Abstract
The JAK2V617F mutation has emerged as an essential molecular determinant of myeloproliferative neoplasms (MPNs). The aim of this study was to evaluate the analytical and clinical performances of a real-time PCR (qPCR) assay using a combination of hydrolysis probes and a wild-type blocking oligonucleotide, all containing locked nucleic acid (LNA) bases. Moreover, we validated a procedure for precise quantification of the JAK2V617F allele burden. We used DNA samples from patients suspected to suffer from MPN and dilutions of HEL cells, carrying the mutation, to compare the LNA-qPCR assay to two previously published methods. All assays detected the same 36 JAK2V617F positive patients of 116 suspected MPN diagnostic samples. No amplification of normal donor DNA was observed in the LNA-qPCR, and the assay was able to detect and reproducibly quantify as few as 0.4% of the JAK2V617F allele in wild-type alleles. Quantification of the JAK2V617F allele burden showed similar proportion levels among the different MPN entities as described by other groups. In conclusion, the LNA-qPCR is a rapid, robust, sensitive, and highly specific assay for quantitative JAK2V617F determination that can be easily implemented in clinical molecular diagnostic laboratories. Moreover, precise quantification allows determination of JAK2V617F burden at diagnosis as well as the evaluation of response to JAK2 inhibitors.
Myeloproliferative neoplasms (MPNs) are clonal stem cell disorders encompassing a heterogeneous group of entities characterized by an increased and effective proliferation of one to three hematopoietic cell lineages in the bone marrow, resulting in increased peripheral blood counts. The differential diagnosis of the three Philadelphia chromosome (Ph) negative MPNs (i.e., polycythemia vera [PV], essential thrombocythemia [ET], and primary myelofibrosis [PMF]) is hampered by mutual morphological similarities and overlap with reactive proliferations. An even more heterogeneous spectrum of clinical presentations follows the continuous progress of disease, eventually resulting in ineffective hematopoiesis, bone marrow failure attributable to myelofibrosis, or transformation into acute leukemia.1
Since the first publications early 2005, the JAK2V617F mutation has emerged as an essential molecular determinant of MPNs, occurring in the majority of patients with PV (>95%) and approximately half of ET or PMF cases. This acquired mutation corresponds to a single-nucleotide change of the nucleotide 1849 in exon 14 of the Janus tyrosine kinase 2 (JAK2) gene, leading to a valine (V) to phenylalanine (F) substitution at amino acid position 617. The mutation in the JH2 pseudokinase autoinhibitory domain results in a constitutive activation of JAK2 tyrosine kinase, responsible for a cytokine-independent activation of the JAK-STAT signaling pathway. Consequently, the hematopoietic precursors bearing this mutation acquire a proliferation and survival advantage.2,3,4,5
The high frequency of this mutation, together with the 100% specificity for clonal disease, resulted in both simplification and accuracy of the diagnostic process in MPNs. Therefore, the presence of the JAK2V617F mutation is now considered as a major diagnostic criterion in the revised World Health Organization diagnostic criteria for PV, whereas it is considered as a clonal marker for the diagnosis in ET and PMF.6,7
Although the precise pathogenic contribution of JAK2V617F to MPNs is not fully elucidated, a certain degree of genotype–phenotype relationship has been described in ET or PV patients.8,9,10,11 Homozygosity for the JAK2V617F mutation as a result of mitotic recombination is rarely observed in ET (2 to 4%), whereas it is now established that the majority of patients with PV and PMF harbor a subpopulation of JAK2V617F homozygous cells, which however do not always predominate.2,3,4,5,12,13 In PV, this status is associated with higher hemoglobin levels and leukocyte count, lower platelet count, and presence of pruritus.12 Not merely the presence of the mutation but also the allele burden of JAK2V617F is likely to be an independent parameter with clinical significance in ET and PV.8,14,15,16,17 Although current evidence is inconclusive regarding the prognostic relevance of either the presence of JAK2V617F or its allele burden, most studies concluded that a higher mutant allele burden is an adverse prognostic factor for fibrotic transformation and thrombosis in ET and PV patients.8,10,11,18 In contrast, low JAK2V617F allele burden in PMF is associated with an inferior overall and leukemia-free survival.19,20
Therefore, quantification of mutant allele burden might be clinically relevant. Moreover, clinical studies using JAK2V617F targeted therapies are ongoing and expected to become available in the near future.21 As the acquired mutation may be present in only a small proportion of hematopoietic cells at diagnosis, and monitoring of the allele burden in response to JAK2 inhibitors will be necessary, sensitive and accurate quantification is needed. Several groups developed assays to enable detection of JAK2V617F mutation. Some are inappropriate for the routine diagnostic laboratories because they are too labor-intensive or complex.2,3,4,22,23 Other approaches, although sensitive, are not quantitative.24,25,26,27,28,29,30 Therefore, real-time PCR (qPCR) methods have been introduced with mutation-specific primers or probes.31,32,33,34,35 Unfortunately, qPCR assays can suffer from nonspecific amplification. In addition, maintaining specificity and sensitivity when adapting qPCR assays to different PCR equipment and reagents may be difficult and time-consuming.
In this study, we improved and evaluated a qPCR assay36 including a locked nucleic acid (LNA)-modified wild-type blocking oligonucleotide and dual-labeled hydrolysis probe for sensitive and specific detection of the JAK2V617F mutation. LNA is a nucleic acid analog that contains an internal 2′-O, 4′-C methylene bridge, which locks the ribose ring into a C3′-endo conformation. Introduction of LNA analogs into oligonucleotides increases thermal stability of heteroduplexes with + 3 to + 8°C per modification.37 Because of high affinity hybridization of the LNA oligonucleotide to the wild-type target template, amplification is limited in the mutation-specific reaction. Moreover, the mutation-specific hydrolysis probe contains LNA monomers leading to increased specificity of allelic discrimination. We validated our method by comparing the results with conventional sequencing for accuracy determination and with two previously published methods in terms of analytical and clinical sensitivity and specificity. We have also introduced a control PCR to normalize the obtained results, allowing precise quantification of the JAK2V617F allele burden. Finally, this quantification approach was clinically validated by comparing the V617F allele proportions between the different MPN entities.
Materials and Methods
Samples
We included peripheral blood or bone marrow samples from 116 consecutive patients, suspected to suffer from a MPN, received in our laboratory from May 2007 until March 2008. Patients were classified based on their diagnosis into five categories: PV (n = 13), ET (n = 20), PMF (n = 11), post-polycythemic myelofibrosis (post-PV MF) (n = 3), and non-MPN (n = 69), encompassing a heterogeneous group of reactive thrombocythemia, secondary polycythemia, chronic myeloid leukemia (CML) patients, three cases with chronic myelomonocytic leukemia and one refractory anemia with ringed sideroblasts (RARS). Diagnosis was based on World Health Organization 2001 criteria.38 Consent was given by the Ethics Committee of our hospital for saving sample leftovers for further analysis. Peripheral blood samples were obtained from 20 healthy individuals. All samples were collected into EDTA-containing blood sampling tubes (Terumo, Leuven, Belgium) and immediately transported to the laboratory at room temperature. The processing of the samples was initiated within 12 hours after collection.
Sample Preparation
Genomic DNA was isolated from samples using the QIAamp DNA mini kit (Qiagen, Paisley, UK) according to the manufacturer's instructions. DNA concentrations were measured by UV spectrophotometry.
LNA-Based qPCR
Both qPCR reactions were performed in a final volume of 25 μl using an iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA). The reaction mixtures contained a maximum of 250 ng genomic DNA and final concentration of reagents were: 300 nmol/L of each primer, 100 nmol/L of both dual-labeled probes in a separate reaction, 1 μmol/L LNA oligonucleotide (mutation-specific reaction only), 400 μmol/L dNTPs, 4 mmol/L MgCl2, and 0.025 U/μL TaqDNA polymerase in PCR reaction buffer (Eurogentec, Seraing, Belgium). The sequences of the primers and probes are provided in Table 1. The PCR cycle parameters were as follows: an initial denaturing step at 95°C for 10 minutes and 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. All samples, including positive and negative controls, were run in duplicate.
Table 1.
PCR Primers/Probes and Sequencing Primers Used in this Study
| Primer set | Primer sequence |
|---|---|
| LNA-based qPCR | |
| JAK2 forward primer | 5′-AGCAGCAAGTATGATGAGCAAG-3′ |
| JAK2 reverse primer | 5′-GAGAAAGGCATTAGAAAGCCTGTAG-3′ |
| Mutation probe | 5′-TGGAGTATGTTTCTGTGGA-3′ |
| LNA oligonucleotide | 5′-TATGTGTCTGT−3′ |
| Control probe | 5′-ACAAGCATTTGGTTTTAAATTATGGAG-3′ |
| AS-PCR25 | |
| JAK2 WT forward primer | 5′-GGTTTTAAATTATGGAGTATGTG-3′ |
| JAK2V617F forward primer | 5′-GGTTTTAAATTATGGAGTATGTT-3′ |
| JAK2 reverse primer | 5′-TACACTGACACCTAGCTGTGA-3′ |
| Larsen-qPCR35 | |
| JAK2 forward primer | 5′-CTTTCTTTGAAGCAGCAAGTATGA-3′ |
| JAK2 WT reverse primer | 5′-GTAGTTTTACTTACTCTCGTCTCCACATAC-3′ |
| JAK2V617F reverse primer | 5′-GTAGTTTTACTTACTCTCGTCTCCACATAA −3′ |
| JAK2 probe | 5′-TGAGCAAGCTTTCTCACAAGCATTTGGTTT-3′ |
| Sequencing primers | |
| JAK2 forward primer | 5′-TTCCTTAGTCTTTCTTTGAAGCA-3′ |
| JAK2 reverse primer | 5′-GTGATCCTGAAACTGAATTTTCT-3′ |
The nucleotide at bp position 1849 is bold. The intended mismatches for the Larsen-qPCR are italic. Capital letters indicate normal bases, and underlined capital letters are LNA bases. AS indicates allele-specific; LNA, locked nucleic acid; WT, wild-type.
JAK2V617F Quantification
The amount of JAK2V617F allele was reported as the proportion of JAK2V617F versus all JAK2 (mutated and wild-type in control PCR reaction) (i.e., JAK2V617F/JAK2V617F + JAK2WT). This JAK2V617F percentage was calculated using plasmid standards for calibrations. PCR fragments of wild-type or JAK2V617F DNA, cloned into the pGEM T easy vector plasmids carrying AmpR selection gene (Promega, Madison, WI), were kindly provided by Dr. Serge Carillo from the Laboratoire de Cytologie Clinique et Cytogénétique (CHU, Nimes, France).39 The % JAK2V617F was corrected for the % neutrophils in whole blood to estimate the % JAK2V617F in the granulocyte fraction according to the following formula: % JAK2V617F corrected = (observed % JAK2V617F / % neutrophils) × 100.
Determination of Analytical Sensitivity and Reaction Efficiency
The analytical sensitivity of the methods compared was measured with a 1:2 serial dilution of HEL cells into JAK2 wild-type peripheral blood white blood cells. Because of chromosomal amplification, the JAK2V617F copy number depends on the cell line clone.40 Plasmid dilution experiments indicated that our HEL cell line clone carries 18 copies of JAK2V617F (data not shown). After correction for copy number, the proportion of mutant DNA in HEL dilutions ranged from 100% to 0.01%. Moreover, to prove an increased sensitivity with 250 ng DNA input, different DNA amounts of the HEL dilutions were tested. The limit of quantification was defined as the lowest dilution that generated an unequivocal and consistently positive JAK2V617F result within mean ± 2SD (see results interassay variation). The limit of detection was defined as the maximum obtained sensitivity.
Efficiency of the LNA-based qPCR reactions were determined by constructing a standard curve using a 1:10 serial dilution of HEL cell DNA into water.
Accuracy Determination
To validate the LNA-based qPCR assay, 77 randomly selected patient samples of 116 were genotyped for the JAK2V617F mutation using conventional sequencing. The target DNA sequence of the JAK2 gene was amplified using a set of sequencing primers (Table 1) to generate a 177-bp amplicon. PCR was performed using a maximum of 250 ng of genomic DNA in 25 μl reaction with final concentrations of 200 nmol/L each of the primers, 200 μmol/L of dNTPs, 4 mmol/L MgCl2, and 0.06 U/μL TaqDNA polymerase in PCR buffer (Applied Biosystems, Foster City, CA). PCR conditions were: 1 cycle at 95°C for 3 min followed by 40 cycles at 95°C for 15 s, 55°C for 30 s and 72°C for 30 s, and final extension at 72°C for 10 min. Amplicons were purified (Wizard SV Gel and PCR Clean-Up System, Promega, Madison, WI) and used as a template for the cycle sequence reaction. Forward and reverse sequencing were performed using Big Dye Terminator version 3.1 kit (Applied Biosystems) with the same primers above mentioned as follows: 1 cycle at 96°C for 1 min followed by 25 cycles at 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. After cycle sequencing, products were ethanol precipitated and, after addition of Hi-Di formamide (Applied Biosystems), analyzed on an ABI 3130 XL sequencer (Applied Biosystems).
Statistical Analysis
The intra- and interassay variation was obtained by calculating the coefficient of variation on allelic burden values.
The nonparametric Mann–Whitney test for independent samples was used to compare JAK2V617F allele burden between MPN categories. Data were analyzed using MedCalc Version 9.3.2.0 (MedCalc Software, Mariakerke, Belgium). A P value of <0.05 was considered as statistically significant.
Results
LNA-Based qPCR
We introduce a qPCR assay that combines the LNA and fluorescent hydrolysis probe technologies for sensitive JAK2V617F point mutation detection. This qPCR, modified from the molecular beacon technology as described by Sidon et al, enhances allelic discrimination by limiting wild-type JAK2 sequences from amplification using a LNA oligonucleotide.36 A mutation-specific probe is labeled with a 5′ 6-carboxyfluorescein (FAM) reporter dye and 3′ black hole quencher (BHQ1) dye (probes and primers sequences are given in Table 1). During PCR cycling, amplification of wild-type allele is limited by the high affinity of the LNA oligonucleotide for this sequence. In contrast, a mutation detection probe in the PCR reaction reveals a signal when V617F alleles are present, not being affected by the LNA oligonucleotide (Figure 1).
Figure 1.
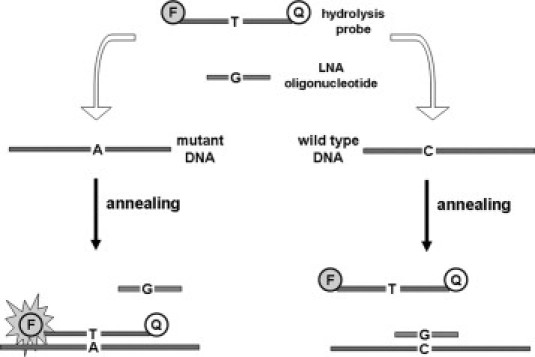
Schematic overview of the mutation-specific LNA-based qPCR reaction. At the left part of the figure, the perfect matching between the hydrolysis probe and the mutated DNA sequence allows annealing of the probe to the target DNA and consequently the Taq polymerase can hydrolyze the probe. In contrast, the single bp mismatch between the LNA oligonucleotide sequence and the mutated DNA sequence prevents specifically its annealing. At the right part of the figure, the LNA oligonucleotide anneals to wild-type target DNA, thereby preventing the detection of wild-type JAK2 sequences. The LNA oligonucleotide thus allows preferential amplification of the mutated DNA if present. Moreover, the mutated specific hydrolysis probe cannot efficiently hybridize to wild-type target because of a single bp mismatch between both. LNA indicates locked nucleic acid.
Additionally, a control PCR reaction was performed for all samples for total JAK2 DNA quantification and to exclude false-negative results by an inhibition or attributable to insufficient quality or quantity. This control amplification was performed in a separate reaction, using the same forward and reverse primer as in the mutation-specific PCR reaction but with a probe located upstream of nucleotide 1849, able to detect both wild-type and mutant alleles.
Comparison of Analytical Specificity of the Detection Methods Used
Sensitive detection of a point mutation in the presence of abundant wild-type sequences is a challenge. A qPCR method according to Larsen et al (Larsen-qPCR)35 that uses a hydrolysis probe and two sets of primers (a common forward primer and wild-type or mutation-specific reverse primers containing an intended mismatch at the third base from the 3′ end to improve hybridization stringency) was not specific in our hands, even when using SDS-PAGE purified primers. Therefore, we optimized a LNA-based qPCR to tackle the specificity problem and compared it to an allele-specific (AS) semiquantitative PCR followed by capillary electrophoresis, according to McClure et al25 and the Larsen-qPCR. The sequences of all primers and probes are provided in Table 1.
Twenty peripheral blood samples from normal volunteers were analyzed. All DNA samples did not contain the JAK2V617F mutation according to the AS-PCR and did not show any amplification in the mutation-specific LNA-based qPCR reaction. In contrast, 19 of 20 showed an amplification signal with the V617F mutation-specific Larsen-qPCR SDS-PAGE purified primer set (see supplemental Figure S1A at http://jmd.amjpathol.org). This underlines the need to define a Ct cut-off value to differentiate between JAK2V617F-positive and negative samples when using this method. The mean Ct value of the 19 positive healthy individuals was 42.4 ± 2.1 (mean ± SD). Based on a 3-SD confidence interval (CI), a threshold Ct value of 36 was defined, below which a sample was considered JAK2V617F positive with the mutation-specific Larsen-qPCR.
Comparison of Analytical Sensitivity
The sensitivity of the assay was determined by measuring a dilution series of JAK2V617F mutant cells in wild-type cells, expressed as % mutant alleles. The LNA-based qPCR showed a limit of JAK2V617F quantification equal to 0.4% and reached a maximum sensitivity of 0.2%. Moreover, different DNA amounts of the serial dilution series were tested, achieving the highest sensitivity using 250 ng template DNA (data not shown). The AS-PCR yielded a reproducible mutation peak on capillary electrophoresis for samples with 3.6% mutant alleles. Further dilutions diminished this peak, visible in only 1 of 6 repeats at 1.8% mutant DNA. Because of nonspecificity, the limit of detection could not be determined for the Larsen-qPCR. However, based on a Ct cut-off point of 36 (see results analytical specificity) a limit of quantification of 0.8% could be demonstrated, because this dilution of mutant DNA yielded all four Ct values below 36.
Reproducibility and Efficiency Determination of the LNA-Based qPCR
To determine the intraassay variability, DNA obtained from a patient carrying JAK2V617F was analyzed 20 times within one run. The SD of the mutation-specific and control qPCR reaction were 0.086 and 0.070, respectively. The mean JAK2V617F allelic ratio was 44.5% with a coefficient of variation value of 6.7%. To evaluate the interassay variability, another mutant patient DNA (mean JAK2V617F allele burden of 81%) was analyzed in duplicate over four different runs. This resulted in an inter-run coefficient of variation value of 7.0% for the JAK2V617F allelic ratio according to plasmid standards.
Assessment of the efficiency of the LNA-based qPCR method with a LNA oligonucleotide concentration of 1 μmol/L showed an efficiency of amplification of 97% and 96% for the mutation-specific and control LNA-based qPCR reaction, respectively (data not shown).
JAK2V617F Genotype Distribution in Clinical Samples
Next, we evaluated 116 samples from patients suspected to suffer from a MPN. All samples were tested for the presence of the JAK2V617F mutation by the AS-PCR, Larsen-qPCR, and LNA-based qPCR. A summary of these results according to the MPN categories is given in Table 2. Using the LNA-based qPCR approach, 36/116 (31%) of the samples gave a positive result and were concordant with the results obtained by the AS-PCR, resulting in a 100% overall agreement. All 36 samples were also detected by the Larsen-qPCR method. However, 79 of the remaining 80 samples, found negative by both AS-PCR and LNA-based qPCR, showed an amplification signal in the mutation-specific Larsen-qPCR (see supplemental Figure S1B at http://jmd.amjpathol.org). Consequently, using a Ct cut-off value of 36 (see results analytical specificity), 5 of those 79 patients were reported to carry the JAK2V617F mutation, and may be considered as false positive results when compared with LNA-based qPCR and AS-PCR (Table 2).
Table 2.
Overview of Parallel JAK2V617F Results by the Different Mutation Detection Techniques in the Four MPN Categories
| Patient classification | AS-PCR25 | Larsen-qPCR*35 | LNA-based qPCR | Sequencing |
|---|---|---|---|---|
| PV | 11/13 | 11/13 | 11/13 | 9/12 |
| ET | 14/20 | 14/20 | 14/20 | 11/16 |
| PMF | 8/11 | 9/11 | 8/11 | 8/9 |
| Post-PV MF | 3/3 | 3/3 | 3/3 | 3/3 |
| Non-MPN | 0/69 | 4/69 | 0/69 | 0/37 |
| Total | 36/116 (31%) | 41/116 (35%) | 36/116 (31%) | 31/77 (40%) |
The table shows positive samples over all patients tested per classification versus technique applied. Asterisk indicates using the defined CT cut-off of 36 to differentiate between positive and negative results; MPN, chronic myeloproliferative disorder; AS, allele-specific; LNA, locked nucleic acid; PV, polycythemia vera; ET, essential thrombocythemia; PMF, primary myelofibrosis; post-PV MF, post-polycythemic myelofibrosis.
To confirm the accuracy of the assay, 77 samples were sequenced and the results were compared with those obtained by the LNA-based qPCR and the AS-PCR. This showed a good correlation between the PCR and sequencing data (Table 2). However, a discrepancy was observed for 5 samples for which the mutation could not be discerned from the background JAK2 sequence. Knowing that the allele burden for JAK2V617F can be low, the limited analytical sensitivity of sequencing, reported to be 10 to 20%,4,26 may explain these discordant results.
Quantitative JAK2V617F Allele Burden Determination by the LNA-Based qPCR
The corrected JAK2V617F allelic ratio was calculated for the 36 positive samples. Box-and-Whisker plot presentation of the JAK2 mutant allele burden in the different disease entities is presented in Figure 2. Of the 13 patients with PV, 11 carried the JAK2V617F mutation (Table 2) with a median burden of 58.2% (95% CI: 25.4% to 71.8%). In total, 7 of 11 patients (64%) displayed >50% mutant alleles and could be considered homozygous. The 14 JAK2V617F-positive ET patients (Table 2) had a lower median JAK2V617F allele burden as compared with the PV group, although not reaching statistical significance (P value = 0.14). A median mutant JAK2 allele ratio of 36.3% (95% CI: 28.6% to 52.3%) was observed. Moreover, the level found in ET patients was significantly lower than the level found in patients with PMF or post-PV MF (P value of 0.003 and 0.008, respectively). The median proportion of mutated alleles in patients with PMF (n = 8) was 69.4% (95% CI: 48.2% to 90.5%), and increased to 99.7% (n = 3) when restricted to post-PV MF.
Figure 2.
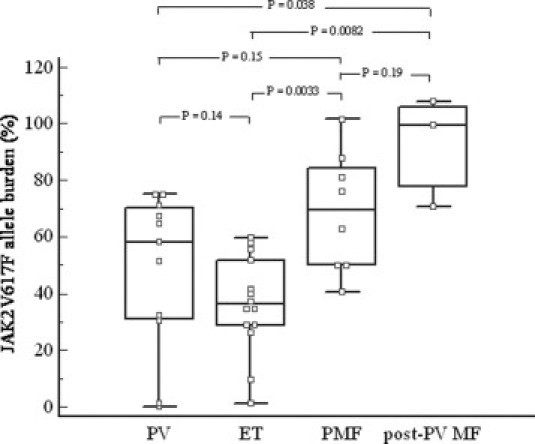
Proportion of JAK2V617F allele burden of the 36 JAK2 mutant patients included in this study. Box-and-Whisker presentation of the results from patients with PV (n = 11), ET (n = 14), PMF (n = 8), and post-PV MF (n = 3). The central box represents 25th to 75th percentile, and the middle line the median. The black vertical line extends from the minimum to the maximum value, excluding “outside” and “far out” values. P values are represented for comparisons between all disease entities. PV indicates polycythemia vera; ET, essential thrombocythemia; PMF, primary myelofibrosis; post-PV MF, post-polycythemic myelofibrosis.
Discussion
Specific quantification of JAK2V617F mutational burden provides an opportunity to evaluate the prognostic significance of the mutation burden at diagnosis and raises the possibility to monitor disease progression or efficacy of therapy. Although several real-time PCR methods for quantitative detection of JAK2V617F have been published,31,32,33,34,35 diagnostics labs can face nonspecific amplification, leading to false positive results. This could have significant clinical consequence because JAK2V617F is being incorporated into diagnostic algorithms for the identification and classification of MPNs.6 False positive results may be explained by the inadequate adaption of published qPCR protocols to local PCR equipment and reagents. Therefore, to ensure specificity, we tested a novel qPCR method for sensitive and specific quantification of JAK2V617F that uses a wild-type blocking LNA-oligonucleotide. The wild-type blocking LNA-oligonucleotide used allows, in combination with a mutation-specific detection probe, to detect very low levels of JAK2V617F alleles with 100% specificity in peripheral blood and in bone marrow samples. This method has been adapted from the assay described by Sidon et al with molecular beacon technology.36 However, compared with hydrolysis probes, molecular beacon probes yield less fluorescence, as no physical separation of fluorophore from quencher occurs. Our experience shows that it is very difficult to obtain good reproducibility between batches with these probes. In addition, hydrolysis probes are more widespread and more accessible because of less complex synthesis.
Moreover, we developed a procedure for accurate quantification of the allele burden. By adding a control JAK2 PCR reaction, we express the results as a proportion of JAK2V617F to total JAK2. The study of Lippert et al39 underlined the necessity to use a universal expression of results and reported the lowest lab-to-lab variation when JAK2V617F allelic burden results were expressed as a percentage of total JAK2 DNA and not, for example, in relation to other genes. Additionally, this control JAK2 PCR reaction allows exclusion of false-negative results attributable to, for example, PCR inhibition.
As B and T lymphocytes typically do not express JAK2V617F,2,3,4,33,34 the JAK2 tumor burden is likely to be underestimated in whole blood assays. In routine analyses, granulocyte purification is time-consuming and whole blood extraction is preferred. Whole blood is, however, only suitable for the diagnosis of MPNs when the JAK2V617F allelic ratio is adjusted using the percentage neutrophils, like we did in our study. Nonetheless, for the purpose of monitoring disease progression and of efficacy of newly developed therapies, purification of blood granulocytes remains necessary.41
Kroger et al31 showed the importance of monitoring patients with myelofibrosis after allogenic stem cell transplantation by JAK2V617F quantification. Broad application of the JAK2V617F mutation as a molecular marker for minimal residual disease detection to monitor disease progression in all MPN entities or in response to novel JAK2 inhibitors will require highly sensitive PCR methods. With an analytical quantitative sensitivity of 0.4%, the LNA-based qPCR assay resulted in the highest sensitivity as compared with the established AS-PCR method validated by McClure et al25 and the qPCR method according to Larsen et al,35 evaluated in this study. Furthermore, the multicenter comparative study of JAK2V617F assays published by Lippert et al39 demonstrated that only two TaqMan AS-qPCR assays with primer-based specificity had a reproducible quantitative sensitivity of 0.2%. However, in our hands, one assay was prone to nonspecific amplification (data not shown). This may be attributable to suboptimal adaptation of the assay to our qPCR equipment, which illustrates the advantage of using a wild-type LNA oligonucleotide to block wild-type JAK2 sequences from amplification. Moreover, the other assay showed a false positive result in the study of Lippert et al.39 Furthermore, we reported a limit of detection of 0.4% with reproducible assessment of the JAK2V617F allele burden, defined as the limit of quantification. However, our assay reached a maximum sensitivity of 0.2%, however without consistent positive results.
As part of the assay validation, JAK2V617F genotype distribution was determined for patients from the various diagnostic MPN categories and for the non-MPN patients. A parallel evaluation of the LNA-based qPCR assay with the AS-PCR revealed that these methods yielded concordant results. Moreover, the quantitative results showed a distribution of V617F allele burden among the different MPN entities as described by other groups.15,16 Figure 2 depicts that the lowest and highest values are expressed by patients with ET and PMF or post-PV MF, respectively, whereas an intermediate allele burden is found in patients with PV.
Finally, the relatively low intra- and interassay variabilities indicate the robustness of our assay, which renders it suitable for quantitative analyses of JAK2V617F in routine clinical laboratories. Moreover, this method is also convenient for quick and accurate identification of patients who carry the mutation because this test is less time consuming than, for example, the AS-PCR that requires electrophoresis postamplification.
In conclusion, the LNA-based qPCR is a rapid and highly specific assay. This technique is suitable for JAK2V617F mutation detection in the routine diagnostic laboratory, because it uses standard equipment and is technically easy to perform with few sample manipulation steps. Our optimized method allows quantification of the JAK2V617F allele burden on DNA extracted from peripheral whole blood samples for diagnostic purposes with a reproducible quantitative sensitivity of 0.4%.
Acknowledgements
We thank Dr. Serge Carillo (Laboratoire de Cytologie Clinique et Cytogénétique, CHU, Nimes, France) for kindly providing the plasmid standards. Moreover, we thank Kelly Heyns and fellow technicians of the molecular lab for the technical support. We are also indebted to Dr. Jan Dierick (AZ Maria Middelares, Ghent) for referring patient samples for analysis to our laboratory and to Dr. Kristien Van Pelt for helping with the validation set-up.
Footnotes
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Web Extra Material
Amplification curves of 20 normal peripheral blood samples (A) and some clinical samples included in this study (B) according to the mutation specific Larsen-qPCR35
Nineteen out of 20 DNA samples of normal peripheral blood samples (A) gave an amplification curve above the threshold Ct value. This implicated the use of a Ct cut-off value of 36 (dark vertical line). The aspecific hybridization of the mutant probe to wild-type sequence creates an overlap between low JAK2V617F positive patients and Ct values of negative samples (circle) (B).
References
- 1.Haferlach T, Bacher U, Kern W, Schnittger S, Haferlach C. The diagnosis of BCR/ABL-negative chronic myeloproliferative diseases (CMPD): a comprehensive approach based on morphology, cytogenetics, and molecular markers. Ann Hematol. 2008;87:1–10. doi: 10.1007/s00277-007-0403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 3.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, Barosi G, Verstovsek S, Birgegard G, Mesa R, Reilly JT, Gisslinger H, Vannucchi AM, Cervantes F, Finazzi G, Hoffman R, Gilliland DG, Bloomfield CD, Vardiman JW. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood. 2007;110:1092–1097. doi: 10.1182/blood-2007-04-083501. [DOI] [PubMed] [Google Scholar]
- 7.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2008. [Google Scholar]
- 8.Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22:1299–1307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]
- 9.Antonioli E, Guglielmelli P, Pancrazzi A, Bogani C, Verrucci M, Ponziani V, Longo G, Bosi A, Vannucchi AM. Clinical implications of the JAK2 V617F mutation in essential thrombocythemia. Leukemia. 2005;19:1847–1849. doi: 10.1038/sj.leu.2403902. [DOI] [PubMed] [Google Scholar]
- 10.Tefferi A. JAK and MPL mutations in myeloid malignancies. Leuk Lymphoma. 2008;49:388–397. doi: 10.1080/10428190801895360. [DOI] [PubMed] [Google Scholar]
- 11.Barosi G, Bergamaschi G, Marchetti M, Vannucchi AM, Guglielmelli P, Antonioli E, Massa M, Rosti V, Campanelli R, Villani L, Viarengo G, Gattoni E, Gerli G, Specchia G, Tinelli C, Rambaldi A, Barbui T. JAK2 V617F mutational status predicts progression to large splenomegaly and leukemic transformation in primary myelofibrosis. Blood. 2007;110:4030–4036. doi: 10.1182/blood-2007-07-099184. [DOI] [PubMed] [Google Scholar]
- 12.Vannucchi AM, Antonioli E, Guglielmelli P, Rambaldi A, Barosi G, Marchioli R, Marfisi RM, Finazzi G, Guerini V, Fabris F, Randi ML, De Stefano V, Caberlon S, Tafuri A, Ruggeri M, Specchia G, Liso V, Rossi E, Pogliani E, Gugliotta L, Bosi A, Barbui T. Clinical profile of homozygous JAK2 617V>F mutation in patients with polycythemia vera or essential thrombocythemia. Blood. 2007;110:840–846. doi: 10.1182/blood-2006-12-064287. [DOI] [PubMed] [Google Scholar]
- 13.Scott LM, Scott MA, Campbell PJ, Green AR. Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood. 2006;108:2435–2437. doi: 10.1182/blood-2006-04-018259. [DOI] [PubMed] [Google Scholar]
- 14.Vannucchi AM, Antonioli E, Guglielmelli P, Longo G, Pancrazzi A, Ponziani V, Bogani C, Ferrini PR, Rambaldi A, Guerini V, Bosi A, Barbui T. Prospective identification of high-risk polycythemia vera patients based on JAK2(V617F) allele burden. Leukemia. 2007;21:1952–1959. doi: 10.1038/sj.leu.2404854. [DOI] [PubMed] [Google Scholar]
- 15.Antonioli E, Guglielmelli P, Poli G, Bogani C, Pancrazzi A, Longo G, Ponziani V, Tozzi L, Pieri L, Santini V, Bosi A, Vannucchi AM. Influence of JAK2V617F allele burden on phenotype in essential thrombocythemia. Haematologica. 2008;93:41–48. doi: 10.3324/haematol.11653. [DOI] [PubMed] [Google Scholar]
- 16.Larsen TS, Pallisgaard N, Moller MB, Hasselbalch HC. The JAK2 V617F allele burden in essential thrombocythemia, polycythemia vera and primary myelofibrosis–impact on disease phenotype. Eur J Haematol. 2007;79:508–515. doi: 10.1111/j.1600-0609.2007.00960.x. [DOI] [PubMed] [Google Scholar]
- 17.Kittur J, Knudson RA, Lasho TL, Finke CM, Gangat N, Wolanskyj AP, Li CY, Wu W, Ketterling RP, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F allele burden in essential thrombocythemia. Cancer. 2007;109:2279–2284. doi: 10.1002/cncr.22663. [DOI] [PubMed] [Google Scholar]
- 18.Tiedt R, Hao-Shen H, Sobas MA, Looser R, Dirnhofer S, Schwaller J, Skoda RC. Ratio of mutant JAK2–V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111:3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- 19.Tefferi A, Lasho TL, Huang J, Finke C, Mesa RA, Li CY, Wu W, Hanson CA, Pardanani A. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia. 2008;22:756–761. doi: 10.1038/sj.leu.2405097. [DOI] [PubMed] [Google Scholar]
- 20.Guglielmelli P, Barosi G, Specchia G, Rambaldi A, Lo Coco F, Antonioli E, Pieri L, Pancrazzi A, Ponziani V, Delaini F, Longo G, Ammatuna E, Liso V, Bosi A, Barbui T, Vannucchi AM. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood. 2009;114:1477–1483. doi: 10.1182/blood-2009-04-216044. [DOI] [PubMed] [Google Scholar]
- 21.Pardanani A. JAK2 inhibitor therapy in myeloproliferative disorders: rationale, preclinical studies and ongoing clinical trials. Leukemia. 2008;22:23–30. doi: 10.1038/sj.leu.2404948. [DOI] [PubMed] [Google Scholar]
- 22.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlmann R, Hochhaus A, Oscier D, Silver RT, Reiter A, Cross NC. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 23.Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility: a paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8:397–411. doi: 10.2353/jmoldx.2006.060007. quiz 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton BC, Allen RA, Zhao ZJ, Dunn ST. Detection of the JAK2V617F mutation by asymmetric PCR and melt curve analysis. Cancer Biomark. 2007;3:315–324. doi: 10.3233/cbm-2007-3605. [DOI] [PubMed] [Google Scholar]
- 25.McClure R, Mai M, Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia. 2006;20:168–171. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- 26.Frantz C, Sekora DM, Henley DC, Huang CK, Pan Q, Quigley NB, Gorman E, Hubbard RA, Mirza I. Comparative evaluation of three JAK2V617F mutation detection methods. Am J Clin Pathol. 2007;128:865–874. doi: 10.1309/LW7Q3739RBRMBXXP. [DOI] [PubMed] [Google Scholar]
- 27.Tan AY, Westerman DA, Dobrovic A. A simple, rapid, and sensitive method for the detection of the JAK2 V617F mutation. Am J Clin Pathol. 2007;127:977–981. doi: 10.1309/1U61JVXTLPPQ7YP1. [DOI] [PubMed] [Google Scholar]
- 28.Murugesan G, Aboudola S, Szpurka H, Verbic MA, Maciejewski JP, Tubbs RR, Hsi ED. Identification of the JAK2 V617F mutation in chronic myeloproliferative disorders using FRET probes and melting curve analysis. Am J Clin Pathol. 2006;125:625–633. doi: 10.1309/TK0X-L917-XK2V-LRPQ. [DOI] [PubMed] [Google Scholar]
- 29.Olsen RJ, Tang Z, Farkas DH, Bernard DW, Zu Y, Chang CC. Detection of the JAK2(V617F) mutation in myeloproliferative disorders by melting curve analysis using the LightCycler system. Arch Pathol Lab Med. 2006;130:997–1003. doi: 10.5858/2006-130-997-DOTJMI. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Lu P, Jones AV, Cross NC, Silver RT, Wang YL. Amplification refractory mutation system, a highly sensitive and simple polymerase chain reaction assay, for the detection of JAK2 V617F mutation in chronic myeloproliferative disorders. J Mol Diagn. 2007;9:272–276. doi: 10.2353/jmoldx.2007.060133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhauser M, Reiter A, Zabelina T, Zander AR, Fehse B. Monitoring of the JAK2–V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109:1316–1321. doi: 10.1182/blood-2006-08-039909. [DOI] [PubMed] [Google Scholar]
- 32.Wolstencroft EC, Hanlon K, Harries LW, Standen GR, Sternberg A, Ellard S. Development of a quantitative real-time polymerase chain reaction assay for the detection of the JAK2 V617F mutation. J Mol Diagn. 2007;9:42–46. doi: 10.2353/jmoldx.2007.060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond E, Shaw K, Carnley B, P'ng S, James I, Herrmann R. Quantitative determination of JAK2 V617F by TaqMan: an absolute measure of averaged copies per cell that may be associated with the different types of myeloproliferative disorders. J Mol Diagn. 2007;9:242–248. doi: 10.2353/jmoldx.2007.060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poodt J, Fijnheer R, Walsh IB, Hermans MH. A sensitive and reliable semi-quantitative real-time PCR assay to detect JAK2 V617F in blood. Hematol Oncol. 2006;24:227–233. doi: 10.1002/hon.800. [DOI] [PubMed] [Google Scholar]
- 35.Larsen TS, Christensen JH, Hasselbalch HC, Pallisgaard N. The JAK2 V617F mutation involves B- and T-lymphocyte lineages in a subgroup of patients with Philadelphia-chromosome negative chronic myeloproliferative disorders. Br J Haematol. 2007;136:745–751. doi: 10.1111/j.1365-2141.2007.06497.x. [DOI] [PubMed] [Google Scholar]
- 36.Sidon P, Heimann P, Lambert F, Dessars B, Robin V, El Housni H. Combined locked nucleic acid and molecular beacon technologies for sensitive detection of the JAK2V617F somatic single-base sequence variant. Clin Chem. 2006;52:1436–1438. doi: 10.1373/clinchem.2006.066886. [DOI] [PubMed] [Google Scholar]
- 37.Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- 38.Jaffe ES, Harris NL, Stein H, Vardiman JW. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2001. World Health Organization Classification of Tumours. [Google Scholar]
- 39.Lippert E, Girodon F, Hammond E, Jelinek J, Reading NS, Fehse B, Hanlon K, Hermans M, Richard C, Swierczek S, Ugo V, Carillo S, Harrivel V, Marzac C, Pietra D, Sobas M, Mounier M, Migeon M, Ellard S, Kroger N, Herrmann R, Prchal JT, Skoda RC, Hermouet S. Concordance of assays designed for the quantification of JAK2V617F: a multicenter study. Haematologica. 2009;94:38–45. doi: 10.3324/haematol.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quentmeier H, MacLeod RA, Zaborski M, Drexler HG. JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia. 2006;20:471–476. doi: 10.1038/sj.leu.2404081. [DOI] [PubMed] [Google Scholar]
- 41.Hermouet S, Dobo I, Lippert E, Boursier MC, Ergand L, Perrault-Hu F, Pineau D. Comparison of whole blood vs purified blood granulocytes for the detection and quantitation of JAK2(V617F) Leukemia. 2007;21:1128–1130. doi: 10.1038/sj.leu.2404588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amplification curves of 20 normal peripheral blood samples (A) and some clinical samples included in this study (B) according to the mutation specific Larsen-qPCR35
Nineteen out of 20 DNA samples of normal peripheral blood samples (A) gave an amplification curve above the threshold Ct value. This implicated the use of a Ct cut-off value of 36 (dark vertical line). The aspecific hybridization of the mutant probe to wild-type sequence creates an overlap between low JAK2V617F positive patients and Ct values of negative samples (circle) (B).


