gene probes both yielding a disease detection level of ∼5%. In the context of a classical or a complex translocation involving partnering of MLL with one of these five common fusion partners, our D-FISH method has a calculated analytical sensitivity to detect leukemic cells at levels as low as 0.6% when 500 nuclei are evaluated. In addition, similar to the FISH MRD studies we have previously published using the BCR/ABL1 D-FISH probe set in detecting only three cells of 6000 nuclei (0.05%), we have extended our application of these five MLL D-FISH assays using the automated BioView spot counter for evaluation of MRD in posttherapeutic bone marrow specimens.37 For each probe set, we have demonstrated a FISH MRD cutoff of 0.05% for the 1R1G2F signal pattern when 6000 nuclei are scored (data not shown). Although RT-PCR would be the method of choice for MRD detection, these MLL translocations are not common enough for industry to develop and market these specific assays, and they are currently not available. Thus, the application of FISH MRD in this patient cohort is currently the best evaluation of genetic MRD.
In conclusion, we present the development of five novel D-FISH probe sets for the detection of MLL rearrangements involving the most frequently observed pairing partner genes, AFF1, MLLT4, MLLT3, ELL, and MLLT1. These five assays proved to be effective in detecting classic and variant translocations of MLL and the most common translocation partners in both pediatric and adult acute leukemia samples. We identified a statistically significant association for atypical FISH signal patterns in pediatric leukemia samples versus adult leukemia samples (P = 0.02) and identified a statistical trend toward more complex karyotypic abnormalities in pediatric patients versus adult patients (P = 0.06). In addition, these assays should allow for a greater sensitivity in MRD detection than chromosome studies or the currently available commercial probes for the detection of MLL rearrangements in acute leukemia.
Acknowledgments
Rhett P. Ketterling dedicates this manuscript to his wife Judith and their children Morgan, Trevor, Graham, and Spencer for their love and support.
Footnotes
Supported by the Mayo Clinic Foundation.
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.Meyer C, Schneider B, Jakob S, Strehl S, Attarbaschi A, Schnittger S, Schoch C, Jansen MW, van Dongen JJ, den Boer ML, Pieters R, Ennas MG, Angelucci E, Koehl U, Greil J, Griesinger F, Zur Stadt U, Eckert C, Szczepanski T, Niggli FK, Schafer BW, Kempski H, Brady HJ, Zuna J, Trka J, Nigro LL, Biondi A, Delabesse E, Macintyre E, Stanulla M, Schrappe M, Haas OA, Burmeister T, Dingermann T, Klingebiel T, Marschalek R. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–784. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 2.Forestier E, Heim S, Blennow E, Borgstrom G, Holmgren G, Heinonen K, Johannsson J, Kerndrup G, Andersen MK, Lundin C, Nordgren A, Rosenquist R, Swolin B, Johansson B. Cytogenetic abnormalities in childhood acute myeloid leukaemia: a Nordic series comprising all children enrolled in the NOPHO-93-AML trial between 1993 and 2001. Br J Haematol. 2003;121:566–577. doi: 10.1046/j.1365-2141.2003.04349.x. [DOI] [PubMed] [Google Scholar]
- 3.Felix CA, Hosler MR, Winick NJ, Masterson M, Wilson AE, Lange BJ. ALL-1 gene rearrangements in DNA topoisomerase II inhibitor-related leukemia in children. Blood. 1995;85:3250–3256. [PubMed] [Google Scholar]
- 4.Super HJ, McCabe NR, Thirman MJ, Larson RA, Le Beau MM, Pedersen-Bjergaard J, Philip P, Diaz MO, Rowley JD. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- 5.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 6.Secker-Walker LM, Moorman AV, Bain BJ, Mehta AB. Secondary acute leukemia and myelodysplastic syndrome with 11q23 abnormalities. EU Concerted Action 11q23 Workshop. Leukemia. 1998;12:840–844. doi: 10.1038/sj.leu.2401021. [DOI] [PubMed] [Google Scholar]
- 7.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 8.Carbonell F, Swansbury J, Min T, Matutes E, Farahat N, Buccheri V, Morilla R, Secker-Walker L, Catovsky D. Cytogenetic findings in acute biphenotypic leukaemia. Leukemia. 1996;10:1283–1287. [PubMed] [Google Scholar]
- 9.Owaidah TM, Al Beihany A, Iqbal MA, Elkum N, Roberts GT. Cytogenetics, molecular and ultrastructural characteristics of biphenotypic acute leukemia identified by the EGIL scoring system. Leukemia. 2006;20:620–626. doi: 10.1038/sj.leu.2404128. [DOI] [PubMed] [Google Scholar]
- 10.Pui CH, Chessells JM, Camitta B, Baruchel A, Biondi A, Boyett JM, Carroll A, Eden OB, Evans WE, Gadner H, Harbott J, Harms DO, Harrison CJ, Harrison PL, Heerema N, Janka-Schaub G, Kamps W, Masera G, Pullen J, Raimondi SC, Richards S, Riehm H, Sallan S, Sather H, Shuster J, Silverman LB, Valsecchi MG, Vilmer E, Zhou Y, Gaynon PS, Schrappe M. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17:700–706. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 12.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 13.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 15.Nilson I, Lochner K, Siegler G, Greil J, Beck JD, Fey GH, Marschalek R. Exon/intron structure of the human ALL-1 (MLL) gene involved in translocations to chromosomal region 11q23 and acute leukaemias. Br J Haematol. 1996;93:966–972. doi: 10.1046/j.1365-2141.1996.d01-1748.x. [DOI] [PubMed] [Google Scholar]
- 16.Reichel M, Gillert E, Angermuller S, Hensel JP, Heidel F, Lode M, Leis T, Biondi A, Haas OA, Strehl S, Panzer-Grumayer ER, Griesinger F, Beck JD, Greil J, Fey GH, Uckun FM, Marschalek R. Biased distribution of chromosomal breakpoints involving the MLL gene in infants versus children and adults with t(4;11) ALL. Oncogene. 2001;20:2900–2907. doi: 10.1038/sj.onc.1204401. [DOI] [PubMed] [Google Scholar]
- 17.Shih LY, Liang DC, Fu JF, Wu JH, Wang PN, Lin TL, Dunn P, Kuo MC, Tang TC, Lin TH, Lai CL. Characterization of fusion partner genes in 114 patients with de novo acute myeloid leukemia and MLL rearrangement. Leukemia. 2006;20:218–223. doi: 10.1038/sj.leu.2404024. [DOI] [PubMed] [Google Scholar]
- 18.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, Delabesse E, de Oliveira MP, Cave H, Clappier E, van Dongen JJ, Balgobind BV, van den Heuvel-Eibrink MM, Beverloo HB, Panzer-Grumayer R, Teigler-Schlegel A, Harbott J, Kjeldsen E, Schnittger S, Koehl U, Gruhn B, Heidenreich O, Chan LC, Yip SF, Krzywinski M, Eckert C, Moricke A, Schrappe M, Alonso CN, Schafer BW, Krauter J, Lee DA, Zur Stadt U, Te Kronnie G, Sutton R, Izraeli S, Trakhtenbrot L, Lo Nigro L, Tsaur G, Fechina L, Szczepanski T, Strehl S, Ilencikova D, Molkentin M, Burmeister T, Dingermann T, Klingebiel T, Marschalek R. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 19.Behm FG, Raimondi SC, Frestedt JL, Liu Q, Crist WM, Downing JR, Rivera GK, Kersey JH, Pui CH. Rearrangement of the MLL gene confers a poor prognosis in childhood acute lymphoblastic leukemia, regardless of presenting age. Blood. 1996;87:2870–2877. [PubMed] [Google Scholar]
- 20.Chen CS, Sorensen PH, Domer PH, Reaman GH, Korsmeyer SJ, Heerema NA, Hammond GD, Kersey JH. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81:2386–2393. [PubMed] [Google Scholar]
- 21.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 22.Heerema NA, Sather HN, Ge J, Arthur DC, Hilden JM, Trigg ME, Reaman GH. Cytogenetic studies of infant acute lymphoblastic leukemia: poor prognosis of infants with t(4;11)—a report of the Children's Cancer Group. Leukemia. 1999;13:679–686. doi: 10.1038/sj.leu.2401413. [DOI] [PubMed] [Google Scholar]
- 23.Pui CH, Behm FG, Downing JR, Hancock ML, Shurtleff SA, Ribeiro RC, Head DR, Mahmoud HH, Sandlund JT, Furman WL. 11q23/MLL rearrangement confers a poor prognosis in infants with acute lymphoblastic leukemia. J Clin Oncol. 1994;12:909–915. doi: 10.1200/JCO.1994.12.5.909. [DOI] [PubMed] [Google Scholar]
- 24.Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, Beverloo HB, Chang M, Creutzig U, Dworzak MN, Forestier E, Gibson B, Hasle H, Harrison CJ, Heerema NA, Kaspers GJ, Leszl A, Litvinko N, Nigro LL, Morimoto A, Perot C, Pieters R, Reinhardt D, Rubnitz JE, Smith FO, Stary J, Stasevich I, Strehl S, Taga T, Tomizawa D, Webb D, Zemanova Z, Zwaan CM, van den Heuvel-Eibrink MM. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114:2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 26.Mrozek K, Heinonen K, Lawrence D, Carroll AJ, Koduru PR, Rao KW, Strout MP, Hutchison RE, Moore JO, Mayer RJ, Schiffer CA, Bloomfield CD. Adult patients with de novo acute myeloid leukemia and t(9;11)(p22;q23) have a superior outcome to patients with other translocations involving band 11q23: a cancer and leukemia group B study. Blood. 1997;90:4532–4538. [PubMed] [Google Scholar]
- 27.Rubnitz JE, Raimondi SC, Tong X, Srivastava DK, Razzouk BI, Shurtleff SA, Downing JR, Pui CH, Ribeiro RC, Behm FG. Favorable impact of the t(9;11) in childhood acute myeloid leukemia. J Clin Oncol. 2002;20:2302–2309. doi: 10.1200/JCO.2002.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Webb DK, Harrison G, Stevens RF, Gibson BG, Hann IM, Wheatley K. Relationships between age at diagnosis, clinical features, and outcome of therapy in children treated in the Medical Research Council AML 10 and 12 trials for acute myeloid leukemia. Blood. 2001;98:1714–1720. doi: 10.1182/blood.v98.6.1714. [DOI] [PubMed] [Google Scholar]
- 29.Daniel A, Arbor RDB, Michelle M, LeBeau, Brunangelo Falini, James S. Vardiman, Anna Porwit, Jurgen Thiele, Clara D. In: Bloomfield: Acute myeloid leukaemia with recurrent genetic abnormalities. Swerdlow SH, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. IARC Press; Lyons, France: 2007. pp. 114–115. [Google Scholar]
- 30.Spurbeck JL, Carlson RO, Allen JE, Dewald GW. Culturing and robotic harvesting of bone marrow, lymph nodes, peripheral blood, fibroblasts, and solid tumors with in situ techniques. Cancer Genet Cytogenet. 1988;32:59–66. doi: 10.1016/0165-4608(88)90312-3. [DOI] [PubMed] [Google Scholar]
- 31.Pardanani A, Ketterling RP, Brockman SR, Flynn HC, Paternoster SF, Shearer BM, Reeder TL, Li CY, Cross NC, Cools J, Gilliland DG, Dewald GW, Tefferi A. CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 2003;102:3093–3096. doi: 10.1182/blood-2003-05-1627. [DOI] [PubMed] [Google Scholar]
- 32.Dewald GW. In: Interphase FISH studies of chronic myeloid leukemia. Fan Y-S, editor. Humana Press; Totowa NJ: 2002. pp. 334–338. [DOI] [PubMed] [Google Scholar]
- 33.Brockman SR, Paternoster SF, Ketterling RP, Dewald GW. New highly sensitive fluorescence in situ hybridization method to detect PML/RARA fusion in acute promyelocytic leukemia. Cancer Genet Cytogenet. 2003;145:144–151. doi: 10.1016/s0165-4608(03)00061-x. [DOI] [PubMed] [Google Scholar]
- 34.Krauter J, Wagner K, Schafer I, Marschalek R, Meyer C, Heil G, Schaich M, Ehninger G, Niederwieser D, Krahl R, Buchner T, Sauerland C, Schlegelberger B, Dohner K, Dohner H, Schlenk RF, Ganser A. Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: individual patient data-based meta-analysis of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2009;27:3000–3006. doi: 10.1200/JCO.2008.16.7981. [DOI] [PubMed] [Google Scholar]
- 35.Pui CH, Gaynon PS, Boyett JM, Chessells JM, Baruchel A, Kamps W, Silverman LB, Biondi A, Harms DO, Vilmer E, Schrappe M, Camitta B. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359:1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 36.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes. FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–2402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 37.Knudson RA, Shearer BM, Ketterling RP. Automated duet spot counting system and manual technologist scoring using dual-fusion fluorescence in situ hybridization (D-FISH) strategy: comparison and application to FISH minimal residual disease testing in patients with chronic myeloid leukemia. Cancer Genet Cytogenet. 2007;175:8–18. doi: 10.1016/j.cancergencyto.2006.12.006. [DOI] [PubMed] [Google Scholar]


