Abstract
We identified a novel breakpoint cluster region-ABL rearrangement in a chronic myeloid leukemia (CML) patient. The e14/a2 (b3/a2) type BCR-ABL mRNA incorporated a 42-nucleotide intronic insertion of ABL intron Ib between BCR exon e14 and ABL exon a2. As we hypothesized that the rearrangement between BCR and ABL genes occurred near the inserted sequence and because of the relative small size of BCR intron 14, we determined the BCR-ABL breakpoint at the genomic DNA level. Using a PCR-based method, this analysis revealed that i) BCR intron 14 brought a potential lariat branch point and the polypyrimidine tract, ii) the BCR-ABL breakpoint created a chimeric acceptor site, and iii) the inserted sequence of ABL intron Ib carried at its 3′ end a well-conserved donor splice site. Therefore, the inserted sequence was flanked by canonical consensus splice sites and recognized as a pseudo-exon (as shown by splice site prediction and exon finder software). Moreover, the insertion did not disrupt the reading frame between BCR and ABL and did not produce a premature stop codon. Instead, this novel BCR-ABL chimeric transcript encoded a functional oncoprotein with an in-frame insertion of 15 new amino acids.
Philadelphia chromosome (Ph1) results from the reciprocal t(9;22)(q34;q11) translocation that fuses the BCR and ABL genes into a BCR-ABL chimeric gene, which encodes a constitutively activated tyrosine kinase oncoprotein.1 Breakpoints in ABL generally occur upstream of exon a2 (rarely exon a3). Depending on the location of the breakpoint in the BCR gene, three main types of BCR-ABL transcripts can be produced.2 In chronic myeloid leukemia (CML), most of the breakpoints in BCR occur in the 5.8-kb major breakpoint cluster region and result in the fusion at the mRNA level of BCR exon e13 (b2) or e14 (b3) to ABL exon a2, known as the e13/a2 or e14/a2 rearrangements. In Ph1-positive acute lymphoblastic leukemia, breakpoints in BCR are generally located in the minor major breakpoint cluster region and result in the e1/a2 BCR-ABL rearrangement. A third breakpoint in BCR (μ-BCR) is observed in typical CML or neutrophilic leukemia and leads to the e19/a2 rearrangement. Finally, e6/a2 and e8/a2 BCR-ABL rearrangements are uncommonly observed in typical CML. Thus, in the great majority of CML cases, BCR-ABL rearrangements result in the joining of BCR exons e13 or e14 to ABL exon a2.
We report here a case of CML with an e14/a2 fusion mRNA containing a 42-bp sequence from ABL intron Ib inserted between BCR exon e14 and ABL exon a2. The insertion was characterized by sequencing at the mRNA level. The location of the BCR-ABL breakpoint was determined on genomic DNA, and the consensus donor and acceptor splice sites as well as a potential lariat branch point were identified. We demonstrated that, in this BCR-ABL rearrangement, the consensus splice sites were brought by BCR intron 14 (branch point and polypyrimidine tract), BCR-ABL junction (chimeric acceptor splice site), and ABL intron Ib (donor splice site).
Materials and Methods
Clinical Report
A 44-year-old man was admitted to the hospital for a screening colonoscopy to explore chronic mild rectal bleeding. The patient had no medical history and no symptoms except for a moderate fatigue and weight loss. During the visit preceding the endoscopic examination, a spleen easily palpable 18 cm below the left costal margin was observed. Blood cell count revealed white blood cells at 214.0 × 109/L (6% metamyelocytes, 22% myelocytes, 8% promyelocytes, and 4% myeloblasts), hemoglobin at 11.7g/dl, and platelets at 427.0 × 109/L. Moreover, renal and liver functions were normal with increased lactate dehydrogenase levels. Bone marrow aspirate revealed granulocytic hyperplasia consistent with chronic-phase CML. Some giant and abnormally shaped metamyelocytes were also observed on the bone marrow smear. The patient was classified as high risk according to Sokal prognosis score (1.67). He received hydroxyurea before entering a therapeutic trial using second generation tyrosine kinase inhibitor as first-line treatment after informed consent was obtained.
RNA Isolation and Multiplex RT-PCR for BCR-ABL Transcript Detection
Total RNA was isolated from peripheral blood using RNAble, according to the manufacturer's protocol (Eurobio, Les Ulis, France). Around 1 μg of total RNA was retrotranscribed using the High-Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). BCR-ABL transcript detection was performed using a multiplex RT-PCR based on primers previously described in the literature3 and the additional primer BCR-e17/18 (5′-CGCCATGAATGGGATCGAAGTA-3′), which enhances the detection of a rearrangement at the μ-BCR locus. The PCR products were analyzed on a 1.2% agarose gel electrophoresis together with controls resulting from the amplification of e1/a2 (483 bp), e13/a2 (312 bp), e13/a3 (138 bp), e14/a2 (387 bp), and e19/a2 (545 bp) BCR-ABL rearrangements.
Genomic DNA Isolation and Genomic Breakpoint Characterization
Genomic DNA was isolated from peripheral blood with BioSprint 15 DNA blood kit (Qiagen, Hilden, Germany) using a KingFisher instrument (Thermo Scientific, Hudson, NH). The genomic BCR-ABL breakpoint was characterized using five forward primers located in BCR introns 13 and 14, BCR-a (5′-CTTGTCACCTGCCTCCCTT-3′), BCR-b (5′-CACCTTCACCCCACAGCA-3′), BCR-c (5′-AGCAAGACTCCGCCTCAAA-3′), BCR-d (5′-CTTTGAGGGGCACCACCA-3′), BCR-e (5′-GGCTTCCCACATCCCCCA-3′), and a reverse primer, ABL-z (5′-GCGTCTTTTCTTGGTTGCTT-3′), located in ABL intron Ib.
Direct Sequencing
PCR products were purified using ExoSAP-it (USB, Cleveland, OH) and double strand sequenced with the Big Dye Terminator version 3.1 reagents (Applied Biosystems), according to the manufacturer's recommendations. Sequencing products were purified on DyeEx 2.0 Spin kit columns (Qiagen) and analyzed through an ABI 3100 Genetic Analyzer (Applied Biosystems).
Results and Discussion
Cytogenetic and molecular analyses were performed at diagnosis. A classical t(9;22)(q34;q11) translocation without any additional abnormality was found, and a chimeric BCR-ABL transcript was identified by multiplex RT-PCR. However, the PCR fragment was distinct in size from those obtained from typical BCR-ABL rearrangements such as e1/a2, e13/a2, or e14/a2. As seen in Figure 1, the apparent size of this fragment in agarose gel electrophoresis was between 387 bp (e14/a2 amplicon) and 483 bp (e1/a2 amplicon). Direct sequencing of the PCR product revealed a 42-nucleotide insertion between BCR exon e14 and ABL exon a2. A basic local alignment search tool analysis (National Center for Biotechnology Information, Bethesda, MD) of the inserted sequence showed 100% identity with a genomic DNA sequence located near the middle of ABL intron Ib (GenBank accession number NG_012034). Analysis of this sequence at the genomic DNA level by the Human Splicing Finder software (Human Splicing Finder; Montpellier University, Montpellier, France; version 2.4. 2008; available at http://www.umd.be/HSF/; accessed September 25, 2009)4 revealed at the 3′ end of the insertion a highly conserved donor site for splicing (ACAgtaagt). Neither lariat branch point nor polypyrimidine tract or complete acceptor site were identified in the ABL genomic DNA upstream of the insertion.
Figure 1.
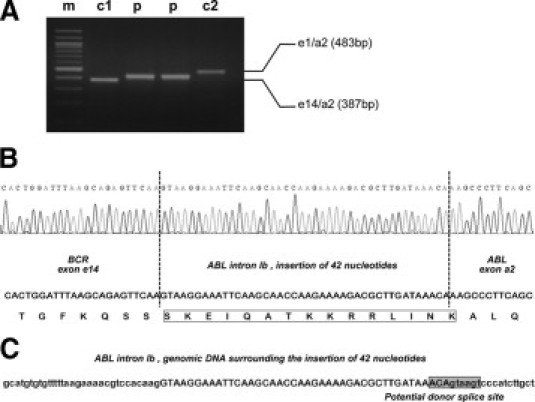
Detection of a novel intronic insertion within an e14/a2 BCR-ABL rearrangement. A: Using multiplex RT-PCR, an atypical BCR-ABL transcript was observed in the patient sample (p) as compared with e14/a2 (c1) and e1/a2 (c2) controls. Lane m: 100-bp ladder. B: Direct sequencing of the RT-PCR product revealed a 42-nucleotide insertion of ABL intron Ib within an e14/a2 BCR-ABL rearrangement. The resulting in-frame insertion of 15 amino acids was shown. C: A well-conserved donor splicing site was detected, at the genomic DNA level, in the 3′ end of the insertion. Uppercase and lowercase nucleotides corresponded respectively to pseudo-exonic and intronic sequences.
Because this sequence of 42 nucleotides was recognized as an exon, we hypothesized that potential branch point and acceptor splicing site were brought by BCR intron 14 during the translocation process and that the genomic breakpoint between BCR and ABL genes was located nearby the 5′ end of ABL intron Ib inserted sequence. To examine this hypothesis and because of the small size of BCR intron 14 (2127bp), the precise location of the breakpoint in BCR was determined (Figure 2A and 2B). For this, PCR experiments were performed using five forward primers (BCR-a to BCR-e) overlapping the BCR genomic region from the 3′ end of intron 13 to the 3′ end of intron 14 and a reverse primer (ABL-z) located in ABL intron Ib (at the 3′ end of the 42-nucleotide insertion). Clear and specific PCR amplifications were obtained with primers BCR-e and BCR-d demonstrating that the rearrangement in BCR was located downstream primer BCR-e (at the 3′ end of intron 14). Direct sequencing of PCR products revealed that the genomic BCR-ABL breakpoint occurred after the thymine at position 126,523 for BCR (95 bp upstream of BCR exon e15; GenBank accession number U07000) and before the guanine at position 75,101 for ABL intron Ib (GenBank accession number NG_012034). The BCR-ABL breakpoint was located nine nucleotides upstream of the intronic inserted ABL sequence.
Figure 2.
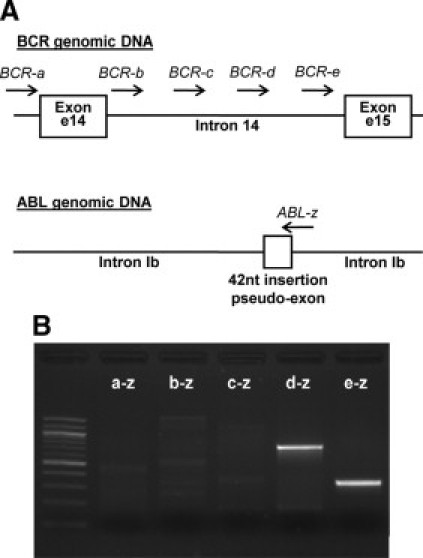
Identification of the BCR-ABL breakpoint on genomic DNA. A: As we hypothesized that the BCR-ABL breakpoint occurred near the 5′ end of the inserted sequence, five primers located in BCR introns 13 and 14 and a reverse primer in ABL intron Ib (at the 3′ end of the insertion) were designed. B: Specific PCR amplifications were observed with two pairs of primers (d-z and e-z).
The genomic sequence surrounding the BCR-ABL breakpoint was then analyzed using Human Splicing Finder and SROOGLE (Splicing RegulatiOn Online GraphicaL Engine; Tel Aviv University, Ramat Aviv, Israel; 2008; available at http://sroogle.tau.ac.il/; accessed October 14, 2009)5 software to identify splicing signals. In addition to the donor site already identified in ABL intron Ib, a consensus acceptor splice site (tatgtccacaagGT), a polypyrimidine tract and a potential lariat branch point (ttacaac) were identified (Figure 3). Finally, GeneMark (Eukaryotic GenMark.hmm. Georgia Institute of Technology, Atlanta, GA; version 3.0; 2005; available at http://exon.gatech.edu/GeneMark/; accessed October 14, 2009),6 and FGENES (SoftBerry, Mount Kisco, NY; 2007; available at http://linux1.softberry.com/berry.phtml; accessed October 14, 2009) software clearly recognized the 42-nucleotide sequence of ABL intron Ib as an exon. Thus, in our observation, the BCR-ABL rearrangement was at the origin of a chimeric splicing acceptor site. The BCR genomic DNA sequence (intron 14) contributed to the polypyrimidine tract and an upstream potential branch point, whereas the ABL DNA sequence (intron Ib) brought a part of the acceptor site and a well-conserved splicing donor site. Consequently, the BCR-ABL rearrangement generated the recognition of an intronic sequence of 42 nucleotides derived from ABL intron Ib as a pseudo-exon. At the mRNA level, the rearrangement consisted in the following sequence: BCR exon e14/ins 42 nucleotides from ABL intron Ib/ABL exon a2. This insertion maintained the reading frame between BCR and ABL mRNA and did not generate a premature stop codon. Therefore, it encoded an in-frame novel 15-amino acid sequence (SKEIQATKKRRLINK) inserted between amino acid 927 of BCR and amino acid 28 of ABL type 1a (Figure 1), leading to a functional protein slightly larger than the classical BCR-ABL oncoprotein of 210 kDa.
Figure 3.

Characterization of the consensus splice sites surrounding the 42-nucleotide insertion. Uppercase and lowercase nucleotides corresponded respectively to pseudo-exonic and intronic sequences. The BCR-ABL breakpoint occurred nine nucleotides upstream of the insertion. Well-conserved consensus splice sites were recognized in the sequence: donor site in ABL intron Ib, acceptor site at the BCR-ABL breakpoint, potential lariat branch point and polypyrimidine tract in BCR intron 14.
Although infrequent, several nonclassical BCR-ABL rearrangements incorporating intronic sequences recognized as a pseudo-exon have been previously reported in CML patients at presentation. In most of the cases, the inserted intronic sequence derived from ABL intron Ib (120.1 kb),7,8,9,10,11 from ABL intron Ia (18.5 kb),12,13 and more rarely from BCR intron 8 (10.2 kb).14 The insertion of intronic sequences within a BCR-ABL rearrangement may have three major consequences: the conservation, the disruption, or the restoration of an open reading frame depending on the location of the insertion and the number of nucleotides inserted (Figure 4). BCR exons e1, e6, e13, e14, and e19 (and theoretically exons e12 and e20) can be fused to ABL exon a2 (rarely a3) without a reading frame shift generating a functional oncoprotein. Because molecular breakpoints in BCR and ABL genes generally occurred in introns, an inserted sequence should be a multiple of three nucleotides to conserve the reading frame (Figure 4A). Concerning the e13/a2 and e14/a2 rearrangements characteristic of CML, very few cases of in-frame insertion of a pseudo-exon between coding BCR and ABL sequences were described. Apart from the case mentioned here, an insertion of 12 nucleotides between BCR exon e13 and ABL exon a2 was reported previously.15 Moreover, it should be noted that several studies described, in addition to a classical e13/a2, e14/a2, or e19/a2 mRNA, aberrant BCR-ABL transcripts harboring a frameshift insertion (3n + 1 or 3n + 2 nucleotides) disrupting the reading frame and leading to a premature stop codon (Figure 4B).7,16,17 Concerning rearrangements involving BCR exons out-of-frame with ABL exon a2 (or a3), an intronic insertion could restore the reading frame between the two coding sequences (Figure 4C). As a consequence, the resulting BCR-ABL protein could then be functional. The BCR exon 8 was most closely involved in such rearrangements with the identification of e8/intronic insertion/a2 BCR-ABL transcripts. To restore an artificial reading frame between BCR and ABL sequences, two mechanisms were mainly described: i) an intronic insertion of 3n + 1 nucleotides between BCR exon e8 and ABL exon a2,10,14 or ii) an intronic insertion within exon 8 (formation of an e8 partial/insertion/a2 rearrangement).11,12,13 Concerning the e8/a2 rearrangement, the same ABL intron Ib inverted insertion has been reported in two CML cases.10,18
Figure 4.
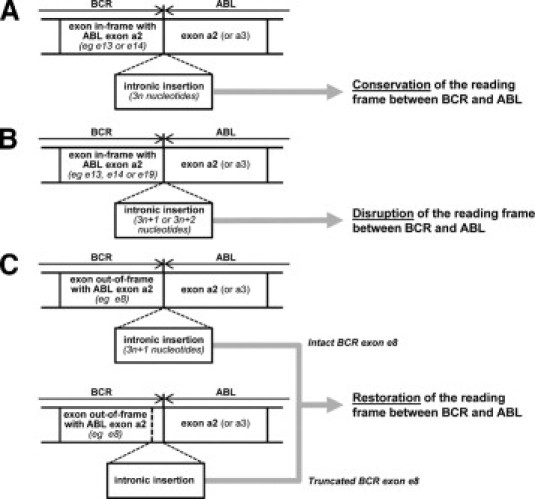
Consequences of an intronic insertion between BCR and ABL exons. A: The insertion conserves the reading frame between BCR and ABL exons. B: The inserted sequence disrupts the reading frame and leads to a premature stop codon. C: The insertion restores a reading frame with intact or truncated last BCR exon e8.
From a technical standpoint, the very low incidence of small sequences inserted within typical e13/a2 or e14/a2 rearrangement in CML could be the consequence of a misidentification of these types of transcript in agarose gel electrophoresis (because of a too small difference in size). Conversely, a rearrangement involving BCR exon 8 (in whole or in part) and an intronic insertion (restoring the reading frame) should be easier to detect because of a theoretical size clearly distinguishable from classical amplicons. Regarding the BCR-ABL rearrangement described here, the insertion of 42 nucleotides, increasing the amplicon length (190 bp as compared with the 148 bp of the typical e14/a2 transcript), could affect the efficiency of standard quantitative RT-PCR.18 Consequently, to correctly monitor the response to tyrosine kinase inhibitor therapy, PCR primers (especially the forward primer in BCR) should be redesigned to reduce the amplicon size.
In conclusion, we identified a novel BCR-ABL transcript produced by the fusion of BCR exon e14 to ABL exon a2 with a 42-bp insertion from ABL intron Ib. Because of the rearrangement process, this pseudo-exon was flanked by canonical consensus splice sites, did not disrupt the reading frame between BCR and ABL, and did not generate a premature termination codon. The encoded BCR-ABL protein contains an in-frame insertion of 15 amino acids. However, the full biological and clinical implications of the rearrangement reported here remain unknown. Therefore, no reliable relationship between this unusual BCR-ABL transcript and an apparent more aggressive form of CML can be established. These results highlight the usefulness of a robust method to theoretically detect all types of BCR-ABL mRNA. Finally, the detection of an atypical transcript (as the BCR exon e14/ins 42 nucleotides from ABL intron Ib/ABL exon a2 rearrangement reported here) requires the adjustment of quantitative RT-PCR protocols for an optimized monitoring of the residual disease.
Footnotes
Supported by grants from the Ligue Contre le Cancer.
References
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 2.Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- 3.Cross NC, Melo JV, Feng L, Goldman JM. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994;8:186–189. [PubMed] [Google Scholar]
- 4.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz S, Hall E, Ast G. SROOGLE: webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Res. 2009;37:W189–W192. doi: 10.1093/nar/gkp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirota M, Hidaka E, Ueno I, Ishikawa M, Asano N, Yamauchi K, Ishida F, Tozuka M, Katsuyama T. Novel BCR-ABL transcript containing an intronic sequence insert in a patient with Philadelphia-positive acute lymphoblastic leukaemia. Br J Haematol. 2000;110:867–870. doi: 10.1046/j.1365-2141.2000.02205.x. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto K, Karasawa M, Sakai H, Ogura H, Morita K, Naruse T. A novel acute lymphoid leukaemia type BCR/ABL transcript in chronic myelogenous leukaemia. Br J Haematol. 1997;96:611–613. doi: 10.1046/j.1365-2141.1997.d01-2066.x. [DOI] [PubMed] [Google Scholar]
- 9.Roman J, Parziale A, Gottardi E, De Micheli D, Cilloni D, Tiribelli M, Gonzalez MG, del Carmen RoM, Torres A, Saglio G. Novel type of BCR-ABL transcript in a chronic myelogenous leukaemia patient relapsed after bone marrow transplantation. Br J Haematol. 2000;111:644–646. doi: 10.1046/j.1365-2141.2000.02394.x. [DOI] [PubMed] [Google Scholar]
- 10.Branford S, Rudzki Z, Hughes TP. A novel BCR-ABL transcript (e8a2) with the insertion of an inverted sequence of ABL intron 1b in a patient with Philadelphia-positive chronic myeloid leukaemia. Br J Haematol. 2000;109:635–637. doi: 10.1046/j.1365-2141.2000.02042.x. [DOI] [PubMed] [Google Scholar]
- 11.Martinelli G, Terragna C, Amabile M, Montefusco V, Testoni N, Ottaviani E, de Vivo A, Mianulli A, Trabacchi E, Saglio G, Tura S. Translisin recognition site sequences flank translocation breakpoints in a Philadelphia chromosome positive chronic myeloid leukemia patient expressing a novel type of chimeric BCR-ABL transcript (E8-INT-A2) Leukemia. 1999;13:1635–1637. doi: 10.1038/sj.leu.2401547. [DOI] [PubMed] [Google Scholar]
- 12.Qin YZ, Jiang B, Jiang Q, Zhang Y, Jiang H, Li JL, Zhu HH, Li LD, Liu YR, Chen SS, Huang XJ. Imatinib mesylate resistance in a chronic myeloid leukemia patient with a novel e8a2 BCR-ABL transcript variant. Acta Haematol. 2008;120:146–149. doi: 10.1159/000178145. [DOI] [PubMed] [Google Scholar]
- 13.Park IJ, Lim YA, Lee WG, Park JS, Kim HC, Lee HJ, Cho SR. A case of chronic myelogenous leukemia with e8a2 fusion transcript. Cancer Genet Cytogenet. 2008;185:106–108. doi: 10.1016/j.cancergencyto.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Tchirkov A, Couderc JL, Perissel B, Goumy C, Regnier A, Uhrhammer N, Verrelle P, Berger M. Major molecular response to imatinib in a patient with chronic myeloid leukemia expressing a novel form of e8a2 BCR-ABL transcript. Leukemia. 2006;20:167–168. doi: 10.1038/sj.leu.2404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubinstein R, Purves LR. A novel BCR-ABL rearrangement in a Philadelphia chromosome-positive chronic myelogenous leukaemia variant with thrombocythaemia. Leukemia. 1998;12:230–232. doi: 10.1038/sj.leu.2400917. [DOI] [PubMed] [Google Scholar]
- 16.Shiratsuchi M, Muta K, Minami R, Motomura S, Suehiro Y, Abe Y, Shiokawa S, Umemura T, Fukui T, Nishimura J, Nawata H. Aberrant BCR-ABL transcript with intronic insertion in a patient with Philadelphia chromosome-positive chronic myeloid leukemia: implications for disease progression. Leuk Lymphoma. 2001;41:411–415. doi: 10.3109/10428190109057996. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Tanaka H, Tanaka K, Ito K, Kyo T, Dohy H, Kamada N, Kimura A. Insertion of a genomic fragment of chromosome 19 between BCR intron 19 and ABL intron 1a in a chronic myeloid leukaemia patient with micro-BCR-ABL (e19a2) transcript. Br J Haematol. 2004;126:752–753. doi: 10.1111/j.1365-2141.2004.05119.x. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto T, Ijima K, Hisatomi H, Murayama T, Mizuno I, Hato A, Imoto S, Nishimura R, Koizumi T. Second case of CML with aberrant BCR-ABL fusion transcript (e8/a2) with insertion of an inverted ABL intron 1b sequence. Am J Hematol. 2004;77:164–166. doi: 10.1002/ajh.20138. [DOI] [PubMed] [Google Scholar]
Uncited reference
- 19.Gabert J, Beillard E, van der Velden V, Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela JM, Cave H, Pane F, Aerts JL, De Micheli D, Thirion X, Pradel V, Gonzalez M, Viehmann S, Malec M, Saglio G, van Dongen J. Standardization and quality control studies of “real-time” quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]


