Abstract
Accumulating evidence indicates that miRNA expression can be used as a diagnostic and prognostic marker for human cancers. We report that the expression level of miR-182 was markedly up-regulated in glioma cell lines and in human primary glioma specimens. Quantitative PCR analysis showed that miR-182 was significantly increased by up to 32-fold in glioma tumors compared with the adjacent nontumor brain tissues obtained from the same patient. Elevated expression of miR-182 was further identified by in situ hybridization in 248 of 253 (98%) archived human glioma biopsies tested. Statistical analysis revealed a significant correlation between miR-182 expression and World Health Organization glioma grading (P < 0.001). The cumulative 5-year survival rate of glioma patients was 51.54% (95% confidence interval, 0.435 to 0.596) in the low miR-182-expression group, whereas it was only 7.23% (95% confidence interval, 0.027 to 0.118) in the high miR-182-expression group (P = 0.001), and multivariate Cox regression analysis indicated that miR-182 expression was an independent prognostic indicator for the survival of glioma patients. Moreover, the correlations of miR-182 level with the clinical features of glioma suggested in the in situ hybridization analysis were further verified by the real-time RT-PCR analysis. Taken together, our results suggest that miR-182 could be a valuable marker of glioma progression and that high miR-182 expression is associated with poor overall survival in patients with malignant glioma.
Gliomas, which represent approximately 70% of all brain tumor cases, are the most common and malignant tumors in the brain of humans, and the overall prognosis for patients inflicted with malignant gliomas is poor.1 The cumulative 1-year survival rate of glioma patients is less than 30%. The median survival time for glioblastoma multiforme (GBM), the grade IV gliomas, is only 15 months.2 Although significant improvements have been made in neurosurgical techniques, development of new chemotherapeutic agents, and exploitation of accurate radiotherapy, extremely poor prognosis of malignant gliomas remains unchanged over the last three decades.3,4 Therefore, it is an urgent clinical challenge to identify sensitive and specific early biomarkers for the diagnosis and prognosis of this malignancy, as well as to develop new therapeutic strategies for this deadly disease.
MicroRNAs (miRNAs) are single stranded and noncoding RNAs that play important roles in many biological processes.5 During the initiation and progression of human cancers, miRNAs have been shown to modulate cell proliferation, survival, and tumor angiogenesis, invasion, and metastasis.6,7,8,9 Dysregulation of miRNA expression has been found in various types of human cancers including cancers occurring in breast, colon, lung, liver, pancreas, chronic lymphocytic leukemia, and malignant gliomas.10,11,12,13,14,15,16 While the molecular mechanisms of miRNA-mediated gene regulation are still under investigation, emerging studies suggest that miRNA expression signatures can be diagnostically and/or prognostically indicative for human cancers.17 For example, miRNA profiles of acute lymphocytic leukemia could categorize patients with different molecular pathologies into different subgroups.18 A unique miRNA signature was found to be associated with prognostic factors and disease progression in chronic lymphocytic leukemia.19 A statistical miRNA profile that differed with tumor histology distinguished lung adenocarcinomas from noncancerous lung tissues and correlated with the prognosis of patients with lung cancers.20 A five-miRNA signature was identified as a potential independent predictor of cancer relapse and survival of non-small-cell lung cancer patients.12 An onco-miRNA, miR-21, was drastically up-regulated in various types of human cancers, including gliomas, and associated with prognosis and therapeutic outcome in colon and breast cancers.15,20,21,22 Acute myeloid leukemia patients with high expression of miR-191 and miR-199a had significantly lower overall and event-free survival rates than AML patients with low expression of the miRNAs.18 Reduced expression of let-7 miRNA in human lung cancer and ovarian cancer was associated with shortened postoperative survival.23,24 Taken together, a body of evidence has suggested that miRNA expression might be a clinically useful marker for analyzing molecular pathogenesis of human cancers and developing targeted therapies or selecting high-risk cancer patients for adjuvant chemotherapies.
Recently, we examined the levels of 450 miRNAs in human primary glioma tissues of varying World Health Organization grades by microarray analyses and found that miR-182 was significantly up-regulated in glioma tissues as compared with normal brain tissues. In the present study, we characterized the miR-182 expression in a cohort of 253 gliomas specimens including World Health Organization grade I to IV tumors. Our results suggested that expression of miR-182 was associated with the progression of human gliomas with significant prognostic value and thus might represent a useful prognostic marker and a potential target for gliomas therapies.
Materials and Methods
Cell Lines
Primary normal human astrocytes (NHA) were purchased from the Sciencell Research Laboratories (Carlsbad, CA) and cultured as recommended by the manufacturer. Glioma cell lines LN18, U87, U118, T98 (American Type Culture Collection, Manassas, VA), SNB19 (from Y.-H. Zhou, University of California, Irvine, CA), A172, U138, U251, and U373 (our own collections25) were grown in the Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 100 units penicillin-streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator.
Patients and Tissue Specimens
A total of 253 paraffin-embedded glioma specimens (only astrocytomas were included and no oligodendrogliomas were used), which had been clinically and histopathologically diagnosed at the First Affiliated Hospital of Sun Yat-Sen University from 2000 to 2005, included World Health Organization grade I to IV tumors. The grade I gliomas used in the study were pilocytic astrocytomas. Among the 253 cases collected, survival information for 119 cases was available. For the use of these clinical materials for research purposes, patient’s consents and approval from the Institutional Research Ethics Committee were obtained. Clinical information of these samples is described in detail in Table 1. The purity in sections adjacent to the regions used for in situ hybridization (ISH) analysis and RNA extraction was validated through routine histopathological analysis. Three normal brain tissue samples were obtained by collecting donations with consents from individuals who died in traffic accidents and were confirmed to be free of any prior pathological lesions.
Table 1.
Clinicopathological Characteristics of Studied Patients and Expression of miR-182 in Gliomas
| Characteristics | No. of cases | % |
|---|---|---|
| Sex | ||
| Male | 172 | 67.98 |
| Female | 81 | 32.02 |
| Age, years | ||
| ≤45 | 154 | 60.87 |
| >45 | 99 | 39.13 |
| Gliomas histopathology (World Health Organization grading) | ||
| Grade I | 29 | 11.46 |
| Grade II | 79 | 31.23 |
| Grade III | 99 | 39.13 |
| Grade IV | 46 | 18.18 |
| Patient survival (n = 119) | ||
| Alive | 41 | 34.45 |
| Deceased | 78 | 65.55 |
| Survival time for low miR-182 expression group, months | ||
| Median | 42.00 | |
| Survival time for high miR-182 expression group, months | ||
| Median | 23.00 | |
| miR-182 expression (in situ hybridization) | ||
| Negative | 5 | 1.98 |
| Positive | 248 | 98.02 |
| Low expression (SI < 6) | 123 | 48.62 |
| High expression (SI ≥ 6) | 130 | 51.38 |
RNA Extraction and Real-Time RT-PCR
Total miRNA from cultured cells, fresh surgical glioma tissues, and paraffin-embedded, archived clinical glioma specimens was extracted by using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer’s instructions. CDNA was synthesized from 5 ng of total RNA by using the Taqman miRNA reverse transcription kit (Applied Biosystems, Foster City, CA), and the expression levels of miR-182 were quantified by using miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems). Real-time RT-PCR was performed by using the Applied Biosystems 7500 Sequence Detection system. The expression of miRNA was defined based on the threshold cycle (Ct), and relative expression levels were calculated as 2−[(Ct of miR-182) − (Ct of U6)] after normalization with reference to expression of U6 small nuclear RNA.
In Situ Hybridization and Data Analyses
In situ hybridization analysis was performed as previously described.11 Briefly, thin sections in 4-μm thickness of paraffin-embedded specimens were deparaffinized with xylene and rehydrated with an ethanol dilution series from 100% to 25%. Sections were treated with 40 μg/ml proteinase K in 0.2% glycine at 37°C for 5 minutes and 30 seconds and re-fixed in 4% paraformaldehyde. Slides were prehybridized in a hybridization solution (50% formamide, 5× standard saline citrate, 0.1% Tween, 9.2 mmol/L citric acid for adjustment to pH 6.0, 50 μg/ml heparin, 500 μg/ml yeast RNA) at 49.5°C for 2 hours. Subsequently, 20 nmol/L of a Locked Nucleic Acid-modified, 5′digoxigenin (DIG) labeled oligonucleotide probe complementary to miR-182 or a scrambled control probe was added to 100 μl of the hybridization solution and hybridized at a temperature of 49.5°C overnight. Sections were rinsed twice in 5× standard saline citrate, followed by three washes of 20 minutes at 50°C in 50% formamide/2× standard saline citrate and subsequently five times washes in PBS/0.1% Tween-20, and blocked in blocking solution (2% goat serum, 2 mg/ml bovine serum albumin in Phosphate Buffered Saline with Tween-20) at room temperature for 1 hour. An anti-DIG antibody (1:1000; Roche, Mannheim, Germany) was applied, and the sections were incubated at 4°C overnight. After washing in a staining solution (10 mmol/L Tris-HCl pH 9.0, 50 mmol/L MgCl2, 100 mmol/L NaCl, 0.1% Tween-20), the sections were incubated with the Nitro-Blue Tetrazolium Chloride/5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine (NBT/BCIP) developing solution (50 ml staining solution, 240 μl of 50 mg/ml NBT, 175 μl of 50 mg/ml BCIP) and rinsed in PBST followed by double distilled water. The sections were then dehydrated and mounted with Entellan.
The expression of miR-182 in a total of 253 paraffin-embedded glioma specimens, randomly divided into two cohorts, were examined by the same group of four investigators, using in situ hybridization analyses. Furthermore, the signals of in situ hybridization of tissue sections were examined and scored separately by two independent investigators blinded to the histopathological features and patient data of the samples. The scores were determined by combining the proportion of positively stained tumor cells and the intensity of staining. The proportion of positively stained tumor cells was graded as follows: 0, no positive tumor cells; 1, <10% positive tumor cells; 2, 10% to 50% positive tumor cells; and 3, >50% positive tumor cells. The cells at each intensity of staining were recorded on a scale of 0 (no staining), 1 (weak staining, light blue), 2 (moderate staining, blue), and 3 (strong staining, dark blue). For tumors that showed heterogeneous staining, the predominant pattern was taken into account for scoring. The staining index (SI) was calculated as follows: staining index = staining intensity × proportion of positively stained tumor cells. Using this method, the expression of miR-182 in gliomas was evaluated by SI, scored as 0, 1, 2, 3, 4, 6, or 9. Cutoff values to define the high- and low-expression of miR-182 were chosen on the basis of a measurement of heterogeneity with the log-rank test statistic with respect to overall survival. Because the optimal cutoff SIs were identified from the current study as 6, an SI score ≥6 was taken to define tumors as high expression, and SI <6 to define tumors as low expression of miR-182.
In situ hybridization signals for miR-182 expression in tumor and normal tissues were quantified by using the AxioVision Rel.4.6 computerized image analysis system assisted with the automatic measurement program (Carl Zeiss, Oberkochen, Germany). The method of Mean Optical Density (MOD) was preformed as previously reported.26 Briefly, the stained slides were evaluated at 200× magnification by using the SAMBA 4000 computerized image analysis system assisted with Immuno 4.0 quantitative program (Image Products International, Chantilly, VA), and 10 randomly picked fields in each specimen were analyzed to determine the MOD, which represents the strength of staining signals as measured per positive pixels, based on which the mean MOD of a study group of samples was further generated for subsequent intergroup comparative analysis. This program takes into consideration of the areas and numbers of positively stained tumor cells to obviate the impact of cell density on MOD value. A negative control with each batch of staining was used for background subtraction in the quantitative analysis. The representative staining fields of each specimen were analyzed and scored independently by two observers who were blinded to each other and to the diagnosis of the specimens. The MOD data were statistically analyzed by using t-test to compare the MOD differences between groups, and P < 0.05 was considered to be statistically significant.
Vectors and Retroviral Infection
To generate an mir-182 expression vector, approximately 200-bp genomic fragment up and downstream of the pre-mir-182 form was amplified by PCR and cloned into Plasmid of Murine Stem Cell Virus (pMSCV). Retroviral production and infection were performed as described previously.27 After 48 hours infection in NHA, the expression of miR-182 was examined either by real-time RT-PCR analysis or in situ hybridization analysis.
Statistical Analysis
All statistical analyses and the patient age cutoff values were performed by using the SPSS 10.0 statistical software package (SPSS Inc, Chicago, IL). The χ2 test was used to analyze the relationship between miR-182 expression and the clinicopathological characteristics. Bivariate correlations between study variables were calculated via Spearman’s rank correlation coefficients. Survival curves were plotted by the Kaplan-Meier method and compared by the log-rank test. Survival data were evaluated by using univariate and multivariate Cox regression analyses. In all cases, P < 0.05 was considered statistically significant.
Results
Expression of miR-182 Is Increased in Human Gliomas
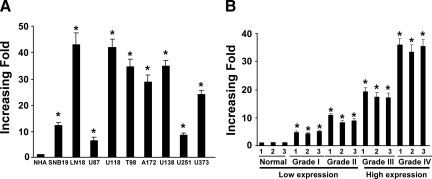
We performed initial screening on the expression profile of a total of 450 miRNAs by using normal brain tissues and primary tumor specimens of four different grades of human gliomas (one specimen per grade). When comparing expression levels of the miRNAs in gliomas samples with those in the normal brain tissues, we found that nine miRNAs displayed more than 1.61-fold increase and levels of eight miRNAs were decreased by 1.28-fold or lower in their expression, among which miR-182 showed the most robust change in expression (3.93-fold increase; Table 2). To further characterize the expression of miR-182 in gliomas, we performed real-time RT-PCR analyses and found that miR-182 was markedly up-regulated to various levels in all nine glioma cell lines examined compared with NHA (Figure 1A). To determine whether miR-182 up-regulation correlated with clinical glioma progression, we assessed the levels of miR-182 expression by real-time RT-PCR analyses in three normal brain tissues and 12 fresh-frozen glioma tissues of varying World Health Organization grades (grades I to IV, with three samples per grade). miR-182 was found to be up-regulated in all 12 human primary glioma tissues compared with the normal brain tissue samples (Figure 1B). Importantly, the trend of miR-182 expression in these human primary glioma tissues correlated with the malignancy of these gliomas.
Table 2.
The Most Significantly Altered MiRNAs in Varying World Health Organization Grades Gliomas Compared with Normal Brain Tissues
| No. | MiRNA | Log2* | P |
|---|---|---|---|
| 1 | miR-182 | 3.93 | 0.023 |
| 2 | miR-10b | 2.91 | 0.035 |
| 3 | miR-630 | 2.57 | 0.043 |
| 4 | miR-486 | 2.50 | 0.024 |
| 5 | miR-451 | 2.08 | 0.032 |
| 6 | miR-671 | 1.98 | 0.016 |
| 7 | miR-130a | 1.80 | 0.022 |
| 8 | miR-188 | 1.73 | 0.043 |
| 9 | miR-25 | 1.61 | 0.041 |
| 10 | miR-124a | −3.50 | 0.023 |
| 11 | miR-129 | −3.01 | 0.026 |
| 12 | miR-139 | −2.96 | 0.034 |
| 13 | miR-33 | −2.51 | 0.036 |
| 14 | miR-7 | −2.47 | 0.038 |
| 15 | miR-136 | −1.36 | 0.042 |
| 16 | miR-219 | −1.35 | 0.044 |
| 17 | miR-218 | −1.28 | 0.029 |
Positive and negative log2 scores indicate up-regulation and down-regulation of miR-182, respectively, in gliomas.
Figure 1.
miR-182 is up-regulated in glioma cell lines and glioma tissues. Real-time RT-PCR analysis of miR-182 expression in NHA and glioma cell lines (SNB19, LN18, U87, U118, T98, A172, U138, U251, and U373; A) and in normal brain tissues and glioma tissue samples of different World Health Organization grades (B) is shown. Expression levels were normalized for U6, and the average ratio of miR-182 expression was quantified. Error bars (SD) were calculated from triplicate samples. Statistical significance of *P < 0.05.
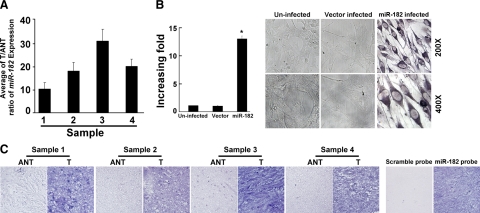
To validate our finding of miR-182 up-regulation, we performed a comparative analysis of miR-182 expression by real-time RT-PCR by using paired glioma tissues and their corresponding adjacent nontumor tissues (ANT) from the four individual glioma patients. As shown in Figure 2A, miR-182 was overexpressed in all four human primary glioma samples compared with the counterparts of paired adjacent nontumorous brain tissues. Importantly, all four tumors displayed greater than fivefold increases of miR-182 compared with adjacent nontumor tissues. In a GBM specimen, a 32-fold increase of the tumor/adjacent nontumorous tissue (T/ANT) ratio of miR-182 expression was found (Figure 2A).
Figure 2.
Expression of miR-182 is elevated in primary gliomas as compared with tumor-adjacent tissues of the same individuals with gliomas. A: Real-time RT-PCR analyses of miR-182 expression in paired primary glioma tissues (T) and glioma ANT of four individual patients. Expression levels were normalized for U6, and the average ratio of miR-182 expression was quantified. B: Ectopic expression of miR-182 in NHA cells analyzed by real-time RT-PCR (left) and ISH (right). Statistical significance of *P < 0.05. C: miR-182 expression level up-regulated in the paired primary T and glioma ANT examined by ISH. Validation for the specificity of the in situ probe of miR-182 is shown. Sister sections of an anaplastic astrocytoma, World Health Organization grade III were hybridized with the miR-182 probe or a scrambled control probe. Error bars (SD) were calculated from triplicate samples.
Increased Expression of miR-182 Correlates With the Malignancy of Gliomas
To further evaluate whether miR-182 up-regulation was linked to the clinical progression of gliomas, we sought to examine the expression of miR-182 in a large cohort of clinical glioma samples by using ISH. The specificity of the miR-182 probe was first examined by ectopically overexpressing miR-182 by retroviral-mediated transduction in NHA that had been shown to express miR-182 at very low level (Figure 1A), followed by in situ hybridization analysis. As shown in Figure 2B (right panel), miR-182 staining was clearly shown in the miR-182-infected NHA, which was further confirmed by real-time RT-PCR analysis (Figure 2B, left panel), but was undetectable in the vector infected-NHA or the uninfected NHA. Consistent with the abovementioned result obtained from real-time RT-PCR, in situ hybridization analysis also showed miR-182 overexpression in four human primary glioma samples compared with the paired nontumor adjacent tissues, whereas a scramble control probe did not yield any signal in sister sections of these tumors (Figure 2C and data not shown), validating the specificity of the miR-182 probe used in the study (Figure 2C).
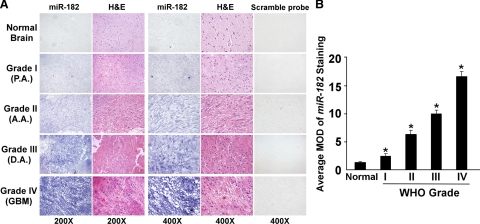
Two hundred fifty-three paraffin-embedded, archived clinical glioma specimens including 29 World Health Organization grade I, 79 grade II, 99 grade III, and 46 grade IV tumors were examined for miR-182 expression by using in situ hybridization analyses (Table 2). The study population of gliomas consisted of 253 patients (172 male patients; 81 female patients) with a median age at diagnosis of 45 years (range, 21 to 72 years). Figure 3A showed representative data of the in situ hybridization results. miR-182 was found to be up-regulated in these glioma samples of all grades compared with normal brain tissues. As summarized in Table 2, miR-182 expression was detected in 248 of 253 (98%) tumors. In contrast, expression of miR-182 was marginally or not detectable in normal brain tissues (Figure 3A). Quantitative analyses showed that MODs of miR-182 staining in grades I to IV tumors were significantly higher than those in normal brain tissues, and that MODs of miR-182 staining were also significantly different among various clinical grades (P < 0.05, Figure 3B).
Figure 3.
miR-182 expression correlates with glioma progression. A: Representative images of ISH analyses of a total of three normal brain tissues and 253 primary glioma specimens, including World Health Organization (WHO) grade I to IV tumors, were stained by ISH by using an miR-182 probe. Sister sections were also stained with a scrambled control probe. ISH analyses were performed two independent times on sections of each specimen with similar results. B: statistical analyses of the average MOD of miR-182 staining between normal brain tissues (three cases) and glioma specimens of different World Health Organization grades (29 random cases per grade). Average MOD of miR-182 staining increases as gliomas progress to higher grades. Statistical significance of *P < 0.05.
Levels of miR-182 expression by in situ hybridization were further analyzed to determine its relationship with the clinical features of gliomas. The median expression of miR-182, using real-time PCR analysis, of 119 patient samples was used as the threshold, and patients were categorized into groups of high (above median) or low (below median) expression. As shown in Table 3, miR-182 expression strongly correlated with the clinicopathological grades of glioma specimens (P < 0.001), whereas no difference was seen among ages (P = 0.137) or between genders (P = 0.480) of these patients. Spearman correlation analysis further validated the correlation coefficient between miR-182 expression level and the histological grading of glioma as 0.373 (P < 0.001; Table 4). Taken together, these results support the notion that the level of miR-182 expression increases as glioma clinically progresses.
Table 3.
Correlation between the Clinicopathological Features and Expression of miR-182
| Patient characteristics | miR-182 in situ hybridization
|
miR-182 real-time RT-PCR
|
||||
|---|---|---|---|---|---|---|
| SI < 6 | SI ≥ 6 | P | <median | >median | P | |
| Sex | ||||||
| Male | 81 | 91 | 0.480 | 37 | 52 | 0.260 |
| Female | 42 | 39 | 9 | 21 | ||
| Age, years | ||||||
| ≤45 | 91 | 85 | 0.137 | 39 | 42 | 0.002 |
| >45 | 32 | 45 | 7 | 31 | ||
| Gliomas histology (World Health Organization) | ||||||
| I | 18 | 11 | <0.001 | 10 | 3 | <0.001 |
| II | 51 | 28 | 21 | 9 | ||
| III | 44 | 55 | 13 | 41 | ||
| IV | 10 | 36 | 2 | 20 | ||
Table 4.
Spearman Correlation Analysis between miR-182 and Clinicopathological Factors
| Variables | miR-182 in situ hybridization
|
miR-182 real-time RT-PCR
|
||
|---|---|---|---|---|
| Spearman correlation | P | Spearman correlation | P | |
| Sex | 0.061 | 0.335 | 0.103 | 0.264 |
| Age | 0.198 | 0.002 | 0.341 | <0.001 |
| Gliomas histology (World Health Organization) | 0.373 | <0.001 | 0.512 | <0.001 |
miR-182 Expression Is Associated with the Prognosis of Patients with Gliomas
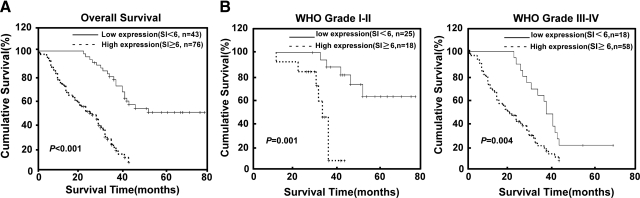
The Somers Dxy Rank Correlation analysis showed the correlation coefficient between miR-182 and patient survival as 0.67 (c = 0.835; Table 5). Furthermore, when a log-rank test and Kaplan-Meier analysis were applied to calculate the effect of miR-182 expression and histological staging of glioma on the patient survival, the expression level of miR-182 in the gliomas significantly displayed a correlation with the patients’ survival time (P < 0.001) with a correlation coefficient of −0.539 (Figure 4 and Table 6). Specifically, the median survival time of patients whose tumors expressed high levels of miR-182 was only 23 months (95% confidence interval, 19.20 to 24.41), whereas the median survival time of those with low levels of miR-182 expression was 42 months (95% confidence interval, 40.61 to 49.44). As shown in Figure 4, A and B, the cumulative 5-year survival rate was 51.54% (95% confidence interval, 0.435 to 0.596) in the low miR-182 expression group (n = 43), whereas it was only 7.23% (95% confidence interval, 0.027 to 0.118) in the high miR-182 expression group (n = 76).
Table 5.
Somers’ Dxy Rank Correlation Analysis between miR-182 and Survival
| Somers’ Dxy Rank Correlation
| |
|---|---|
| Statistics | Value |
| C | 0.835 |
| Dxy | 0.6695 |
| n | 119 |
| Missing | 0 |
Figure 4.
Kaplan-Meier curves with univariate analyses (log-rank) for patients with low miR-182-expressing tumors (bold line) versus high miR-182-expressing tumors (dotted line). A: The cumulative 5-year survival rate was 51.54% (95% confidence interval, 0.435 to 0.596) in the low miR-182 expression group (n = 43), whereas it was only 7.23% (95% confidence interval, 0.027 to 0.118) in the high miR-182 expression group (n = 76). B: The statistical significance of the difference between curves of miR-182 high-expressing and low-expressing patients was compared within subgroups of World Health Organization (WHO) grades I and II (A) and III and IV (B). P values were calculated by using the log-rank test.
Table 6.
Univariate and Multivariate Analysis of Different Prognostic Parameters in Patients with Gliomas by Cox-Regression Analysis
| Patient no. | In situ hybridization analysis | Real time RT-PCR analysis
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis
|
Multivariate analysis
|
Univariate analysis
|
Multivariate analysis
|
||||||||
| P | Regression coefficient (SE) | P | Relative risk | 95% confidence interval | P | Regression coefficient (SE) | P | Relative risk | 95% confidence interval | ||
| Age | <0.001 | 0.033 (0.007) | 0.387 | 1.007 | 0.991–1.022 | <0.001 | 0.033 (0.007) | 0.792 | 1.002 | 0.986–1.019 | |
| ≤45 | 81 | ||||||||||
| >45 | 38 | ||||||||||
| Gliomas histology (World Health Organization grade) | <0.001 | 1.191 (0.167) | <0.001 | 2.531 | 1.755–3.649 | <0.001 | 1.191 (0.167) | <0.001 | 2.759 | 1.929–3.947 | |
| I | 13 | ||||||||||
| II | 30 | ||||||||||
| III | 54 | ||||||||||
| IV | 22 | ||||||||||
| miR-182 | <0.001 | 1.500 (0.274) | 0.009 | 2.190 | 1.213–3.955 | <0.001 | 1.609 (0.287) | <0.001 | 3.099 | 1.714–5.601 | |
| Low | 46 | ||||||||||
| High | 73 | ||||||||||
The expression level of miR-182 in gliomas was significantly correlated with patients’ survival time (P < 0.001); the correlation coefficient was −0.539, indicating that higher levels of miR-182 expression was correlated with shorter survival time.
Univariate and multivariate analyses were performed and determined whether the miR-182 expression level was an independent prognostic factor of patient outcomes. As shown in Table 6, miR-182 expression and glioma World Health Organization histology were identified as independent prognostic factors. Moreover, the prognostic value of miR-182 expression in selective patient subgroups according to the histological grade was evaluated. A significant correlation between high miR-182 expression and shorter overall survival time was found in World Health Organization grading subgroups of glioma. Patients with tumors exhibiting high miR-182 expression had significantly shorter overall survival periods than those with low expression of miR-182, both in the grades I and II subgroup (n = 43; log-rank, P = 0.001; Figure 4B, left panel) and in grades III and IV subgroup (n = 76; log-rank, P = 0.004; Figure 4B, right panel). Taken together, our results suggest that miR-182 might represent a novel and potentially useful independent biomarker for the prognosis of patients with gliomas.
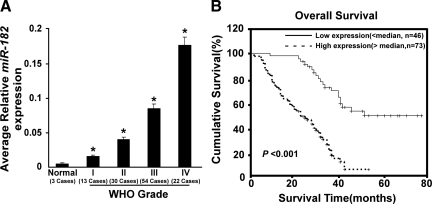
In the attempt to provide independent verification on the correlation study based on our in situ hybridization data, levels of miR-182 expression were verified with real-time RT-PCR. Totally, three paraffin-embedded normal brain tissues and 119 paraffin-embedded, archived clinical glioma specimens (World Health Organization grade I tumors, 13; grade II, 30; grade III, 54; and grade IV, 22), for which the survival information was available (Table 2), were examined for miR-182 expression by using real-time RT-PCR. As shown in Figure 5A, miR-182 was found to be up-regulated in all 119 glioma specimens examined compared with that in the normal brain tissues, and relative increases in miR-182 expressions well correlated to the intensity elevations of miR-182 signals detected by the ISH method. Quantitative analyses showed that the average miR-182 expression in grades I to IV tumors was significantly higher than those in normal brain tissues. Furthermore, statistical analysis results derived from real-time RT-PCR assays revealed that miR-182 expression strongly correlated with the clinicopathological grades of glioma specimens (P < 0.001), whereas no difference was seen between genders (P = 0.260) of these patients (Table 3). Spearman correlation analysis corroborated the correlation coefficient between miR-182 expression level and the histological grading of glioma as 0.512 (P < 0.001; Table 4). When a log-rank test and Kaplan-Meier analysis was applied to calculate the effect of miR-182 expression and histological staging of glioma on the patient survival, the expression level of miR-182 in the gliomas displayed a significant correlation with the patients’ survival time (n = 119; log-rank, P < 0.001; Figure 5B). Finally, univariate and multivariate analyses were performed to determine whether the miR-182 expression level was an independent prognostic factor of patient outcomes. As shown in Table 6, miR-182 expression and the World Health Organization histology were identified as independent prognostic factors for gliomas. Meanwhile, we examined the information of glioma patients and did not find the difference in demographics or glioma grade or other characteristics in patients with follow-up compared with those without. Taken together, our results suggest that miR-182, as demonstrated in both the ISH and the real-time PCR analyses, could be a valuable marker of glioma progression, and that high miR-182 expression is associated with poor overall survival in patients with malignant gliomas.
Figure 5.
Clinical correlation of gliomas with miR-182 confirmed by real-time RT-PCR analysis. A: Statistical analyses of the average relative miR-182 expression between normal brain tissues (three cases) and glioma specimens of different World Health Organization (WHO) grades (WHO grade I tumors, 13; grade II, 30; grade III, 54; and grade IV, 22; *P < 0.05). B: The cumulative 5-year survival rate was 54.12% (95% confidence interval, 0.432 to 0.650) in the low-miR-182 expression group (n = 46), whereas it was only 9.08% (95% confidence interval, 0.032 to 0.144) in the high-miR-182 expression group (n = 73).
Discussion
The key finding of the current study is that up-regulation of miR-182 expression is associated with the progression of malignant gliomas. Our results show that miR-182 is up-regulated in both glioma cell lines and primary glioma specimens. Results of in situ hybridization analysis and real-time RT-PCR demonstrate that the expression level of miR-182 significantly correlates with the World Health Organization histological grades of the glioma, and statistical analyses reveal that patients with high expression of miR-182 have a poorer prognosis. Taken together, these data suggest that miR-182 is a significant predictor of poor prognosis for glioma patients and could be a useful prognostic marker and potential target for glioma therapy.
Several recent studies have demonstrated that the expression of miRNAs is deregulated in gliomas. Ciafrè et al15 examined the alterations of 245 miRNAs in World Health Organization grade IV GBM, in which miR-221 was up-regulated, whereas miR-128, miR-181a, miR-181b, and miR-181c were down-regulated in the GBM specimens. Chan et al20 showed that expression of miR-21 was markedly up-regulated in primary GBMs and glioma cell lines compared with normal brain tissues and nontumor glial cells. In contrast, miR-124 and miR-137 were found to be significantly decreased in grade III anaplastic gliomas and grade IV GBMs compared with adjacent nontumor brain tissues.28 Additionally, miR-128, miR-181a, miR-181b, and miR-451 were also found to be down-regulated in glioma tissues and glioma cell lines compared with normal brain tissues.29,30,31 These findings suggest that miRNAs are involved in glioma development and progression. In our present study, we examined the global expression levels of miRNAs in glioma tissues of different World Health Organization grades and found that miR-182 was significantly up-regulated in both glioma cell lines and glioma tissues as compared with normal astrocytes and paired adjacent nontumorous tissues, respectively. In situ hybridization analyses of 253 paraffin-embedded archival glioma specimens showed that 248 of 253 (98%) specimens displayed positive miR-182 staining in tumor cells, among which 130 specimens (51%) displayed moderate to strong miR-182 signals, whereas no significant staining of miR-182 was detected in the adjacent noncancerous astrocytes. Furthermore, quantitative analysis of both the in situ hybridization and real-time RT-PCR results demonstrated that levels of miR-182 expression increased with glioma progression, and that miR-182 staining strongly correlated with the World Health Organization grading and survival time of the patients of gliomas. Patients with low-level miR-182 expression had a cumulative 5-year survival rate of 51.54%, whereas it was only 7.23% in high miR-182 expression patients. These data, together with further univariate and multivariate analyses, suggest that high expression of miR-182 is a significant predictor of poor prognosis for glioma patients. Our study also demonstrates that miR-182 might be a new prognostic marker in gliomas for all World Health Organization grades. Apparently, further evaluating miR-82 expression with established grading parameters for gliomas, including mitotic activity, microvascular proliferation, and necrosis, as well as other histological or molecular parameters that may be associated with outcome, would be helpful for better understanding the effect of miR-182 on the biological behavior of gliomas.
Our finding of up-regulation of miR-182 in primary glioma specimens and its correlation with glioma progression and poor prognosis could have high clinical and pathogenetical significance in glioma biology. The miR-182 gene is located in chromosome 7q32.1, where the fragile site FRA7H also locates and the MET gene locus is nearby. During glioma progression, multiple oncogenes, such as EGFR and MET that are also on chromosome 7 are frequently amplified.32,33 Interestingly, two other miRNAs, miR-96 and miR-183, which are clustered in tandem with miR-182,34 were also found to be up-regulated in our initial analyses (data not shown), suggesting that the miR-96-183-182 cluster might be involved in the development and progression of gliomas. In the current study, we found that 248 of 253 (98%) archived human glioma specimens showed elevated expression of miR-182 by using the ISH method. However, amplification of miR-182 was reported in 28.9% of epithelial ovarian cancer by using microRNA microarrays.35 The discrepancy between our data in glioma and miR-182 amplification in epithelial ovarian cancer is probably due to distinct properties of these two types of human cancers that were originated from distinct types of cells as well as the differences in sensitivities of ISH analysis and the miRNA microarray analysis. Additionally, ectopic expression of miR-182 in an epithelial ovarian cancer cell line significantly promoted tumor growth in vivo, suggesting a role of miR-182 as a putative oncogene.35 Recently, Segura et al36 found that miR-182 up-regulation correlated with gene copy number in a subset of melanoma cell lines and that miR-182 overexpression promoted cell migration and survival by directly repressing microphthalmia-associated transcription factor-M and FOXO3, whereas enhanced expression of either microphthalmia-associated transcription factor-M or FOXO3 blocked the pro-invasive effect of miR-182. During preparation of this article, Guttilla and White37 reported that miR-182 could target the 3′ untranslated region (UTR) of FOXO1, another FOXO family member, and that antisense inhibitor to miR-182 in breast cancer MCF-7 cells led to a significant increase in endogenous FOXO1 expression and a decrease in cell number. Thus, the role of miR-182 in the development and progression of gliomas warrants further investigations because the phenotypic changes in miR-182-overexpressing and -knocked down models could provide mechanistic insight and lead to the development of a new anti-gliomas strategy.
Interestingly, we found that all four tumor-adjacent nontumorous brain tissues displayed slightly higher levels of miR-182 compared with that in normal brain tissues, indicating that the different microenvironments in which tumor-adjacent and normal human astrocytes are present might result in the difference in miR-182 expression. In support of this speculation, Li et al38 have reported recently that stress could induce the up-regulation of miR-182 in primary cultures of human diploid fibroblast and human trabecular meshwork cells.
In summary, the results of our study indicate that expression of miR-182 is strongly correlated with histological grades and overall survival times of patients inflicted with malignant gliomas, providing evidence in support of the possibility that up-regulation of miR-182 might play an important role in the progression of this deadly disease. Multiple studies have demonstrated that several miRNAs such as miR-21, miR-181a, miR181b, miR-221, and miR-222 are up-regulated in human gliomas and function as tumor promoters or suppressors by modulating various key regulators critical for gliomagenesis and progression.24,25,31,39,40,41 Further study of the molecular mechanisms by which miR-182 contributes to the initiation and progression of gliomas is warranted.
Acknowledgments
We thank Dr. Yi-Hong Zhou for SNB19 cells and Mrs. Naama Balass for her proofreading of this article.
Footnotes
Address reprint requests to Jun Li, Ph.D., Zhongshan School of Medicine, Sun Yat-sen University, 74 Zhongshan Road II, Guangzhou, Guangdong 510080, China. E-mail: lijun37@mail.sysu.edu.cn.
Supported by the Ministry of Science and Technology of China (973)2005CB724605; the Natural Science Foundation of China (numbers 30670803, 30770836, 30771110, 30870963 and 30831160517); Program for New Century Excellent Talents in Universities (number NCET-07-0877); the Science and Technology Department of Guangdong Province, China (numbers 07001503, 8251008901000006 and 2008A030201006); Ministry of Education of China (number (2008)890 and number 200805580047); the Science and Technology Department of Zhuhai Municipality; Guangdong Provincial Natural Science Foundation (to J.L. and L.S.); and grants US NIH CA102011, CA130966, and ACS RSG CSM-107111 (S.-Y.C.).
L.J., P.M., and L.S. contributed equally to this work.
References
- Deorah S, Lynch CF, Sibenaller ZA, Ryken TC. Trends in brain cancer incidence and survival in the United States: surveillance, epidemiology, and end results program, 1973 to 2001. Neurosurg Focus. 2006;20(4):E1. doi: 10.3171/foc.2006.20.4.E1. [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organization for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol. 2008;18:122–129. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1919. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foà R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R, Cheng SY. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. MiR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14–30. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, Rechavi G, Givol D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Liu W, Qiang B, Zhao J, Yuan J, Peng X. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. Hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins V. P: gene amplification in human gliomas. GLIA. 1995;15:289–296. doi: 10.1002/glia.440150309. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Bützow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–741. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]