Abstract
Molecular diagnostic tools capable of identifying Shiga toxin-specific genetic determinants in stool specimens permit an unbiased approach to detect Shiga toxin-producing Escherichia coli (STEC) in clinical samples and can indicate when culture-based isolation methods are required. It is increasingly recognized that clinically relevant STEC are not limited to the singular O157 serotypes, and therefore diagnostic assays targeting toxin-encoding determinants must be able to account for any genetic variation that exists between serotypes. In this study conventional PCR and four real-time PCR assays (HybProbe, TaqMan, SYBR Green, and LUX) targeting the stx1 and stx2 Shiga toxin coding sequences were used to identify STEC in enriched stool samples (n = 36) and a panel of O157 and non-O157 strains (n = 64). PCR assays targeting stx1 and stx2 had variable specificity and sensitivity values with enriched stool samples. Molecular assays using DNA from pure cultures revealed that some primers were not sensitive to all stx2 variants. This evaluation concluded that the TaqMan-based probes were most appropriate in high throughput clinical diagnostic laboratories in consideration of cost, turn around time, and assay performance.
Shiga toxin-producing Escherichia coli (STEC) that belong to serotype O157:H7 are recognized globally as important causative agents of enterocolitis food poisoning. Several prominent outbreaks of this STEC serotype have also gained significant public health attention, leading to the overall appreciation of O157:H7 as a major public health concern in the food production industry.1,2,3 STEC can lead to a wide array of clinical manifestations ranging from barely noticeable acute gastritis to bloody diarrhea, and occasionally to complications such as hemolytic uremic syndrome (HUS).4,5,6 These infections, hospitalizations, and subsequent complications have been shown to result in dramatic costs to both the health care system and the state infection control departments involved in their management.7 Typically, STEC infection is most severe in pediatric and elderly patients, whereas adults do not present with severe symptoms in most cases.8 Additionally, Shiga toxins, encoded by stx1 and stx2, are the primary virulence factor possessed by STEC and contribute to the manifestation of both bloody diarrhea and HUS.5 Although a great deal of attention (both in the general public and the scientific communities) has been focused on the O157:H7 serotype, several studies suggest that up to 50% of STEC illness is caused by serotypes other than O1579,10,11,12,13 of which there are over 100.14
O157 STEC are routinely screened for in the clinical microbiology laboratory by selective plating on sorbitol MacConkey media, which exploits the nonsorbitol fermenting phenotype of most O157 strains. As a result, O157 can readily be distinguished from normal E. coli flora. However, the recent identification of sorbitol fermenting O157 strains associated with HUS calls for additional screening methods to be used in addition to sorbitol MacConkey.15 BBL CHROMagar O157 (BD Canada, Mississauga, ON, Canada) plated medium is an alternative method for identifying O157 growth based on differential colorimetric colony growth.16 However, these plating methods only select for sorbitol fermenting strains of O157 or simply O157 strains of STEC, respectively, whereas commensal E. coli and non-O157 STEC are indistinguishable on both media and therefore require additional molecular diagnostic assays. To date, no widely used standard method for identifying non-O157 STEC infections is used by clinical diagnostic laboratories.
There are several research methods reported for the detection of non-O157 STEC, most of which focus on the detection of Shiga toxins or Shiga toxin-encoding genetic determinants. A standardized method for detecting the presence of toxin in stool involves filtration of stool for purification of toxin with subsequent incubation with Vero tissue culture cells. This assay requires a highly skilled technician to maintain the cell lines as well as monoclonal antibodies for confirmation of Shiga toxin. More importantly, the Vero cell assay is considered time and labor intensive, not for practical clinical diagnostic purposes. However, the Meridian Premier EHEC kit (Meridian Diagnostics, Inc., Cincinnati, OH) can detect Shiga toxin in stool samples via an immunoassay and can aid in the identification of non-O157 STEC. Alternatively, a myriad of PCR-based assays designed to detect stx1 and stx2 in E. coli has been described.6,17,18,19,20,21,22,23,24,25,26 Recently, the Centers for Disease Control and Prevention released updated guidelines for the detection of STEC in relation to acute community-acquired diarrhea, which included specific testing for Shiga toxins or their genetic determinants in addition to traditional culture (http://www.cdc.gov/mmwr/preview/mmwrhtml/rr5812a1.htm, updated Oct 16, 2009).
We compared four real-time PCR assays (two described in this study) as well as a conventional PCR method for the detection of STEC stx1 and stx2 genes. The assays were compared for sensitivity, specificity, detection threshold, as well as cost and time requirements. These methods were compared for their ability to detect various STEC serotypes from pure cultures as well as from stool specimens and optimized for the respective amplification platforms located in four different laboratories. Extraction of DNA from cultures or spiked stool samples was performed in one selective laboratory and distributed to the other laboratories for testing. The results of this study provide guidelines for diagnostics laboratories to adopt a methodology according to available platform and budget.
Materials and Methods
Enrichment of STEC in Stool Samples and DNA Extraction
A total of 36 stool samples previously identified27 to contain the following 41 STEC strains (co-infection was found in five samples) were included in the study: O157:H7 (n = 18); O26:H11 (n = 10); O121:H19 (n = 3); O26:NM (n = 1); O5:NM (n = 1); O111:NM (n = 1); O145:NM (n = 1); O103:H2 (n = 1); O177:NM (n = 1); O1:H7 (n = 1); O8:H19 (n = 1); O2:H4 (n = 1); and O25:H1 (n = 1).
An aliquot of 200 μl of watery stool or a “pea” size was inoculated into 5 ml of Trypticase Soy Broth and incubated at 37°C overnight. After incubation, 200 μl of the enriched culture was transferred into a 1.5-ml screw cap centrifuge tube, centrifuged for 3 minutes at 14,000 × g, and the supernatant was removed and the pellet was washed with 1 ml of 12 mmol/L Tris buffer, pH 7.4. After centrifuging for 3 minutes at 14,000 × g, the wash buffer was removed and the pellet was resuspended in 200 μl of rapid lysis buffer (100 mmol/L NaCl; 10 mmol/L Tris-HCl, pH 8.3; 1 mmol/L EDTA, pH 9.0; 1% Triton X-100) and boiled for 15 minutes. After centrifugation, the supernatant was removed and retained to be used as a template in PCR assays, and then subsequently stored at −70°C. For extraction control, an STEC-negative stool sample was spiked with an O157:H7 STEC (American Type Culture Collection 43895) at a concentration of 100 colony forming units/ml, and 200 μl was inoculated into 5 ml of Trypticase Soy Broth, incubated overnight, and extracted along with the clinical sample. A STEC-negative stool sample was also used as a negative extraction control. DNA prepared from O157:H7 STEC (American Type Culture Collection 43895) was used as positive control for all subsequent PCR assays, whereas water was included as the PCR negative control.
DNA was extracted from STEC cultures of known serotypes containing stx1 (n = 21), stx2 (n = 22), stx1 and stx2 (n = 14), and seven non-Shiga toxin-producing E. coli isolates by rapid lysis method. A subset of these isolates was obtained from the aforementioned Stx-positive stool samples. Single colony was touched and dispensed into 200 μl of lysis buffer (100 mmol/L NaCl; 10 mmol/L Tris-HCl, pH 8.3; 1 mmol/L EDTA, pH 9.0; 1% Triton X-100), boiled for 15 minutes, and the supernatant after centrifugation was used as template for PCR.
Real-Time and Conventional PCR Protocols for the Detection of the Stx Genes
Sets of primers and probes using HybProbe (TIB Molbiol, Adephia, NJ), TaqMan (Applied Biosystems Inc., Foster City, CA), SYBR Green (Invitrogen Life Science, Burlington, Ontario, Canada), and LUX (Invitrogen Life Science) real-time assays as well as conventional PCR for the detection of the stx genes on different amplification platforms were evaluated. Target location of primers and probes in respect to each other is illustrated in Figure 1, and coordinates are provided in Table 1. Oligonucleotide primers and fluorescent probes, conditions, and amplification platforms are described in Tables 1and 2. The previously published assays (LUX,28 [Light Upon Extension] conventional PCR,22 and HybProbe29) were performed without modification. The SYBR Green and LUX assays for stx1 and stx2 were each performed in separate reactions, whereas the conventional PCR, HybProbe, and TaqMan assays were performed as duplex assays in a single reaction, respectively. Duplex PCR assays were selected because they saved labor time in the set up as well as template DNA, reagents and consumables. For the detection of stx1 and stx2 by conventional PCR, amplified products were analyzed by electrophoresis in 1.5% (w/v) agarose gels running at 150 V for 45 minutes by using a 1 kb ladder (Invitrogen Life Science) for size reference. The predicted amplicon sizes for stx1 and stx2 are 614 bp and 779 bp, respectively.
Figure 1.
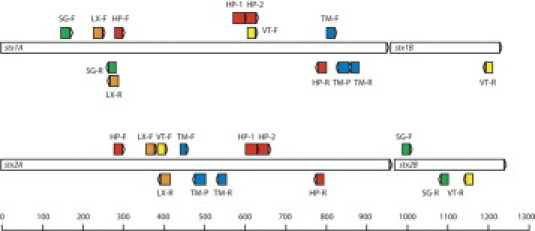
Schematic of PCR and real-time PCR oligonucleotide primers and probes in reference to the stx1 and stx2 loci. HP, HybProbe; LX, Light Upon eXtension; SG, SYBR Green; TM, TaqMan; and VT, conventional PCR primers.
Table 1.
Oligonucleotides Used in this Study
| Oligonucleotide | Target | Sequence | Coordinates |
|---|---|---|---|
| STEC-HP-F | stx1, stx2 | 5′-GARCRAAATAATTTATATGTG-3′ | 1352569–1352589 and (2996681–2996701) |
| STEC-HP-R | stx1, stx2 | 5′-TGATGATGRCAATTCAGTAT-3′ | (1353067–1353086) and 2996181–2996200 |
| STEC 1-HP-1 | stx1 | 5′-TTTACGTTTTCGGCAAATACAGAGGGGAT-[FAM]-3′ | (2996383–2996411) |
| STEC 1-HP-2 | stx1 | 5′-[Red 640]-TCGTACAACACTGGATGATCTCAGTGGG-Ph-3′ | (2996354–2996381) |
| STEC 2-HP-1 | stx2 | 5′-TCAGGCACTGTCTGAAACTGCTCCTGTGTA-[FAM]-3′ | 1352892–1352921 |
| STEC 2-HP-2 | stx2 | 5′-[Red 705]-ACCATGACGCCGGGAGACGTGGACCT-Ph-3′ | 1352923–1352948 |
| STX1-TM-F | stx1 | 5′-CATCGCGAGTTGCCAGAAT-3′ | (2996160–2996178) |
| STX1-TM-R | stx1 | 5′-GCGTAATCCCACGGACTCTTC-3′ | 2996101–2996121 |
| STX1-TM-P | stx1 | 5′-[FAM]-CTGCCGGACACATAGAAGGAAACTCATCA-[TAMRA]-3′ | 2996125–2996153 |
| STX2-TM-F | stx2 | 5′-CCGGAATGCAAATCAGTC-3′ | 1352732–1352749 |
| STX2-TM-R | stx2 | 5′-CAGTGACAAAACGCAGAACT-3′ | (1352826–1352845) |
| STX2-TM-P | stx2 | 5′-[VIC]-ACTGAACTCCATTAACGCCAGATATGA-[TAMRA]-3′ | (1352767–1352793) |
| STX1-SG-F | stx1 | 5′-CATTACAGACTATTTCATCAGGAGGTA-3′ | (2996809–2996835) |
| STX1-SG-R | stx1 | 5′-TCGTTCAACAATAAGCCGTAGATTA-3′ | 2996696–2996720 |
| STX2-SG-F | stx2 | 5′-GCGGTTTTATTTGCATTAGC-3′ | 1353279–1353298 |
| STX2-SG-R | stx2 | 5′-TCCCGTCAACCTTCACTGTA-3′ | (1353374–1353393) |
| LX1-FDOB66 | stx1 | 5′-cggctATTATTTCGTTCAACAATAAGCcG (Alexa 546)-3′ | 2996690–2996713 |
| LX1-RDOB67 | stx1 | 5′-CAGAGGGATAGATCCAGAGGAAGG-3′ | (2996730–2996753) |
| LX2-FGIL290 | stx2 | 5′-cggacaCAGAGTGGTATAACTGCTGTCcG (FAM)-3′ | (1352684–1352706) |
| LX2-RGIL291 | stx2 | 5′-ATATCAGTGCCCGGTGTGACAA-3′ | 1352647–1352668 |
| VT1F | stx1 | 5′-ACACTGGATGATCTCAGTGG-3′ | (2996355–2996374) |
| VT1R | stx1 | 5′-CTGAATCCCCCTCCATTATG-3′ | 2995773–2995792 |
| VT2F | stx2 | 5′-CCATGACAACGGACAGCAGTT-3′ | 1352675–1352695 |
| VT2R | stx2 | 5′-CCTGTCAACTGAGCACTTTG-3′ | (1353435–1353454) |
The coordinates of the primers and probes are described in reference to GenBank accession number AE005174.2 (Escherichia coli O157:H7 EDL933 complete genome), and sequences that match the complement strand are indicated in brackets.
FAM, 6-carboxyfluorescein; TAMRA, Tetramethyl-6-Carboxyrhodamine; Red 640, LightCycler-Red 640-N-hydroxy-succinimide ester; Red 705, LightCycler-Red 705-N-hydroxy-succinimide ester; Ph, 3′-phosphate; O, C-Phosphorothioate; F, A-Phosphorothioate; E, G-Phosphorothioate; R, IUB code for A or G (wobble base). Lowercase bases at the 5′ end indicate those required for LUX primer hairpin formation and are not present in the target sequence; the penultimate 3′ base is tagged with the fluorescent molecule indicated in brackets.
Table 2.
Thermocycling Parameters and Instrumentation for the PCR Methods
| Method | Instrumentation | PCR conditions |
|---|---|---|
| HybProbe real-time PCR29 | Roche LightCycler version 1.5 | 95°C/10 minutes; 40 cycles of 95°C/10 seconds; 50°C/20 seconds; 72°C/30 seconds |
| TaqMan real-time PCR | ABI Prism 7000 SDS | 95°C/10 minutes; 40 cycles of 95°C/15 seconds; 60°C/1 minute; hold at 25°C/1 minute |
| SYBR Green real-time PCR | Stratagene MX4000 | 95°C/10 minutes; 40 cycles of 95°C/30 seconds; 55°C/1 minute; 72°C/30 seconds |
| LUX real-time PCR28 | Cepheid SmartCycler | 95°C/3 minutes; 40 cycles of 95°C/10 seconds; 55°C/15 seconds; 72°C/15 seconds |
| Conventional PCR22 | ABI GeneAmp 9600 | 95°C for 5 minutes; 40 cycles of 94°C/40 seconds; 56°C/40 seconds and 72°C/40 seconds; final extension at 72°C/6 minutes |
An amplification curve is indicative of a positive result for the LUX reaction. For the TaqMan assay, a cycle threshold of 0.2 was used as signal cut-off for inferring a positive amplification result. Both SYBR Green as well as HybProbe real-time PCR used amplification and melting curves for the analysis for both assays. For SYBR Green, the expected melting temperature (Tm) of stx1 is 86.5°C and 83.5°C for stx2. The expected Tm values for stx1 in the HybProbe assay are 62°C, 65°C, or 68°C, depending on the gene variant. Amplicons generated by stx2 variants have Tm values of 51°C, 55°C, 62°C, 63°C, and 65°C.
Specificity and Sensitivity Determination
In this study, a true positive was defined as a sample that yielded a consensus of three positive results for the five tested methods. A true negative was defined similarly as a sample that yielded at least three negative results for the five tested methods.
End-Point Detection Level of the Different PCR Assays
For end-point detection thresholds for the stx amplification assays, total genomic DNA was isolated from liquid cultures of E. coli O157:H7, O26:H11, O121:H19, and O111:NM strains as previously described,28 and the DNA was 10-fold serially diluted from 33 ng/μL to 0.33 fg/μL. A standard amount of DNA was used in each of the assays, and the end point was determined as the last dilution showing a positive result.
Calculation of Cost and Time Requirement for Each Assay
The cost based on 25 samples as per routine submission in our laboratory was calculated in Canadian dollars (CAD) for materials and consumables without taking into account the cost of labor because of the differences in salary in each facility. One hour and 10 minutes was added to each assay in which the following steps were included: sample setup, loading onto each of the dedicated platform, and analysis of results and reporting. Another 20 minutes was added to conventional PCR for preparation of agarose gels and sample loading. Sample lysis and template preparation processes were not included in the total time because the protocols used vary in different laboratories. All of the calculations were based on the availability of a single amplification platform, and these platforms are listed in Table 2.
Results
Evaluation of PCR Platforms
PCR assays examining for Shiga toxin genetic determinants in enriched stool specimens are summarized in Table 3. All these stool samples had been previously identified27 to contain positive cultures of the following: O157:H7; O26:H11; O121:H19; O26:NM; O5:NM; O111:NM; O145:NM; O103:H2; O177:NM; O1:H7; O8:H19; O2:H4; and O25:H1.
Table 3.
Sensitivity and Specificity Using Enriched Stool Specimens
| Conventional |
HybProbe |
TaqMan |
SYBR Green |
LUX |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of stool samples | Stx genotype | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 |
| 14 | stx1 | 14/14 | 0/14 | 13/14 | 0/14 | 14/14 | 0/14 | 14/14 | 0/14 | 14/14 | 0/14 |
| 17 | stx1/stx2 | 15/17 | 16/17 | 15/17 | 16/17 | 17/17 | 17/17 | 17/17 | 15/17 | 17/17 | 17/17 |
| 5 | stx2 | 1/5 | 5/5 | 0/5 | 5/5 | 0/5 | 5/5 | 1/5 | 5/5 | 0/5 | 5/5 |
| Sensitivity, % | 94 | 96 | 91 | 96 | 100 | 100 | 100 | 92 | 100 | 100 | |
| Specificity, % | 84 | 100 | 100 | 100 | 100 | 100 | 84 | 100 | 100 | 100 | |
Only the TaqMan and LUX assays showed 100% sensitivity and specificity for both stx1 and stx2. Amplification of stx1 by conventional PCR, SYBR Green, and HybProbe each showed a sensitivity of 94%, 100%, and 91%, and a specificity of 84%, 84%, and 100%, respectively (Table 3). With stx2 amplification using conventional PCR, SYBR Green, and HybProbe, the sensitivity was at 96%, 92%, and 96%, respectively, but the specificity was at 100% by the three assays (Table 3).
PCR results on the panel of O157 and non-O157 E. coli are summarized in Table 4. Amplification of both stx1 and stx2 genes by conventional PCR, TaqMan, and LUX assays showed 100% sensitivity and specificity. The sensitivity of both SYBR Green and HybProbe assays for stx1 was at 97% with 100% specificity. As for the amplification of stx2 using SYBR Green assay, the sensitivity was at 81% with a specificity of 100%, whereas the HybProbe assay has a sensitivity and specificity of 100%.
Table 4.
Sensitivity and Specificity Using Pure Cultures
| Conventional |
HybProbe |
TaqMan |
SYBR Green |
LUX |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of cultures | Stx genotype | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 |
| 7 | Negative | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 |
| 21 | stx1 | 21/21 | 0/21 | 20/21 | 0/21 | 21/21 | 0/21 | 20/21 | 0/21 | 21/21 | 0/21 |
| 14 | stx1/stx2 | 14/14 | 14/14 | 14/14 | 14/14 | 14/14 | 14/14 | 14/14 | 13/14 | 14/14 | 14/14 |
| 22 | stx2 | 0/22 | 22/22 | 0/22 | 22/22 | 0/22 | 22/22 | 0/22 | 16/22 | 0/22 | 22/22 |
| Sensitivity, % | 100 | 100 | 97 | 100 | 100 | 100 | 97 | 81 | 100 | 100 | |
| Specificity, % | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
For templates prepared from O26:H11, O113:H21, O177:NM, O6:H34, O85:H1, and O91:H21 strains encoding stx2, the SYBR Green assay did not produce an amplification curve.
End-Point Detection Threshold of PCR Platforms
The detection threshold of each of the PCR-based stx detection methods was determined by using diluted DNA template prepared from pure cultures of O157:H7, O26:H11, O121:H19, and O111:NM strains encoding different combinations of stx1 and/or stx2. Results in Table 5 showed the end-point detection of each assay in the dilution series. The TaqMan assay detected the lowest concentration of template DNA for both stx1 and stx2 at a dilution of 10−8 (0.99 fg/reaction) and 10−7 (9.9 fg/reaction). The detection threshold of the conventional PCR assay was the lowest in comparison with all of the real-time PCR assays. For conventional PCR, the detection levels were at 9.9 ng/reaction for stx1 and 0.99 to 9.9 ng/reaction for stx2. End-point titration results of the different assays varied from each other, but no significant difference was observed within the assay from one to another strain.
Table 5.
End-Point Detection Threshold for Stx1 and Stx2 Genes for Conventional and Real-Time PCR Platforms
| Conventional |
HybProbe |
TaqMan |
SYBR Green |
LUX |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Template | Stx genotype | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 | stx1 | stx2 |
| O157:H7 | stx1, stx2 | 10−1 | 10−1 | 10−5 | 10−5 | 10−8 | 10−7 | 10−6 | 10−6 | 10−7 | 10−6 |
| O26:H11 | stx1 | 10−1 | — | 10−4 | — | 10−8 | — | 10−6 | — | 10−7 | — |
| O121:H19 | stx2 | — | 10−2 | — | 10−4 | — | 10−7 | — | 10−6 | — | 10−7 |
| O111:NM | stx1, stx2 | 10−1 | 10−2 | 10−4 | 10−4 | 10−8 | 10−7 | 10−7 | 10−5 | 10−6 | 10−7 |
Tenfold dilutions of DNA at a concentration of 33 ng/μL were made from 10−1 to 10−9. —, not performed.
Cost of Each Assay on Its Dedicated Amplification Platforms
The cost and time requirement to perform each amplification assay is shown in Table 6. HybProbe real-time PCR assay was performed by using the Roche LightCycler (Laval, Quebec, Canada) version 1.5, which has a capacity of running 32 samples. The cost was the highest at $196.11 CAD for 25 samples tested. The performance of LUX real-time PCR on the Cepheid SmartCycler (Sunnyvale, CA) had the second highest cost at $138.08 CAD. This assay cannot be performed as a multiplex and this instrument can accommodate 16 samples at one time. Consequently, four runs of amplification were required to complete testing 25 samples and thus increasing the cost and turn around time of the assay. The remaining three platforms [(ABI GeneAmp 9600, Applied Biosystems Inc., Foster City, CA) ABI Prism 7000 SDS (Applied Biosystems Inc.), and Stratagene MX4000 (Stratagene, Santa Clara, CA)] for conventional PCR, TaqMan, and SYBR Green real-time PCR have the capability of running 96 samples each run. Conventional PCR was the cheapest based on materials and consumables, but additional hands-on time was required for the agarose gel analysis and thus increasing the labor cost.
Table 6.
Cost and Time for Conventional and Real-Time PCR Assays for Stx1 and Stx2 Amplification
| Method | Cost for 25 samples | Turn around time |
|---|---|---|
| Conventional PCR | C$53.35 | 4 hours and 10 minutes |
| TaqMan real-time PCR | C$58.94 | 3 hours and 30 minutes |
| LUX real-time PCR | C$138.08 | 4 hours and 10 minutes |
| HybProbe real-time PCR | C$196.11 | 2 hours |
| SYBR Green real-time PCR | C$73.30 | 2 hour and 40 minutes |
Discussion
Non-O157 STEC have been associated with disease outbreaks worldwide, with select serotypes (such as O26:H11, O103:H2, O103:H25, O111:NM, O121:H19, and O145:NM) associated with severe complications such as HUS.30 Due to clinical screening limitations in many countries, the true worldwide incidence of STEC is not known. Current methods used in most clinical microbiology laboratories rely on classical culture-based techniques using BBL CHROMagar O157 (BD Canada) giving a characteristic color for O157 STEC or sorbitol MacConkey plates to identify only O157 sorbitol nonfermenting STEC. Molecular diagnostic assays such as PCR do not rely on phenotypic traits, but rather screen for the genetic determinants that define STEC. As a result of this broad genotypic screen, patients' stool samples containing non-O157 STEC can be readily identified that otherwise would have gone undetected by using only classical phenotypic-dependent culture methods. With the increasing number of assays and platforms available for amplification detection of stx1 and stx2 genes, it is of interest to compare the multiple methods for their ability to accurately identify the presence of these genes in clinical isolates and specimens.
When DNA extracts from enriched stool culture were used as templates, TaqMan and LUX methods identified every sample accurately, whereas conventional PCR, SYBR Green, and HybProbe assays were found to have a lower specificity and sensitivity. Evaluation of these PCR methods with the broader panel of non-O157 STEC strains indicated that the SYBR Green assay could not detect some stx2 variant alleles due to polymorphisms in the primer binding sites. The forward primer STX2-SG-F has a polymorphic site at the 3′ end compared with the corresponding primer binding site in the O26:H11 strain (GenBank accession number DQ143181), and this variation likely accounted for the amplification failure. The corresponding region of stx2 was sequenced from the non-O157 strains with a false negative SYBR Green real-time PCR result, and all had multiple point mutations in the SYBR Green primer binding sites (data not shown). It is therefore recommended that laboratories do not use primers that target variable regions of the stx loci. This should greatly improve the sensitivity for the SYBR Green assay.
From the examined real-time PCR chemistries used in this study, the TaqMan assay performed on the ABI Prism 7000 SDS is recommended because of the high degree of sensitivity and specificity for both stool culture and pure culture extracts, the ability to duplex both stx1 and stx2 probes into a single reaction, and a higher confidence level due to the use of probes in combination with primers. Like the TaqMan assay, the LUX assay on the Cepheid SmartCycler also provided 100% specificity and sensitivity for both stool samples and cultures. The LUX assay also employs a combination of primers and probes, but unlike the TaqMan assay, this real-time reaction could not be performed as a duplex assay (data not shown). Furthermore, the Cepheid SmartCycler platform has a capacity of running 16 amplification assays in one run as compared with the capacity of 96 samples per run in the ABI Prism 7000 SDS. Consequently, the reagent costs and turn around time would be higher under these circumstances. However, the Cepheid SmartCycler provides a potential advantage to a low volume testing laboratory with a wide variety of testing targets as this platform has 16 independent units, which allow various assays with different cycling parameters to be performed simultaneously. Therefore, there is an advantage of using this platform in a low volume testing laboratory with wide varieties of testing menu.
The TaqMan assay was second only to conventional PCR in terms of lowest cost for testing 25 samples. Conventional PCR, however, has the inherent disadvantage of requiring gels for analysis, which not only requires additional technical handling for analysis but also results in further expenses associated with the disposal of ethidium bromide waste (not reflected in the calculated price in Table 6) and the handling of a potential carcinogenic agent in the laboratory. An alternative to the ethidium bromide is the precast nonethidium Lonza FlashGel system at a cost of $21.00, which can accommodate up to 32 samples including positive and negative controls and size standards. This replacement will increase the cost of the conventional PCR test. As well, conventional PCR did not have 100% sensitivity and specificity for stool samples as was achieved with the TaqMan assay.
The HybProbe and SYBR Green assays had given the shortest turn around time for reporting; however, SYBR Green had the lowest sensitivity and specificity values of the tested methods, and HybProbe is nearly four times more expensive than conventional PCR and TaqMan, making it the least favorable method. At the time of this study, the TaqMan assay was performed by using the ABI 7000 SDS, and the total time for the total amplification program requires approximately 2 hours (3.5 hours including extraction time). Taking into account the extra labor cost for the preparation of agarose gel and loading of the amplified products for using conventional PCR for STEC testing, this makes the TaqMan assay the cheapest test.
The assays for detecting STEC demonstrate the effectiveness of multiple molecular methods for the detection of STEC from various serotypes lineages directly from clinical stool enrichment cultures, and will be useful to direct culture-based isolations of STEC strains. This study also provides the framework for which individual diagnostic centers can adopt real-time PCR assays for stx genes detection according to their available platforms and budgets.
Acknowledgements
We thank Joanne McCrea and Dobryan Tracz for their technical help, and Dr. Lourens Robberts for his valuable feedback.
Footnotes
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.CDC Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach, United States. MMWR, Morb Mortal Wkly Rep. 2006;55(38):1045–1046. [PubMed] [Google Scholar]
- 2.Currie A, MacDonald J, Ellis A, Siushansian J, Chui L, Charlebois M, Peermohamed M, Everett D, Fehr M, Ng LK. Outbreak of Escherichia coli 0157:H7 infections associated with consumption of beef donair. J Food Prot. 2007;70(6):1483–1488. doi: 10.4315/0362-028x-70.6.1483. [DOI] [PubMed] [Google Scholar]
- 3.Denny J, Bhat M, Eckmann K. Outbreak of Escherichia coli O157:H7 associated with raw milk consumption in the Pacific Northwest. Foodborne Pathog Dis. 2008;5(3):321–328. doi: 10.1089/fpd.2007.0072. [DOI] [PubMed] [Google Scholar]
- 4.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 2004. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. 1985. J Infect Dis. 2004;189(3):556–563. doi: 10.1086/jid/189.3.566. [DOI] [PubMed] [Google Scholar]
- 5.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paton AW, Paton JC. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol. 2002;40(1):271–274. doi: 10.1128/JCM.40.1.271-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts JA, Upton PA, Azene G. Escherichia coli O157:H7; an economic assessment of an outbreak. J Public Health Med. 2000;22(1):99–107. doi: 10.1093/pubmed/22.1.99. [DOI] [PubMed] [Google Scholar]
- 8.Eklund M, Leino K, Siitonen A. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J Clin Microbiol. 2002;40(12):4585–4593. doi: 10.1128/JCM.40.12.4585-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fey PD, Wickert RS, Rupp ME, Safranek TJ, Hinrichs SH. Prevalence of non-O157:H7 shiga toxin-producing Escherichia coli in diarrheal stool samples from Nebraska. Emerg Infect Dis. 2000;6(5):530–533. doi: 10.3201/eid0605.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelacic JK, Damrow T, Chen GS, Jelacic S, Bielaszewska M, Ciol M, Carvalho HM, Melton-Celsa AR, O'Brien AD, Tarr PI. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J Infect Dis. 2003;188(5):719–729. doi: 10.1086/376999. [DOI] [PubMed] [Google Scholar]
- 11.Lockary VM, Hudson RF, Ball CL. Shiga toxin-producing Escherichia coli, Idaho. Emerg Infect Dis. 2007;13(8):1262–1264. doi: 10.3201/eid1308.070189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning SD, Madera RT, Schneider W, Dietrich SE, Khalife W, Brown W, Whittam TS, Somsel P, Rudrik JT. Surveillance for Shiga toxin-producing Escherichia coli, Michigan, 2001–2005. Emerg Infect Dis. 2007;13(2):318–321. doi: 10.3201/eid1302.060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson LH, Giercke S, Beaudoin C, Woodward D, Wylie JL. Enhanced surveillance of non-O157 verotoxin-producing Escherichia coli in human stool samples from Manitoba. Can J Infect Dis Med Microbiol. 2005;16(6):329–334. doi: 10.1155/2005/859289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis. 2006;43(12):1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 15.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect Immun. 2008;76(12):5598–5607. doi: 10.1128/IAI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Church DL, Emshey D, Semeniuk H, Lloyd T, Pitout JD. Evaluation of BBL CHROMagar O157 versus sorbitol-MacConkey medium for routine detection of Escherichia coli O157 in a centralized regional clinical microbiology laboratory. J Clin Microbiol. 2007;45(9):3098–3100. doi: 10.1128/JCM.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastian SN, Carle I, Grimont F. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res Microbiol. 1998;149(7):457–472. doi: 10.1016/s0923-2508(98)80001-6. [DOI] [PubMed] [Google Scholar]
- 18.Belanger SD, Boissinot M, Menard C, Picard FJ, Bergeron MG. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the smart cycler. J Clin Microbiol. 2002;40(4):1436–1440. doi: 10.1128/JCM.40.4.1436-1440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beutin L, Jahn S, Fach P. Evaluation of the “GeneDisc” real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O145, and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J Appl Microbiol. 2009;106(4):1122–1132. doi: 10.1111/j.1365-2672.2008.04076.x. [DOI] [PubMed] [Google Scholar]
- 20.Bischoff C, Luthy J, Altwegg M, Baggi F. Rapid detection of diarrheagenic E. coli by real-time PCR. J Microbiol Methods. 2005;61(3):335–341. doi: 10.1016/j.mimet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Chassagne L, Pradel N, Robin F, Livrelli V, Bonnet R, Delmas J. Detection of stx1, stx2, and eae genes of eterohemorragic Escherichia coli using SYBR Green in a real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;64(1):106–109. doi: 10.1016/j.diagmicrobio.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 22.Gannon VP, King RK, Kim JY, Thomas EJ. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58(12):3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J Clin Microbiol. 2008;46(5):1752–1757. doi: 10.1128/JCM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen EM, Andersen MT. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J Clin Microbiol. 2003;41(7):2884–2893. doi: 10.1128/JCM.41.7.2884-2893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40(10):3613–3619. doi: 10.1128/JCM.40.10.3613-3619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziebell KA, Read SC, Johnson RP, Gyles CL. Evaluation of PCR and PCR-RFLP protocols for identifying Shiga toxins. Res Microbiol. 2002;153(5):289–300. doi: 10.1016/s0923-2508(02)01322-0. [DOI] [PubMed] [Google Scholar]
- 27.Gilmour MW, Chui L, Chiu T, Tracz DM, Hagedorn K, Tschetter L, Tabor H, Ng LK, Louie M. Isolation and detection of Shiga toxin-producing Escherichia coli in clinical stool samples using conventional and molecular methods. J Med Microbiol. 2009;58:905–911. doi: 10.1099/jmm.0.007732-0. [DOI] [PubMed] [Google Scholar]
- 28.Gilmour MW, Tracz DM, Andrysiak AK, Clark CG, Tyson S, Severini A, Ng LK. Use of the espZ gene encoded in the locus of enterocyte effacement for molecular typing of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 2006;44(2):449–458. doi: 10.1128/JCM.44.2.449-458.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reischl U, Youssef MT, Kilwinski J, Lehn N, Zhang WL, Karch H, Strockbine NA. Real-time fluorescence PCR assays for detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J Clin Microbiol. 2002;40(7):2555–2565. doi: 10.1128/JCM.40.7.2555-2565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karmali MA. Host and pathogen determinants of verocytotoxin-producing Escherichia coli-associated hemolytic uremic syndrome. Kidney Int. 2009;Suppl 112:S4–S7. doi: 10.1038/ki.2008.608. [DOI] [PubMed] [Google Scholar]


