Abstract
DNA methylation, a key mechanism of repressing gene expression, is of particular relevance in controlling development and cell differentiation. We analyzed the extent and regulation of DNA methylation of the α-smooth muscle actin (α-SMA) gene to elucidate its potential role in myofibroblast differentiation. These experiments revealed the presence of three CpG islands that were methylated at different levels in fibroblasts, myofibroblasts, and alveolar epithelial type II cells. Coordinately, these cells expressed low, high, or no α-SMA, respectively. In addition, inhibition of DNA methyltransferase activity or knock down of DNA methyltransferase using specific small interfering RNA caused significant induction of α-SMA in fibroblasts. In contrast, induced overexpression of DNA methyltransferase suppressed α-SMA gene expression. Transforming growth factor β induced myofibroblast differentiation was enhanced or suppressed by knockdown or overexpression of DNA methyltransferase, respectively. Finally, in vitro DNA methylation of the α-SMA promoter suppressed its activity. These findings suggest that DNA methylation mediated by DNA methyltransferase is an important mechanism regulating the α-SMA gene expression during myofibroblast differentiation.
Expression of α-smooth muscle actin (α-SMA) is a key indicator of myofibroblast differentiation in fibroblasts.1,2 Myofibroblasts have a phenotype intermediate between fibroblasts and smooth muscle cells.1,2 Their accumulation in tissue remodeling and fibrosis leads to excessive deposition of the extracellular matrix, production of profibrogenic cytokines, and altered mechanical properties of affected tissue.2,3 In view of their importance in fibrosis and certain cancers, understanding the mechanism of this differentiation is important for complete elucidation of the pathogenesis of fibrosis, as well as its management and treatment. While a great deal is known about transcriptional regulation of the α-SMA gene,4,5 there is little information regarding the epigenetic regulation of this aspect of myofibroblast differentiation. The importance of histone acetylation is recently suggested in dermal myofibroblast differentiation,6 while inhibition of DNA methylation suppresses hepatic myofibroblast differentiation.7 However whether this is mediated via direct alterations in DNA methylation of the α-SMA gene is uncertain.
DNA methylation is a covalent modification in which cytosine is methylated in a reaction catalyzed by DNA methyltransferases (Dnmts) with S-adenosyl-methionine as the methyl donor.8 In adult somatic tissues, DNA methylation typically occurs in a CpG dinucleotide context while non-CpG methylation is prevalent in embryonic stem cells.9 In mammalian cells, three Dnmt isoforms, namely Dnmt1, Dnmt3a, and Dnmt3b, have been described responsible for DNA methylation.8,9,10,11,12,13,14 It is thought that Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development,10 while Dnmt1 serves as a maintenance type of methyltransferase that is responsible for copying DNA methylation patterns to the daughter strands during DNA replication.11,12,13
The DNA methylation pattern is an important component of the regulatory mechanisms of gene expression.7,8,9,10,11,12,13,14,15,16,17 In many disease processes such as cancer, gene promoter CpG islands acquire abnormal hypermethylation, which results in heritable transcriptional silencing.15,16,17 In an attempt to find out the regulatory mechanism of myofibroblast differentiation, the potential role of DNA methylation was investigated in terms of its impact on α-SMA gene expression. The findings revealed the presence of three CpG islands in the α-SMA gene that were differentially methylated in α-SMA expressing myofibroblasts versus nonexpressing lung alveolar epithelial type II cells. Inhibition of fibroblast DNA methyltransferase with either an inhibitor or specific DNA methyltransferase small interfering (si)RNA leads to significant induction of α-SMA expression, while ectopic expression of Dnmts suppressed its expression. Moreover in vitro DNA methylation of the α-SMA promoter abolished its activity. These data suggested that DNA methylation by Dnmts represented a key mechanism for suppression of myofibroblast differentiation.
Materials and Methods
Animals and Cell Culture
Pathogen-free female Fisher 344 rats (7 to 8 weeks old) were purchased from Charles River Breeding Laboratories, Inc. (Wilmington, MA). Fibroblasts were isolated from rat lungs by enzymatic digestion as before.18,19,20 Cells were then maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% plasma-derived serum (Cocalico Biologicals, Inc., Reamstown, PA), antibiotics, 1% insulin/transferring/selenium (Sigma Chemicals, St. Louis, MO), 5 ng/ml platelet-derived growth factor (R&D Systems, Inc., Minneapolis, MN), and 10 ng/ml epidermal growth factor (R&D Systems, Inc., Minneapolis, MN). The adherent cells were then trypsinized and passaged for at least three times before use. Where indicated, cells were treated with 4 ng/ml of transforming growth factor (TGFβ1; R&D systems, Inc., Minneapolis, MN) for 48 hours or 72 hours to induce myofibroblast differentiation as before.18
Rat alveolar epithelial type II cells were isolated by elastase cell dispersion and IgG panning as before.19 They were cultured on 6-well tissue culture dishes precoated with fibronectin (R&D Systems, Inc., Minneapolis, MN) in Dulbecco’s modified Eagle’s medium supplemented with 10% newborn calf serum (Sigma). The cells were of >90% purity based on assessment using by anti-cytokeratin5/8 (BD Biosciences Inc, San Diego, CA) immunofluorescent staining.
DNA Pyrosequencing Analysis
Genomic DNA were extracted from cells using Wizard genomic DNA extraction kit (Promega Inc., Madison, WI) and 1 μg of the genomic DNA was bisulfite modified using the Zymo Research EZ Methylation gold kit (Zymo Research Corp., Orange, CA) in accordance with the manufacturer’s protocol. The bisulfite modified sample DNA was then 10-fold diluted and 1 μl of diluted DNA was used in PCR reactions with 3 μl 10× PCR buffer, 200 μmol/L of dNTPs, 6 pmol forward primer, 6 pmol reverse primer, and 3 mmol/L MgCl2, 0.75 U Qiagen HotStar Taq polymerase (Qiagen Inc., Valencia, CA. Cat. # 205203) in 30 μl total volume adjusted using double distilled H2O as necessary. The PCR cycling condition is as following: 95°C 15 minutes; 45 × (95°C 30 s; 51°C 30 s; 72°C 30 s); 72°C 10 minutes; 4°C ∞. The PSQ96HS system is used according to standard procedures for the PyrosequencingTM analysis.
Plasmid and Constructs
The mouse Dnmt1, Dnmt3a, and Dnmt3b cDNA constructs were purchased from Thermo Fisher Scientific Inc., Huntsville, AL. They were amplified by PCR, digested, and inserted into vector pLenti6/V5-DEST to form CMV promoter driven constitutively expressed Dnmt constructs pLenti6/V5-DEST-dnmt1, pLenti6/V5-DEST-dnmt3a, and pLenti6/V5-DEST-dnmt3b respectively. The lentivirus based siRNA constructs specific for rat Dnmt1, Dnmt3a and Dnmt3b, respectively, and the negative control siRNA construct were from Open Biosystems and inserted into lentivirus by the Vector Core at the University of Michigan.
The rat −2880 to + 32 (promoter) and −2880 to + 2803 (promoter plus first intron) rat α-SMA gene promoter region sequences (numbered from transcription start site) were amplified by PCR and cloned into promoterless pGL3-basic vector to form the α-SMApro-Luc and α-SMApro-intron-Luc constructs, respectively, wherein the luciferase reporter gene expression was regulated by these promoters.
Transfection and Reporter Gene Assay
All transient transfections of cells were performed using the FuGENE6 reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions as previously described.18 Supercoiled DNA was isolated with an endotoxin-free Qiagen column kit (Qiagen Inc, Valencia, CA). Unless otherwise indicated, 2 μg DNA of the α-SMA promoter-luciferase construct of interest and 100 ng plasmid pRL-SV40 control vector (used for normalization) were co-transfected per culture into rat lung fibroblasts in serum-free Dulbecco’s modified Eagle’s medium. Four hours after transfection, the media were replaced with Dulbecco’s modified Eagle’s medium containing 10% plasma-derived serum and treated with either 4 ng/ml TGFβ or buffer only. The cells were harvested 48 hours after transfection, and the activity of firefly or Renilla luciferase was measured using the dual luciferase assay system from Promega Corporation, Madison, WI. The relative luciferase activity was calculated by normalizing firefly luciferase activity to that of Renilla luciferase. Experiments with each construct were repeated two to four times and results shown as relative light units and expressed as mean ± SE.
RNA Analysis by Quantitative Real-Time PCR
This was undertaken to assess gene expression using a GeneAmp 7500 Sequence Detection System (PE/ABI, Foster City, CA) as before.18,19,20 All required primers and probes were purchased from Applied Biosystems (PE/ABI, Foster City, CA) and the results were expressed as 2−ΔΔCT using 18s RNA as the reference. The specific calibrator for each assay was indicated in the figure legends.
Nuclear Extraction and DNA Methyltransferase Assay
Nuclear extracts were prepared with the nuclear extraction kit from Epigentek (Brooklyn, NY) in accordance with the manufacturer’s protocol. Protein concentration was determined using the bicinchoninic acid assay from Thermo Fisher Scientific Inc. (Huntsville, AL) according to the user manual. DNA methyltransferase activity was assayed using the EpiQuik DNA Methyltransferase Activity/Inhibition Assay Kit from Epigentek. Results were expressed as absorbance units and normalized to the activity in control untreated rat lung fibroblasts (set as 100%).
Flow Cytometry Analysis
Fibroblasts or alveolar epithelial type II cells were infected for 72 hours with the respective lentivirus construct as indicated. After detachment with cell dissociation buffer (Invitrogen Co. Carlsbad, CA) and washing in PBS, they were then fixed with 4% paraformaldehyde for 15 minutes at room temperature. They are stained with phycoerythrin conjugated anti-α-SMA antibody (R&D Systems, Inc., Minneapolis, MN) or corresponding phycoerythrin conjugated IgG control in 0.1% saponin containing 1× PBS for 1 hour at room temperature. After stringent washing with 0.1% Saponin containing 1× PBS, they were analyzed on a Cytomics FC500 machine (Beckman Coulter Inc., Miami, FL). Data collected were then analyzed using Winlist software (Verity Software House Inc., Topsham, ME).
Western Blot Analysis
The anti-α-SMA antibody was purchased from Sigma-Aldrich St. Louis, MO and all of the other antibodies were purchased from Santa Cruz Biotechnology Inc., Santa Cruz, CA. Equal amounts of cell protein extracts were loaded onto 12% SDS polyacrylamide gels and transferred onto Hybond-P membranes for Western blotting as previously described.18,19,20
Statistical Analysis
This was undertaken as before18,19,20 using analysis of variance, followed, where appropriate, by post hoc testing using Scheffé’s test. A value of P < 0.05 was used as a criterion for statistical significance in comparisons between any two groups.
Results
Identification and Methylation Status of CpG Islands
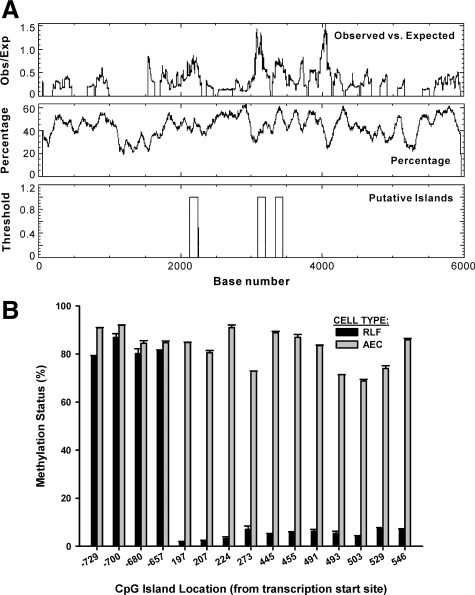
De novo appearance of myofibroblasts with their distinct α-SMA expressing phenotype is characteristic of, and potentially important in, tissue remodeling, fibrosis and cancer.1,2,3,4 To study if DNA methylation was involved in the control of α-SMA gene expression, the gene sequence was first scanned for potential CpG islands using the EMBOSS CpGPlot/CpGReport/Isochore program at http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html. This analysis revealed the presence of three CpG islands residing in the promoter and the first intron (Figure 1A). To determine the methylation status of these islands and its functional significance vis-à-vis expression of α-SMA, genomic DNA isolated from fibroblasts and alveolar epithelial type II cells were pyrosequenced using primers specific for the α-SMA gene. The results revealed that the CpG islands in the α-SMA promoter region were on the average >80% methylated in both fibroblasts and alveolar epithelial cells (Figure 1B). However, the CpG islands in the first intron showed very limited methylation (≤7%) in fibroblasts, but were highly methylated (>59%) in alveolar epithelial cells. This differential methylation of the CpG islands in the intronic region between these two cell types correlated with expression of α-SMA in the former and not in the latter cells, suggesting that methylation of these CpG islands may be of functional significance in repressing α-SMA gene expression. To evaluate this further, the effects of manipulating DNA methylation by altering the expression of Dnmts were examined.
Figure 1.
A: Location of CpG islands in α-SMA gene. DNA sequence of α-SMA gene −2900 to + 3100 from transcriptional start site was submitted to the EMBOSS CpGPlot/CpGReport/Isochore program online. The results revealed three islands between bp 2142 to 2244 (corresponding to −759 to −656 from transcriptional start site, namely promoter region), bp 3095 to 3199 (corresponding to + 195 to + 299 from transcriptional start site, namely first intro region1), and bp 3339 to 3452 (corresponding to + 439 to + 552 from transcriptional start site, namely first intron region 2). B: Methylation status of CpG islands in the α-SMA gene. This was determined by DNA pyrosequencing of the α-SMA gene promoter and the first intron regions 1 and 2 where CpG islands were located. The position of each methylated cytidine from the transcriptional start site was indicated in the x axis. The results were expressed as percentages and shown as the means ± SE from three independent experiments.
Dnmt Expression Profile in Fibroblasts and Epithelial Cells
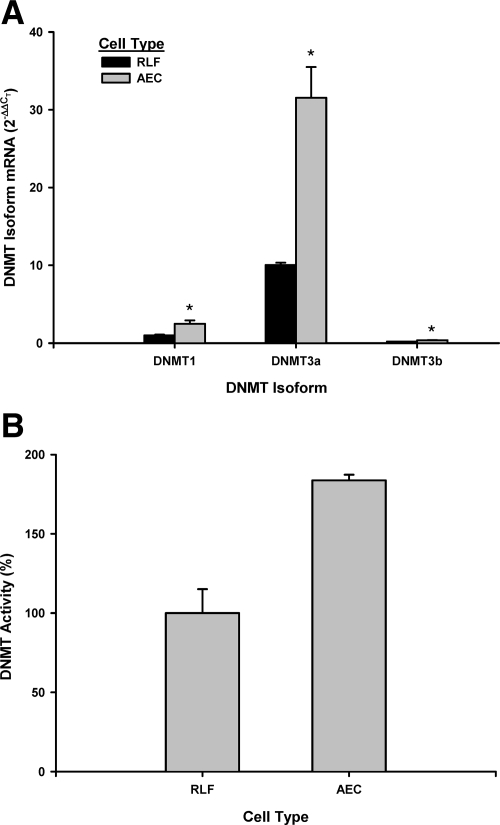
Initially the Dnmt isoform expression profile for these cells was examined. In mammalian cells, three Dnmt isoforms, Dnmt1, Dnmt3a, and Dnmt3b, have been identified and are responsible for methylation of DNA.8,9,10,11,12,13,14 To determine the presence of these Dnmts in fibroblasts and epithelial cells, RNA was isolated from these cells and analyzed for the respective Dnmt isoform mRNA level by real-time PCR. The results indicated that all three Dnmt isoforms were detectable in both fibroblasts and epithelial cells (Figure 2A). Dnmt3a showed the highest expression level in both cells. Interestingly epithelial cells, which do not express α-SMA, generally expressed higher levels of all Dnmts than fibroblasts. To confirm this relative difference in Dnmt expression between these two cell types, fresh nuclear extract from both cells were prepared and assayed for the level of DNA methyltransferase activity. Consistent with the Dnmt mRNA data (Figure 2B), the results showed that DNA methyltransferase activity was significantly higher (>180%) in epithelial cells than fibroblasts. This would be consistent with the finding of increased methylation of the CpG islands in the α-SMA gene of epithelial cells versus that of fibroblasts.
Figure 2.
Dnmt expression in fibroblasts (RLF) and epithelial cells (AEC). A: Total RNA from the indicated cell type were analyzed by real-time PCR for the indicated Dnmt isoform mRNA. The results were expressed as 2−ΔΔCT with 18s rRNA used as the endogenous control. Data were shown as the mean ± SE from triplicate samples. An asterisk indicated statistically significant difference when compared with the corresponding mean value in AECs. B: DNA methyltransferase assay was conducted using equal amounts of freshly prepared nuclear extracts from either fibroblasts (RLF) or epithelial cells (AEC). The results were expressed as a percentage of the mean activity in fibroblasts. Mean ± SE from triplicate samples are shown.
Dnmt Inhibition Induced α-SMA Gene Expression in Fibroblasts
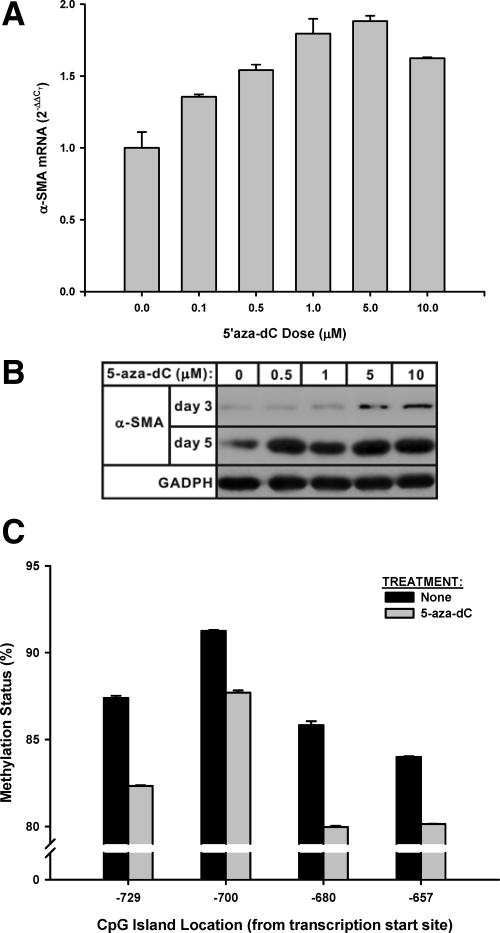
To confirm that DNA methyltransferase activity is an important regulator of myofibroblast differentiation, the effect of a Dnmt inhibitor, 5-aza-2′-deoxycytidine (5-aza-dC), on α-SMA expression was examined. This compound has been used for treating cancers21,22 and recently widely used as a DNA methyltransferase inhibitor. It is incorporated into DNA to form stable complexes with Dnmt resulting in its inactivation.21,22 To study if DNA methyltransferase could regulate myofibroblast differentiation, fibroblasts were treated with 5-aza-dC at the indicated doses and then analyzed for α-SMA gene expression by real-time PCR and Western blot analysis. The results showed that 5-aza-dC significantly induced expression of α-SMA in fibroblasts in a dose dependent manner, at both mRNA (Figure 3A) and protein (Figure 3B) levels. Much higher levels of protein expression were noted after 5 days of treatment relative to those seen at 3 days. To confirm that the treatment with this inhibitor did have an effect on the methylation status of the α-SMA gene, genomic DNA isolated from control and 5-aza-dC treated fibroblasts were pyrosequenced using primers corresponding to the α-SMA gene. The results showed that the methylation ratio of deoxycytidine in the α-SMA gene was decreased by 3.55 to 5.86% depending on the location of the CpG island (Figure 3C). The overall average reduction in all sites was 4.59% in cells treated with the inhibitor for 72 hours relative to that in control cells (Figure 3C). Thus inhibition of DNA methytransferase activity resulted in reduced DNA methylation and induction of α-SMA expression.
Figure 3.
Effect of DNA methyltransferase inhibitor on α-SMA expression. A: Fibroblasts were treated with the indicated doses of DNA methyltransferase inhibitor, 5-aza-dC for three days. Total RNA was then extracted and analyzed for α-SMA mRNA by real-time PCR. The results were expressed as 2−ΔΔCT using 18s rRNA as the endogenous control. Data were shown as the mean ± SE from triplicate samples. All treated samples showed significantly higher levels of α-SMA expression compared with untreated controls. B: Fibroblasts were treated with 5-aza-dC at the indicated dose for three or five days, and then analyzed for α-SMA protein levels by Western blotting. Equal amounts of total protein were loaded onto the gels for separation. The membrane was striped and reblotted with horseradish peroxidase-conjugated anti-glyceraldehyde-3-phosphate dehydrogenase antibody as a loading control. C: The cells were treated with either control buffer (“None”) or 5-aza-dC for 72 hours. The genomic DNA was then isolated and pyrosequenced using primers corresponding to the α-SMA gene promoter region. The extent of methylation (expressed as percentages) was plotted versus the corresponding position of the CpG island from the transcriptional start site. The results were shown as mean ± SE.
Dnmt siRNAs Induced α-SMA Expression
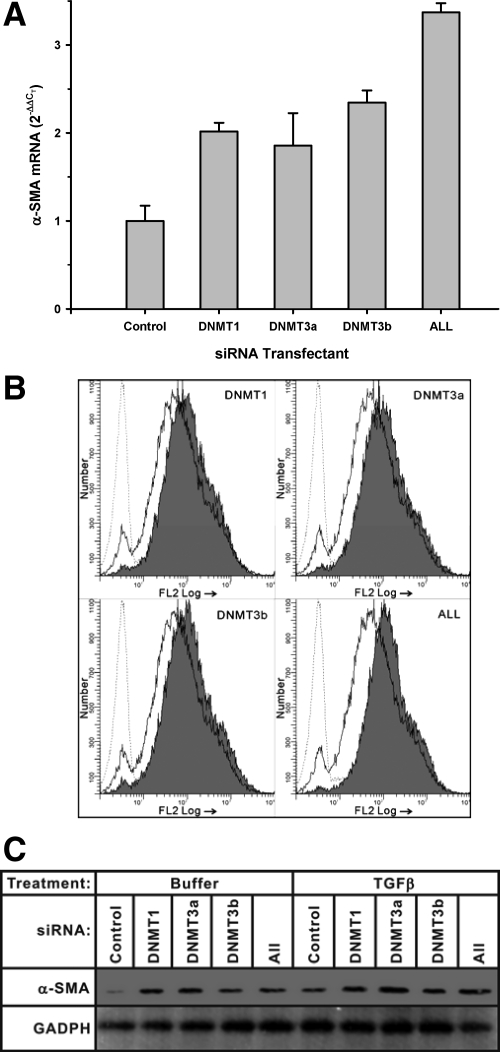
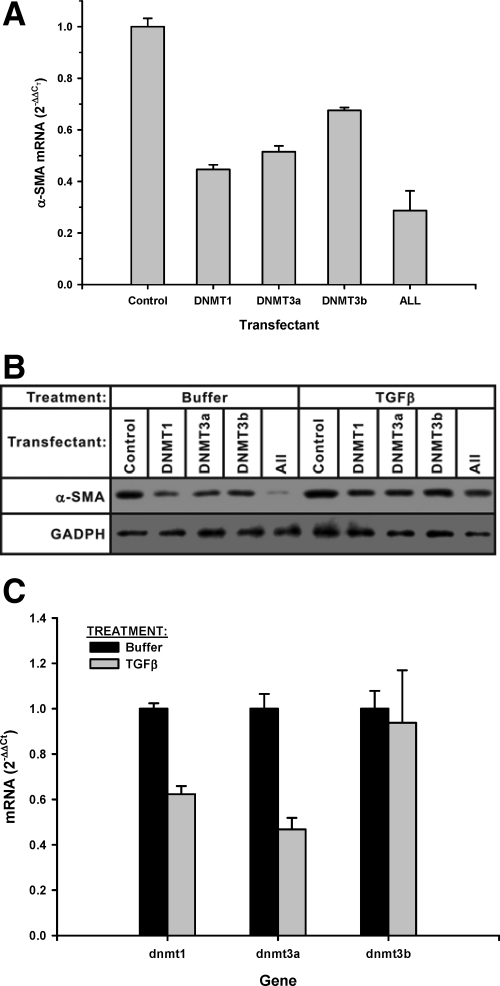
siRNA has been effectively used to inhibit expression of specific gene targets.23,24 This strategy was used to evaluate if DNA methylation mediated by specific Dnmts could regulate α-SMA expression in fibroblasts. Lentivirus siRNA mimics against the indicated Dnmt isoforms were transfected into fibroblasts and 48 hours later were analyzed for α-SMA mRNA levels by real-time PCR. The results showed that consistent with the DNA methyltransferase inhibitor studies, transfection of each of the indicated Dnmt isoform siRNA individually stimulated α-SMA expression (Figure 4A). Addition of all three Dnmt isoform siRNAs caused a further increase in α-SMA expression over the transfection by individual Dnmt isoform siRNA, but not synergistically. Analysis of α-SMA protein levels by flow cytometry (Figure 4B) confirmed the stimulatory effects of these Dnmt siRNAs on α-SMA expression. To put this observation in the context of TGFβ induced myofibroblast differentiation, the effects of these Dnmt siRNAs on α-SMA expression were examined in cells treated with TGFβ. As expected, the results showed that TGFβ treatment alone enhanced α-SMA protein expression (Figure 4C). Control untreated (buffer only) cells exhibited similar stimulation of α-SMA protein expression by Western blot analysis (Figure 4C). In TGFβ-treated cells, similar enhancement of α-SMA expression by Dnmt siRNA was also observed. In both situations, the effect of Dnmt3b was the weakest. Thus all Dnmt isoforms examined could significantly regulate α-SMA expression in an additive manner.
Figure 4.
Effect of Dnmt siRNAs on fibroblast α-SMA expression. A: Fibroblasts were transfected with the indicated siRNA construct or constructs or all three constructs (“ALL”) for 48 hours and then analyzed for α-SMA mRNA by real-time PCR. The results were expressed as 2−ΔΔCT using 18s rRNA as the endogenous control. Data were shown as the mean ± SE from triplicate samples. Mean values of cells treated with Dnmt siRNA were significantly higher than that of cells treated with the control vector. B: The indicated siRNA construct or all three constructs (“ALL”) were transfected into fibroblasts and then stained with phosphatidylethanolamine-conjugated anti-α-SMA antibody followed by flow cytometric analysis. The dashed line indicated the IgG control, while the solid line represented the cells transfected with control siRNA and stained with phosphatidylethanolamine-conjugated anti-α-SMA antibody. The shaded region represented cells transfected with the indicated Dnmt siRNA construct(s) stained with phosphatidylethanolamine-conjugated anti-α-SMA antibody. C: Fibroblasts were transfected with the indicated individual Dnmt siRNA or all three siRNAs together (“ALL”) and treated with either buffer only (“Buffer”) or TGFβ (4 ng/ml) for 72 hours. On harvest, equal amounts of total protein extracts were loaded onto gels for Western blotting. The membrane was striped and reblotted with HRP conjugated anti-glyceraldehyde-3-phosphate dehydrogenase antibody as a loading control.
Dnmt Overexpression Suppressed α-SMA Expression
Thus far inhibition of DNA methyltransferase activity or Dnmt expression effectively induced α-SMA expression in fibroblasts. This would be consistent with the initial finding that increased DNA methylation in the α-SMA gene correlated with suppression of α-SMA expression. To further confirm that Dnmt could regulate α-SMA expression, the complementary experiment to the siRNA studies was undertaken to analyze the effect of Dnmt overexpression on α-SMA expression. Dnmt isoform expression plasmids were constructed and transfected into fibroblasts to achieve overexpression of each of the indicated isoforms. The results showed that overexpression of each of the Dnmt isoforms individually caused significant reduction in α-SMA mRNA levels by real-time PCR analysis, with the greatest effect (>50% inhibition) exhibited by Dnmt1 overexpression (Figure 5A). Combined overexpression of Dnmts 1, 3a, and 3b resulted in a greater inhibitory effect without evidence of synergism. Similar inhibition of α-SMA protein levels was also demonstrated by western blotting analysis, and remained detectable even when the cells were treated with TGFβ to induce myofibroblast differentiation (Figure 5B). As with the siRNA experiments, the effect of Dnmt3b overexpression was the weakest. In view of the noted effects on TGFβ induced differentiation, the possibility that this cytokine could up-regulate α-SMA expression by suppressing Dnmt expression was investigated. Fibroblasts were treated with buffer only or TGFβ and 6 hours later were analyzed for Dnmt mRNA levels by real-time PCR. The results showed that TGFβ treatment significantly inhibited expression of Dnmt1 and Dnmt3a but not Dnmt3b (Figure 5C). Thus consistent with the inhibitor and siRNA studies, overexpression of Dnmts suppressed fibroblast α-SMA expression and TGFβ induced myofibroblast differentiation, while TGFβ had suppressive effects on Dnmt expression.
Figure 5.
Effect of Dnmt overexpression on α-SMA expression. A: Fibroblasts were transfected with indicated the indicated Dnmt isoform expression plasmid or all three plasmids (“ALL”) for 48 hours. They were then analyzed for α-SMA mRNA levels by quantitative real-time PCR. The results were expressed as 2−ΔΔCT using 18s rRNA as the endogenous control. Data were shown as the mean ± SE from triplicate samples. All samples treated with Dnmt plasmid(s) were significantly different from the sample treated with the control vector. B: The cells were transfected with the indicated individual Dnmt isoform expression plasmid or all three plasmids together (“ALL”) and treated with either buffer only (“Buffer”) or TGFβ for 72 hours and then analyzed for α-SMA content by Western blotting. The membrane was stripped and reblotted with HRP conjugated anti-glyceraldehyde-3-phosphate dehydrogenase antibody as a loading control. C: Rat lung fibroblasts were treated with 4 ng/ml TGFβ or buffer only (“Buffer”) for 6 hours and then total RNA was extracted and analyzed for mRNA of the indicated dnmt gene by real-time PCR. The results were expressed as mean ± SE with N = 3. The inhibition by TGFβ treatment was significant only for dnmt1 and dnmt3a mRNA.
Methylation of α-SMA Promoter Inhibits α-SMA Gene Expression
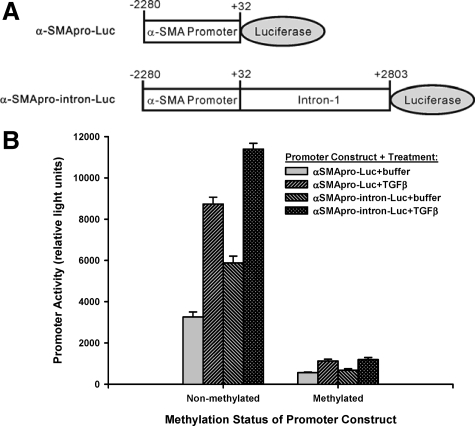
To further confirm the importance of DNA methylation in suppression of α-SMA gene expression, the α-SMA gene promoter region and the α-SMA gene promoter region inclusive of the first intron were cloned with fusion to a luciferase reporter gene, wherein the expression of luciferase was driven by the α-SMA gene promoter (diagrammed in Figure 6A). These constructs were then methylated in vitro with CpG methyltransferase M. SssI isolated from E. Coli, which methylates all cytosine residue (C5) within the double stranded dinucleotide recognition sequence 5′-CG-3′. These methylated and nonmethylated constructs were then transfected into rat lung fibroblasts followed by treatment with TGFβ or buffer to examine the effect of methylation on promoter activity. The results showed the expected promoter activity of both nonmethylated promoter constructs, which was significantly enhanced by TGFβ treatment (Figure 6B). The nonmethylated construct containing the first intron showed higher activity than its intronless counterpart, but exhibited a lower level of stimulation by TGFβ. In contrast, both methylated promoter constructs exhibited essentially baseline levels of activity with minimal stimulation by TGFβ. These results confirmed that methylation of the α-SMA gene promoter region suppressed its expression.
Figure 6.
Effect of DNA methylation on α-SMA promoter activity. α-SMA promoter constructs without the first intronic region (α-SMApro-Luc) and the α-SMA promoter constructs with the first intronic region (α-SMApro-intron-Luc) were methylated in vitro and transfected into rat lung fibroblast (diagrams of constructs in A). B: The transfected cells were treated with either TGFβ or buffer only for 48 hours and tested for the promoter activity in terms of luciferase activity in relative light units. Experiments with each construct were repeated two to four times and the results were expressed as mean ± SE.
Discussion
Myofibroblast differentiation is commonly detected by induction of α-SMA expression in fibroblasts.1,2,3 Regulation of its expression has been extensively studied as a means of understanding the mechanism of differentiation. Understanding this mechanism is of importance because of the key roles they play in tissue fibrosis and more recently in cancer.4 Despite discovery of an increasing number of trans-acting factors and cis-acting elements that control α-SMA gene expression,20,21,22,25,26,27 the epigenetic regulation of its expression has not received this level of attention. In this paper, an attempt was made to determine the potential importance of epigenetic regulation in myofibroblast differentiation from the standpoint of the importance of DNA methylation status of the α-SMA gene on its expression. Highly methylated sequences in CpG islands of many genes are known to be responsible for gene silencing,8,13,14 and especially in the context of cell differentiation and development they may be responsible for maintenance of the undifferentiated state.28 Thus the first step in this study was to determine the extent of DNA methylation in the α-SMA gene in fibroblasts with the capacity for myofibroblast differentiation. Fibroblast cultures routinely contain some myofibroblasts (usually <20%), and thus show a certain level of α-SMA expression even in the absence of an exogenous differentiating stimulus.18,19,20 In these cells, the findings revealed the presence of at least three CpG islands in the promoter and first intron of the α-SMA gene with a distinct methylation pattern. While they revealed a >78% methylation in the promoter sequences, the intronic region was only poorly methylated (<8%). This was in contrast to freshly isolated lung alveolar epithelial type II cells that do not express α-SMA, which showed uniformly high methylation in comparable sequences of the α-SMA gene. Thus consistent with its role in gene silencing, highly methylated DNA sequences in the α-SMA gene correlated with suppression of its expression.
The importance of DNA methylation was further evidenced by stimulation of α-SMA expression by an inhibitor of DNA methyltransferase, 5-aza-dC, which was associated with a reduction in DNA methylation of the α-SMA gene. This stimulatory effect of 5-aza-dC is opposite to that found in hepatic stellate cells, where it was found to inhibit their differentiation to myofibroblasts via an nuclear factor κB dependent mechanism.7 The basis for this difference is unclear, but may be related to the different cell types and/or experimental conditions used. Nevertheless, to confirm the importance of DNA methylation in suppression of myofibroblast differentiation, the effects of suppressing or overexpressing Dnmts on α-SMA expression were examined in fibroblasts. The three known Dnmt isoforms that regulates DNA methylation, Dnmt1, Dnmt3a, and Dnmt3b, are known to be responsible for methylating DNA sequences.8,9,10,11,12,13,14 When Dnmt expression was suppressed using isoform specific siRNAs, there was a significant induction of α-SMA expression. In contrast overexpression of Dnmts by transfection with expression plasmids caused significant repression of α-SMA expression. These observations were noted even in cells treated with TGFβ to induce myofibroblast differentiation. Moreover TGFβ itself inhibited Dnmt expression, suggesting an additional mechanism by which it could regulate α-SMA expression to effect myofibroblast differentiation by suppression of DNA methylation. Thus the totality of the findings revealed significant regulation of α-SMA expression by methylation of its gene sequence mediated in part by Dnmts 1, 3a, and 3b. These results signify the importance of DNA methylation and Dnmts in suppression of myofibroblast differentiation, which may be important in homeostatic prevention of differentiation. Abrogation of this suppression, in addition or in conjunction with transcriptional activation of the α-SMA gene may be involved in the de novo emergence of myofibroblasts in fibrosis or cancer. The findings in this study thus revealed an additional potential target for novel therapeutic approaches for controlling both these processes.
The differential pattern of DNA methylation between fibroblasts and epithelial cells are of some interest, especially since this may account for the absence of α-SMA expression in the latter cell type. Despite expression of all three Dnmt isoforms in both these cell types, this differential methylation pattern suggests additional mechanisms are involved in causing this difference between the two cell types. The consequences of the higher methylation of intronic sequences in epithelial cells on transcriptional activation may contribute to the silencing of the α-SMA gene in these cells. This is supported by the observation that in fibroblasts the α-SMA promoter construct with nonmethylated first intronic region exhibited significantly higher activity than one without the first intronic region, indicative of the potent stimulatory effect of this intron. However, when the constructs were methylated, the presence or absence of the first intron had no significant effect since promoter activity was essentially abolished. Thus additional studies on transcriptional regulation via the first intronic region in myofibroblast differentiation may be warranted in future studies.
Acknowledgments
We thank Matthew Ullenbruch for his assistance in taking care of the animals.
Footnotes
Address reprint requests to Dr. Sem H. Phan, Department of Pathology, University of Michigan Medical School, 109 Zina Pitcher Place, Ann Arbor, MI 48109-2200. E-mail: shphan@umich.edu.
Supported by grants HL28737, HL31963, HL52285, HL77297 and HL91775 from the National Institute of Health.
References
- Gabbiani G, Ryan GB, Majno G. Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction. Experientia. 1971;27:549–550. doi: 10.1007/BF02147594. [DOI] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan SH. The myofibroblast in pulmonary fibrosis. Chest. 2002;122:286S–289S. doi: 10.1378/chest.122.6_suppl.286s. [DOI] [PubMed] [Google Scholar]
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta. 2007;1773:1572–1582. doi: 10.1016/j.bbamcr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: cpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier H-U, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Liu Y, Oakeley EJ, Sun L, Jost J-P. Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res. 1998;26:1038–1045. doi: 10.1093/nar/26.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk E, Defossez PA. The cell biology of DNA methylation in mammals. Biochim Biophys Acta. 2008;1783:2167–2173. doi: 10.1016/j.bbamcr.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Plachot C, Lelièvre SA. DNA methylation control of tissue polarity and cellular differentiation in the mammary epithelium. Exp Cell Res. 2004;298:122–132. doi: 10.1016/j.yexcr.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Poplawski T, Tomaszewska K, Galicki M, Morawiec Z, Blasiak J. Promoter methylation of cancer-related genes in gastric carcinoma. Exp Oncol. 2008;30:112–116. [PubMed] [Google Scholar]
- Lee MP, Dunn BK. Influence of genetic inheritance on global epigenetic states and cancer risk prediction with DNA methylation signature: challenges in technology and data analysis. Nutr Rev. 2008;66 Suppl 1:S69–S72. doi: 10.1111/j.1753-4887.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze-Jun- L, Masato M. Gene promoter methylation–a potential cancer marker. Adv Clin Path. 2002;6:43–46. [PubMed] [Google Scholar]
- Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-beta-induced alpha-smooth muscle actin expression. Am J Respir Cell Mol Biol. 2003;29:397–404. doi: 10.1165/rcmb.2003-0063OC. [DOI] [PubMed] [Google Scholar]
- Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH. FIZZ1 stimulation of myofibroblast differentiation. Am J Pathol. 2004;164:1315–1326. doi: 10.1016/S0002-9440(10)63218-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wu Z, Jin H, Hashimoto N, Liu T, Phan SH. CCAAT/enhancer-binding protein beta isoforms and the regulation of alpha-smooth muscle actin gene expression by IL-1 beta. J Immunol. 2004;173:4661–4668. doi: 10.4049/jimmunol.173.7.4661. [DOI] [PubMed] [Google Scholar]
- Momparler RL, Eliopoulos N, Ayoub J. Evaluation of an inhibitor of DNA methylation, 5-aza-2′-deoxycytidine, for the treatment of lung cancer and the future role of gene therapy. Adv Exp Med Biol. 2000;465:433–446. doi: 10.1007/0-306-46817-4_38. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Zamore P, Tuschl T, Sharp P, Bartel D. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Foster DN, Min B, Foster LK, Stoflet ES, Sun S, Getz MJ, Strauch AR. Positive and negative cis-acting regulatory elements mediate expression of the mouse vascular smooth muscle alpha-actin gene. J Biol Chem. 1992;267:11995–12003. [PubMed] [Google Scholar]
- Kumar MS, Hendrix JA, Johnson AD, Owens GK. Smooth muscle alpha-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ Res. 2003;92:840–847. doi: 10.1161/01.RES.0000069031.55281.7C. [DOI] [PubMed] [Google Scholar]
- Nishida M, Miyamoto S, Kato H, Miwa T, Imamura T, Miwa K, Yasumoto S, Barrett JC, Wake N. Transcriptional repression of smooth-muscle alpha-actin gene associated with human papillomavirus type 16 E7 expression. Mol Carcinog. 1995;13:157–165. doi: 10.1002/mc.2940130305. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]