Abstract
Novel mutations in the isocitrate dehydrogenase 1 (IDH1) and 2 (IDH2) genes have been identified in a large proportion of diffuse gliomas. Tumors with IDH1/2 mutations have distinctive clinical characteristics, including a less aggressive course. The aim of this study was to develop and evaluate the performance of a novel real-time PCR and post-PCR fluorescence melting curve analysis assay for the detection of IDH1 and IDH2 mutations in routine formalin-fixed, paraffin-embedded tissues of brain biopsies. Using the established assay, we tested 67 glial neoplasms, 57 non-neoplastic conditions that can often mimic gliomas (eg, radiation changes, viral infections, infarctions, etc), and 44 noncentral nervous system tumors. IDH1 and IDH2 mutations were detected in 72% of lower grade diffuse gliomas and in 17% of glioblastomas. The IDH1 mutation was the most common (93%), with the most frequent subtype being R132H (88%). These mutations were not identified in non-neoplastic glioma mimickers and in noncentral nervous system tumors including thyroid carcinomas. The results of this assay had a 100% correlation with the results obtained by conventional sequencing. In summary, we report here the real-time PCR/fluorescence melting curve analysis assay that provides rapid and sensitive detection of IDH mutations in formalin-fixed, paraffin-embedded tissues, and is therefore useful as a powerful adjunct diagnostic tool for refining histopathological diagnosis of brain lesions and guiding patient management.
Recently, mutations in the isocitrate dehydrogenase enzyme isoform 1 (IDH1) and 2 (IDH2) genes have been identified in a large proportion of diffusely infiltrative gliomas.1,2,3 Wild-type IDH1 and IDH2 encode isozymes that catalyze the oxidative carboxylation of isocitrate to α-ketoglutarate, in the process yielding NADPH.4,5 Mutations in both genes are restricted to analogous codons, in exon 4 at codon 132 of the IDH1 gene and exon 4 at codon 172 of the IDH2 gene. These heterozygous mutations alter the arginine residue that normally binds to isocitrate, thereby inhibiting normal enzyme activity in a dominant-negative fashion. The mutant enzymes also gain a novel catalytic ability to produce 2-hydroxyglutarate.2,5,6
IDH1 and IDH2 mutations are found in 60% to 90% of World Health Organization grades 2 and 3 astrocytomas and oligodendrogliomas, as well as in secondary glioblastomas (GBMs) that developed from these lower-grade tumors. Interestingly, although mutations in these genes are unusual in primary GBMs, they are completely absent in pilocytic astrocytomas2,7,8 and are not found in non-neoplastic conditions that can often mimic gliomas (eg, radiation changes, viral infections, infarctions, etc). Furthermore, IDH1/2 mutations are fairly specific for gliomas compared with non-central nervous system (CNS) solid tumors, especially those that frequently metastasize to the brain.1,3 Parenthetically, a few nonglial neoplasms have been found to be positive for IDH1 mutations, including some acute myeloid leukemias (9%),9 rare prostate carcinomas, and a few B-cell acute lymphoblastic leukemias.10 In addition to diagnostic value, IDH1/2 mutations have been associated with better outcome and longer survival in patients with low-grade diffuse gliomas, anaplastic astrocytomas, and GBMs, and have been shown to be a powerful independent prognostic factor for prolonged survival.1,2,11,12 Together, these features make IDH1 and IDH2 mutation screening important for both diagnostic and prognostic purposes.
LightCycler (Roche Applied Science, Indianapolis, IN) real-time RT-PCR utilizes fluorescence resonance energy transfer probes, which bind to the PCR product in a head-to-tail fashion. When these two probes bind to the specific PCR product, their fluorophores come into close proximity, allowing energy transfer from a donor to an acceptor fluorophore. This produces an increase in fluorescence that is proportional to the amount of amplified product. Post-PCR fluorescence melting curve analysis (FMCA) exploits the fact that even a single bp mismatch between the labeled probe and the sequence of interest will significantly reduce the specific melting temperature. For example, if no mutation is present in the sample DNA, the probes will bind perfectly and melt at a higher temperature (Tm), showing a single peak on post-PCR FMCA. In contrast, if a heterozygous point mutation is present, probes will bind to the mutant amplicon imperfectly and will melt (dissociate) at a lower temperature. This produces two melting peaks, a lower Tm peak for the mutant allele, and a higher Tm peak for the wild-type allele. Because each nucleotide substitution produces a distinct melting peak at a specific Tm, all possible mutation subtypes are detectable with a single pair of primers and probes.
We previously reported a nucleotide Sanger sequencing method that allows detection of IDH1 and IDH2 mutations in formalin-fixed, paraffin-embedded (FFPE) tissue, even in specimens where glioma was difficult to recognize via light microscopy.13 Although we have used this method for routine clinical testing of biopsy samples, real-time PCR and post-PCR FMCA represents a new generation in clinical testing and allows for fast and reliable detection of mutations in both frozen and FFPE tissues, with higher sensitivity than Sanger sequencing.14 In this study, we report a simple and accurate real-time PCR and post-PCR FMCA assay for detection of IDH1/2 mutations in routine FFPE brain biopsies.
Materials and Methods
Tissue Samples
A total of 168 FFPE tissue samples were used for method development and validation, including 67 malignant gliomas and 57 non-neoplastic brain lesions that can often mimic gliomas (Table 1). In addition, 44 non-CNS tumors were studied (Table 1). All archival surgical gliomas were re-evaluated (by C.H.) to ensure that grading was according to World Health Organization 2007 criteria.15 Blocks that contained adequate tissue were sectioned at a thickness of 5 μm. Sections were stained with H&E to confirm that diagnostic tissue had been retained. All molecular analyses and interpretations were performed while blinded to diagnosis.
Table 1.
Prevalence of IDH1 and IDH2 Mutations in Gliomas, Non-Neoplastic Glioma Mimickers, and Non-CNS Tumors Detected by FMCA Assay
|
IDH1 |
IDH2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Diagnosis | No. | R132H | R132S | R132C | R132G | R132L | R172M | % positive |
| Gliomas | 67 | 37 | 2 | 1 | 1 | 1 | 3 | 67 |
| A2 | 17 | 9 | 0 | 0 | 1 | 0 | 0 | 59 |
| O | 24 | 19 | 0 | 0 | 0 | 0 | 3 | 92 |
| OA | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 50 |
| AA | 10 | 4 | 0 | 0 | 0 | 0 | 0 | 40 |
| AO | 8 | 5 | 1 | 1 | 0 | 0 | 0 | 88 |
| GBM | 6 | 0 | 0 | 0 | 0 | 1 | 0 | 17 |
| Non-neoplastic glioma mimickers | 57 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ischemia/infarct | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Reactive gliosis around metastatic tumor | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Reactive gliosis, idiopathic | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Abscess | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vasculopathy/malformation | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Viral or toxoplasma encephalitis | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Demyelination | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Inflammation NOS | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Radiation-induced damage | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vasculitis | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PML | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hippocampal sclerosis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Shunt-induced reactive gliosis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Non-CNS tumors | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCC | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CRC | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PTC, FV | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FTC | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ATC/PDCA | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (% of all IDH1/2 mutations) | 168 | 37 (82) | 2 (4) | 1 (2) | 1 (2) | 1 (2) | 3 (7) | |
A total of 168 FFPE biopsies were assessed for IDH1/2 mutations, including 67 gliomas, 57 non-neoplastic glioma mimickers, and 44 non-CNS neoplasms. A total of 45 (67%) gliomas were positive for mutations, most commonly R132H on IDH1. A2, diffuse astrocytoma, World Health Organization grade 2; O, oligodendroglioma; OA, oligoastrocytoma; AA, anaplastic astrocytoma; AO, anaplastic oligodendroglioma; GBM, glioblastoma; HCC, hepatocellular carcinoma; CRC, colorectal adenocarcinoma; PTC, FV, papillary thyroid carcinoma, follicular variant; FTC, follicular thyroid carcinoma; ATC/PDCA, anaplastic thyroid carcinoma/poorly-differentiated carcinoma.
DNA Isolation
Tumor targets were manually microdissected from the 5 μm unstained histological sections under the guidance of a corresponding H&E slide by using an Olympus SZ61 stereo microscope (Olympus, Hamburg, Germany). DNA was isolated from each target more than 2 mm in diameter with the DNeasy Blood and Tissue kit on the automated QIAcube (Qiagen, Valencia, CA) instrument or, for targets less than 2 mm, manual DNA isolation was performed by using the QIAamp DNA Micro kit (Qiagen). Both kits were used according to the manufacturer's instructions. The quantity of isolated DNA was assessed by using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Real-Time PCR and FMCA Assay
Detection of IDH1 and IDH2 mutations was performed by using real-time PCR and post-PCR FMCA on the LightCycler (Roche Applied Science). A pair of primers flanking each mutation site, along with two fluorescent probes with the sensor probe spanning the mutational “hot spot,” was designed by using LightCycler Design Software 2.6 (Roche Applied Science; Figure 1). The probes were complementary to the wild-type IDH1 and IDH2 sequences. This design allows detection of all possible mutation subtypes with a single pair of primers and probes, and is based on a difference in Tm between wild-type and mutant amplicons. Each mutation subtype has a distinct Tm, reflecting the thermodynamic stability of both complementary and mismatched probe-target duplexes. If no mutation is present, the probes bind perfectly to the sample DNA and dissociate at a higher Tm, showing a single peak on post-PCR FMCA. In contrast, if a heterozygous point mutation is present, probes will bind to the mutant amplicon imperfectly and dissociate at a lower Tm. This produces two melting peaks, including a lower peak for the mutant allele and a higher peak for the wild-type allele (Figure 2). For IDH1 mutation detection, forward primer (5′-ACGGTCTTCAGAGAAGC-3′), reverse primer (5′-GGTGTAGATACCAAAAGATAAGAAT-3′), and two probes (5′-LC640-ATGATAGGTTTTACCCATCCACTCACAAGC-3′ and 5′-ATCCCCATAAGCATGACGACCTA-FL-3′) were used to generate a 202-bp PCR product. Similarly, for IDH2 detection, forward primer (5′-TGCAGTGGGACCACTATTATC-3′), reverse primer (5′-CTTGACACCACTGCCATC-3′), and two probes (5′-LC640-TGATGGGCTTGGTCCAGCCAGGG-3′ and 5′-TGGGCGTGCCTGCCAATG-FL-3′) were used to generate a 360-bp PCR product. All primers and probes were obtained from TIB Molbiol (Berlin, Germany).
Figure 1.
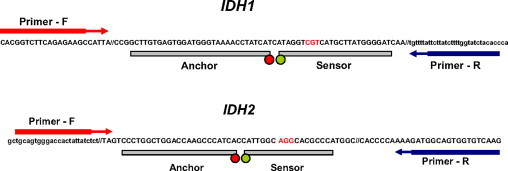
Primer and probe design for the real-time PCR and post-PCR FMCA assay for detection of IDH1 and IDH2 mutations. Probes are designed to be complementary to IDH1 and IDH2 wild-type sequences, which allow detection of all mutational subtypes by using a single pair of primers and probes.
Figure 2.
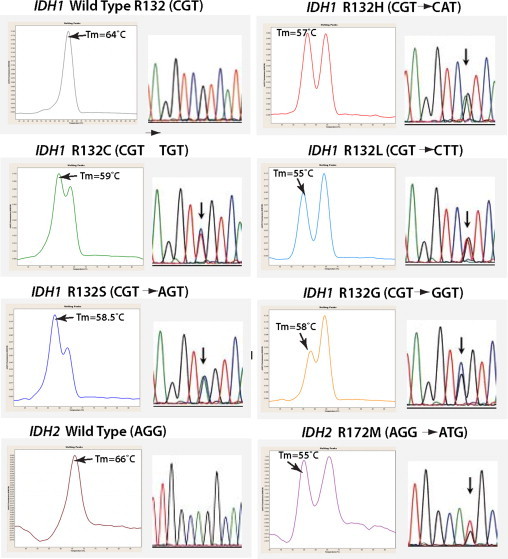
LightCycler FMCA detection of IDH1 and IDH2 point mutations based on distinct Tm of duplexes formed between the wild-type probe and either wild-type (wt) or mutant sequences. For IDH1 locus, the wt sequence Tm was 64°C, the R132H mutation Tm was 57°C, the R132C mutation Tm was 59°C, the R132L mutation Tm was 55°C, the R132S mutation Tm was 58.5°C, and the R132G mutation Tm was 58°C. For IDH2, the wt Tm was 66°C, and the R172M mutation Tm was 55°C. All mutation types were confirmed by conventional sequencing. Arrows indicate mutation sites.
Amplification was performed in a glass capillary tube by using 5 to 50 ng of DNA in a 20-μl reaction volume. In detail, 2 μl of 10X LightCycler Master Mix from the LightCycler FastStart DNA Master HybProbe Kit (Roche Applied Science) containing PCR buffer, deoxynucleotide triphosphates, 10 mmol/L MgCl2, and Taq polymerase, 1.6 μl of 25 mmol/L MgCl2, 40 pmol of each forward primer, 10 pmol of each reverse primer, and 5 pmol of each hybridization probe was used. The reaction mixture was subjected to 40 cycles of rapid PCR consisting of denaturation at 95°C for 5 seconds, annealing at 54°C for 20 seconds, and extension at 72°C for 12 seconds. Post amplification FMCA was performed by gradual heating of samples at a rate of 0.1°C/second from 40°C to 95°C. Fluorescence melting peaks were built by plotting the negative derivative of fluorescent signal corresponding to the temperature (−dF/dT).
Sequencing Analysis
Sanger (dideoxy) sequencing analysis was performed as previously described. In detail, IDH1 forward (5′-ACCAAATGGCACCATACGA-3′) and reverse (5′-GCAAAATCACATTATTGCCAAC-3′) primers, and IDH2 forward (5′-GCTGCAGTGGGACCACTATT-3′) and reverse (5′-TGTGGCCTTGTACTGCAGAG-3′) primers, were used. PCR amplification was performed by using 5 to 50 ng of DNA, 0.2 μmol of each primer, and AmpliTaq Gold PCR Master Mix (Applied Biosystems, Inc., Foster City, CA) in a total volume of 50 μl. The reaction mixture was subjected to an initial denaturation of 95°C for 10 minutes, followed by 40 cycles of amplification consisting of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension 72°C for 60 seconds. The PCR products were sequenced in both sense and antisense directions by using the BigDye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions and were analyzed on ABI 3130 (Applied Biosystems). The sequence electropherograms were analyzed by using Mutation Surveyor software (SoftGenetics, LLC, State College, PA). Each case was classified as either positive or negative for the IDH mutation based on the sequencing results.
Results
We analyzed 168 FFPE tissues for the presence of IDH1 and IDH2 mutations, including 67 gliomas, 57 non-neoplastic pathological conditions of the brain, and 44 non-CNS tumors by real-time PCR/FMCA and conventional sequencing (Table 1). Both methods showed total concordance, ie, all mutations detected by FMCA assay were detected by the sequencing analysis. Analytical sensitivity of real-time PCR/FMCA and conventional sequencing were tested by using a dilution series by mixing DNA from heterozygous positive controls carrying IDH1 R132H or IDH2 R172M mutations with wild-type DNA from peripheral blood. As determined by conventional sequencing, these heterozygous mutations were present in approximately 100% of tumor cells. The results indicated that real-time PCR/FMCA assay allowed detection of as little as 10% of mutant alleles in a background of normal DNA as compared with the detection of 20% of mutant alleles in a background of normal DNA by sequencing analysis. Melting temperatures for each mutation subtype were determined, showing Tm of 64°C for the IDH1 wild-type sequence, whereas Tm for mutant alleles varied from 55°C to 59°C (Figure 2). Similarly, Tm for the IDH2 wild-type was 66°C, whereas the R172M mutant Tm was 55°C (Figure 2). The entire length of the real-time PCR/FMCA assay was only 80 minutes, which is significantly shorter as compared with conventional sequencing, which requires approximately 5 hours of machine time.
With regards to frequency and distribution of IDH1 and IDH2 mutations in gliomas, 45 of the 67 gliomas (67%) were positive for mutations. As expected, most mutations (93%) were found in IDH1, with only three (7%) detected in IDH2 (Table 1). Representative positive mutations detected by real-time PCR and post-PCR/FMCA, including their correlation with sequencing analysis, are shown in Figure 2.
In the neoplastic subset, 72% of the World Health Organization grades 2 and 3 diffuse gliomas were positive for IDH mutations, whereas 17% of the GBMs were positive. Clinically, all positive GBM cases were believed to be of the secondary, progressive type. Based on melting curve profiles that were confirmed by sequencing, the most frequent mutation in IDH1 was R132H (82%), followed by R132S (4%) in grade 2 and grade 3 oligodendrogliomas, R132C (2%) in a grade 3 oligodendroglioma, R132G (2%) in a grade 2 astrocytoma, and R132L (2%) in a secondary GBM (Table 1). Three cases (7%) were positive for R172M IDH2 mutation, all of which were grade 2 oligodendrogliomas. IDH1 and IDH2 mutations were mutually exclusive. These results are in good agreement with prior studies,1,2,3,7,11,16 demonstrating that real-time PCR and post-PCR FMCA reliably detects IDH1/2 mutations in routine FFPE tissue samples.
We previously reported the absence of IDH1 and IDH2 mutations in non-neoplastic brain tissues, including conditions that frequently resemble gliomas on light microscopy (ie, viral infections, radiation-induced changes, or reactive gliosis around metastatic tumors).13 Similar to our previous observation, we did not find any IDH1 or IDH2 mutations in 57 non-neoplastic lesions of brain using real-time PCR and FMCA, which is more sensitive than Sanger sequencing (Table 1). These results suggest that the presence of IDH1/2 mutations is diagnostic for glioma and cannot be attributed to non-neoplastic diseases, which makes them useful markers for rendering pathological diagnoses.
Additionally, we tested 45 non-CNS tumors for presence of IDH1/2 mutations by using this new method. No mutations were identified in colorectal adenocarcinomas, which correlate with previously published reports.2,3 Furthermore, we did not find IDH mutations in other types of cancer, including thyroid follicular carcinoma, follicular variant of papillary carcinoma, anaplastic and poorly differentiated thyroid carcinoma, or in hepatocellular carcinomas (Table 1).
Discussion
The recent discovery of IDH1/2 mutations in gliomas has opened up a new avenue of research in brain tumors, as well as making screening for such mutations an important diagnostic and prognostic tool. In this study, we report a new assay for the detection of IDH1/2 mutations in gliomas by real-time PCR and post-PCR FMCA. The primer and probes we designed allow for detection of all IDH mutational variants with high sensitivity, even in FFPE tissue samples, and provide identical results compared with the “gold standard” conventional Sanger sequencing. However, this method is faster, less laborious, and more sensitive than Sanger sequencing. It can be performed in 80 minutes and allows detection of as little as 10% mutant alleles in a background of normal DNA.
Real-time PCR and post-PCR FMCA have been successfully applied for detection of RAS (including KRAS codons 12 and 13), BRAF, and other mutations as adjunct diagnostic tests in molecular pathology.14,17,18 This approach is based on real-time PCR amplification and on utilization of fluorescence resonance energy transfer probes for post-PCR melting curve analysis, which allows not only an accurate detection of all mutational types, but also provides an additional source of specificity to the assay. As compared with other sequencing techniques, such as pyrosequencing, it benefits from being performed in a closed system without post-PCR processing, which minimizes the length of procedure, errors in sample handling, and risk of contamination.
We evaluated 168 FFPE tissues including gliomas, non-neoplastic CNS conditions mimicking gliomas, and non-CNS tumors for the presence of IDH1/2 mutations by using this novel real-time PCR and FMCA assay, and compared them with conventional sequencing. The assay was able to accurately detect the mutations and did not generate any false-positive results in non-neoplastic glioma mimickers. The presence and distribution of IDH1/2 mutations in gliomas was comparable with previously reported studies.2,7,16 In addition, our results demonstrate the absence of IDH1/2 mutations in non-CNS tumors including hepatocellular carcinomas, colorectal adenocarcinomas, and thyroid carcinomas, supporting the previously described high specificity of IDH1/2 mutations to brain neoplasms. Moreover, we show for the first time that IDH1/2 mutations are not found in multiple types of thyroid cancer besides conventional papillary carcinoma, including follicular variant of papillary carcinoma, follicular carcinoma, poorly differentiated, and anaplastic carcinomas.
One of the advantages of PCR-based diagnostic techniques is their ability to detect tumor-specific mutant DNA even when light microscopic examination is indeterminate for neoplasm. This was demonstrated in our previous study, wherein brain tissue biopsies that were equivocal for glioma still showed the same IDH1/2 mutations as their patient-matched diagnostic counterparts.13 However, Sanger sequencing is less sensitive for detection of mutations, whereas this real-time PCR and FMCA assay can detect as little as 10% mutant alleles. Thus, this method is likely to detect gliomas in “near miss” biopsies even when more conventional molecular methods do not.
Recently, several studies revealed the use of monoclonal antibodies for the detection of IDH1 R132H mutation by immunohistochemical or Western blot analysis.19,20,21 The immunohistochemical approach could conveniently detect the mutation in tissue sections, although the specificity of the detection as well as the limitations imposed by testing for only one mutation type remain to be fully characterized. Based on the current knowledge, 10% to 15% of gliomas carry less common mutants of IDH1 or IDH22,7 and therefore are not yet amenable for immunohistochemical detection at this time.
The lack of IDH1/2 mutations in different types of reactive brain lesions offers a powerful diagnostic tool to distinguish between gliomas and non-neoplastic brain lesions. Moreover, data from several groups suggest that these mutations also carry prognostic significance, as their presence correlates with better survival in patients affected by different types of gliomas.1,2,11,12 Therefore, the detection of IDH1 and IDH2 mutations in brain biopsies is important for routine clinical practice to improve both diagnostic accuracy and tumor prognostication. We report here that rapid and sensitive detection of these mutations in FFPE tissue samples can be achieved by real-time PCR and post-PCR melting curve analysis.
Acknowledgements
We thank Geeta Manta for her helpful discussion of this study.
Footnotes
Supported by NIH grant R01 88041 to Y.E.N.
C.H. and L.K. contributed equally to this work.
Research was performed at the Division of Molecular Anatomic Pathology, Department of Pathology, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, Rodolfo M, Scarpa A, Felicioni L, Buttitta F, Malatesta S, Marchetti A, Bardelli A. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 4.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 8.Korshunov A, Meyer J, Capper D, Christians A, Remke M, Witt H, Pfister S, von Deimling A, Hartmann C. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118:401–405. doi: 10.1007/s00401-009-0550-z. [DOI] [PubMed] [Google Scholar]
- 9.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 11.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, Hallani SE, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 12.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German glioma network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 13.Horbinski C, Kofler J, Kelly LM, Murdoch GH, Nikiforova MN. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 14.Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW 2nd, Tallini G, Kroll TG, Nikiforov YE. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88:2318–2326. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 15.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. In: WHO Classification of Tumors of the Central Nervous System. Ohgaki H, editor. IARC; Lyon: 2007. pp. 90–91. [Google Scholar]
- 16.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 17.Elenitoba-Johnson KS, Bohling SD, Wittwer CT, King TC. Multiplex PCR by multicolor fluorimetry and fluorescence melting curve analysis. Nat Med. 2001;7:249–253. doi: 10.1038/84708. [DOI] [PubMed] [Google Scholar]
- 18.Nikiforova MN, Ciampi R, Salvatore G, Santoro M, Gandhi M, Knauf JA, Thomas GA, Jeremiah S, Bogdanova TI, Tronko MD, Fagin JA, Nikiforov YE. Low prevalence of BRAF mutations in radiation-induced thyroid tumors in contrast to sporadic papillary carcinomas. Cancer Lett. 2004;209:1–6. doi: 10.1016/j.canlet.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 20.Kato Y, Jin G, Kuan CT, McLendon RE, Yan H, Bigner DD. A monoclonal antibody IMab-1 specifically recognizes IDH1R132H, the most common glioma-derived mutation. Biochem Biophys Res Commun. 2009;390:547–551. doi: 10.1016/j.bbrc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, Ackermann U, Tessmer C, Korshunov A, Zentgraf H, Hartmann C, von Deimling A. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2009;2010, 20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


