Abstract
Overdistension of hollow organs evokes pathological changes characterized by smooth muscle remodeling. Mechanical stimuli induce smooth muscle cell (SMC) growth through acute activation of signaling cascades and by increased expression of soluble mitogens. Physical forces have also been implicated in ligand-independent activation of receptor tyrosine kinases, including the platelet-derived growth factor (PDGF) receptor, although the extent to which this occurs in intact tissue is unknown. Previously, we implicated Akt and activator protein-1 (AP-1) as mediators of growth and gene expression in SMC exposed to cyclic stretch or PDGF. Here we show that bladder wall distension leads to PDGFR activation and identify thrombomodulin (TM) as an Akt and AP-1 target in SMC. We demonstrate that TM, also induced by bladder stretch injury, is regulated at the transcriptional level by the AP-1 components c-jun and Fra1. Mutation of an AP-1 motif at −2010/−2004 abolished both AP-1 binding and PDGF responsiveness of the TM promoter. Fra1 silencing diminished PDGF-induced TM expression and SMC cell cycle transit. In contrast, TM knockdown did not affect cell growth but attenuated PDGF-stimulated SMC migration. Taken together, these results reveal new facets of TM regulation in SMC and provide the first demonstration of a role for endogenous TM in PDGF-induced cell migration. Moreover, TM induction on bladder injury suggests that it may be a biomarker for pathological smooth muscle remodeling.
Fibroproliferative remodeling of smooth muscle underlies a wide range of diseases, including asthma, atherosclerosis, and lower urinary tract dysfunction.1,2,3 Smooth muscle mediates contractile functions and is present in multiple organs, where it interacts with diverse cell types and is exposed to a range of physiological and pathological stimuli. Unlike cardiac and skeletal muscle, smooth muscle cells (SMC) are not terminally differentiated and can alter their phenotype in response to environmental perturbations. This so-called phenotypic modulation involves dedifferentiation and proliferation of SMC and is initially beneficial to allow tissues to adapt to increasing pressure or other stress. Despite this initial adaptation, however, sustained exposure to injurious stimuli can lead to aberrant smooth muscle growth and loss of contractility.4 Diverse stimuli can induce smooth muscle remodeling, including mechanical or chemical injury, inflammation, and altered innervation. In hollow organs such as the urinary bladder, sustained increases in wall stretch and/or luminal pressure are known to promote hypertrophy and hyperplasia of smooth muscle (reviewed in Ref. 5). However, the molecular mechanisms underlying these processes are poorly understood.
Previous studies from our group and others have demonstrated that while mechanical stress itself is mitogenic, mechanical stimuli also up-regulate expression of soluble SMC mitogens such as heparin-binding epidermal growth factor-like growth factor and platelet-derived growth factor (PDGF).6,7,8 Moreover, several in vitro studies have implicated physical forces in activation of receptor tyrosine kinases in SMC, including the PDGF receptor (PDGFR),9 the epidermal growth factor receptor/ErbB1,10 and ErbB2.11 However, the extent to which this occurs in intact tissue has not been determined. We also implicated the phosphatidylinositol 3-kinase (PI3K)/Akt pathway and the activator protein-1 (AP-1) transcriptional complex as mediators of DNA synthesis and gene expression, respectively, in primary human bladder SMC (pBSMC) exposed to cyclic stretch relaxation or PDGF.12,13 Although Akt and AP-1 have been studied independently in many different contexts, the extent to which they interact to regulate smooth muscle remodeling remains unknown. In this study, we show that the PDGFR is activated in the bladder wall in response to distension and describe a novel Akt- and AP-1-dependent pathway in pBSMC that regulates expression of thrombomodulin (TM). Using a variety of complementary approaches, we implicate the c-jun∼Fra1 AP-1 dimer as a critical regulator of TM gene expression in response to PDGFR activation. We also provide the first evidence for endogenous TM as a regulator of SMC migration, a critical aspect of smooth muscle remodeling.
Materials and Methods
Cell Culture
pBSMC were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Valley Biomedical, Winchester, VA), 2 mmol/L l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere of 95% air-5% CO2. All experiments were performed on cells between passages 3 and 6. Cells were subjected to serum depletion (DMEM/0.5% FBS) for at least 24 hours or up to 48 hours before treatment.
Preparation of Nuclear Extracts
Nuclear extracts from vehicle and PDGF-treated pBSMC were prepared essentially as described previously.14 Briefly, cells were scraped in PBS. Cells were allowed to swell for 15 minutes at 4°C in hypotonic buffer A (10 mmol/L HEPES, 10 mmol/L KCl, 0.1 mmol/L EDTA, pH 8.0, 0.1 mmol/L EGTA, 2 μg/ml leupeptin, 1 mmol/L dithiothreitol, and protease inhibitors (Complete Mini tablets; Roche Applied Science, Indianapolis, IN)). Nonidet P-40 was added to a final concentration of 3% (v/v), and cells were vortexed for 10 seconds. Samples were centrifuged at 16,000 × g for 1 minute, and supernatants were removed as cytosolic fractions. Pellets were resuspended in buffer B (20 mmol/L HEPES, pH 7.9, 0.42 M NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 2 μg/ml leupeptin, 1 mmol/L dithiothreitol, and protease inhibitors) for 30 minutes at 4°C. The suspension was centrifuged at 16,000 × g for 10 to 15 minutes, and supernatant was used as nuclear extract. Protein was quantified by the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). In selected experiments, cells were preincubated with the Akt inhibitor triciribine (Calbiochem, Gibbstown, NJ) at 10 μmol/L final concentration for 30 minutes before stimulation with PDGF and preparation of nuclear extracts. PDGF-BB was from R&D Systems (Minneapolis, MN).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assays (EMSAs) for AP-1 were performed with 3 to 5 μg of nuclear extract and 0.5 ng of 32P-labeled oligonucleotide probe in a 20-μl reaction volume containing 25 mmol/L Tris-HCl, pH 8, 50 mmol/L KCl, 5 mmol/L MgCl2, 0.5 mmol/L EDTA, 8% glycerol, 2 μg of BSA, and 0.5 μg of poly (dI-dC). The reaction was incubated for 20 minutes at room temperature, and the DNA-protein complexes were resolved on 5% polyacrylamide gels in 0.5× Tris-borate-EDTA buffer at 4°C. The gels were dried, and the complexes visualized by autoradiography using a Typhoon Trio Variable Mode phosphorimager (GE Healthcare Biosciences, Piscataway, NJ). For competition experiments, a 25- to 100-fold molar excess of the cold competitor oligonucleotide was added simultaneously with the probe. For supershift experiments, antibodies to phospho-c-jun (KM-1; sc-822-X), Fra-1 (R-20; sc-605-X), or isotype control (all from Santa Cruz Biotechnology, Santa Cruz, CA) were preincubated on ice for 1 hour with nuclear extract, followed by addition of the other components for 20 minutes at room temperature. The following oligonucleotides were used in EMSAs: AP-1 wild-type (wt), containing the consensus AP-1 binding site (in bold) (5′-CGCTTGATGACTCAGCCGGAA-3′), AP-1 mut containing a mutation (in italics) in the consensus AP-1 site (5′-CGCTTGATGACTTGGCCGGAA-3′), TM AP-1 wt containing the AP-1 binding site (in bold) from the TM promoter (5′-GACCCCCTGATTCAGCCTAGG-3′), and TM AP-1 mut containing a mutation (in italics) in the TM promoter AP-1 motif (5′-GACCCCCTGATTTGGCCTAGG-3′).
Transcription Factor ELISA
To examine activation of multiple AP-1 subunits concurrently, we used a commercially available AP-1 family Transcription Factor ELISA (Active Motif, Carlsbad, CA). Nuclear extracts from pBSMC treated without or with PDGF for 2 hours, in the absence or presence of a 30-minute pretreatment with 10 μmol/L triciribine were assayed according to the manufacturer’s protocol.
RNA Interference in pBSMC
To target Akt expression in pBSMC, oligonucleotides specific for each Akt isoform or scrambled control oligo were designed using the siSearch engine, and specificity was determined using the National Center for Biotechnology Information BLAST tool. Oligos were annealed and ligated into pSUPER.retro.puro (for oligo targeting Akt1/2) or pSUPER.retro.neo (for oligo targeting Akt3) vectors (OligoEngine, Seattle, WA), according to the manufacturer’s protocol. Viral supernatants obtained from transfection of 293GPG cells with either control vector or vectors encoding Akt1/2 or Akt3 short hairpin RNAs were used to infect pBSMC, in the presence of 4 μg/ml polybrene as described previously.15 Infection was repeated once, after which cells were placed in selection medium containing 0.375 μg/ml puromycin and 125 μg/ml G418. Following selection and confirmation of Akt knockdown by immunoblot analysis, pBSMC were subjected to PDGF treatment as described previously.12 In selected experiments, small interfering RNA (siRNA) oligos targeting Fra1 and TM were obtained from Dharmacon (Lafayette, CO) and Santa Cruz Biotechnology, respectively. pBSMC were nucleofected using an Amaxa IIN nucleofector (Amaxa, Gaithersburg, MD) with 1 μmol/L nontargeting or gene-specific siRNA oligos, according to the manufacturer’s protocol. Twenty-four hours after nucleofection, transfected cells were serum depleted in DMEM/0.5% FBS for 24 to 48 hours before stimulation with PDGF. In the Transwell migration assay, cells receiving siRNA oligos were cotransfected with pmaxGFP to monitor transfection efficiency. Knockdown of targeted proteins was confirmed by immunoblot analysis.
Deletion and Mutation Analysis of TM Promoter
To identify regulatory elements within the TM gene, we amplified a 2.1-kb fragment of the TM promoter or deletion mutants of this fragment by PCR and subcloned them into the pGL3 luciferase reporter vector (Promega, Madison, WI) using standard methods. To examine the potential role of an AP-1 site at position −2010 to −2004 relative to the translational start site, we performed site directed mutagenesis (from TGATTCA to TGATTTG) using the QuikChange II mutagenesis kit (Stratagene, La Jolla, CA), according to the manufacturer’s protocol. All constructs were sequenced before use to verify sequence integrity.
Transfection and Luciferase Assays
pBSMC were seeded in 6-cm dishes at a density of 4 × 105 cells/dish and allowed to plate overnight. Cells were transfected with 4 μg of promoter deletion or pGL3-basic plasmids per dish with 12 μl of Mirus reagent (Mirus Bio, Madison, WI). Eight hours later, cells were trypsinized, counted, and seeded at 105 cells/well in 4 wells of a 6-well plate. Twenty-four hours later, cells were serum depleted for 24 hours in DMEM/0.5%FBS and then exposed to PDGF for 12 hours. Cells were harvested in reporter lysis buffer, and luciferase activity was determined as described previously.14 For a given deletion construct, a single population of cells transfected with plasmid was split between dishes receiving vehicle or those treated with PDGF, thereby circumventing concerns about transfection efficiency between treatment conditions.
Semiquantitative Real-Time RT-PCR
Total RNA was isolated from cells with TRIzol reagent (Invitrogen). RNA was reverse transcribed using the iScript cDNA synthesis reagent (Bio-Rad), and cDNAs were amplified using gene-specific primers as described previously.13 The relative abundance of a given transcript was estimated using the 2−ΔΔCt method, following normalization to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase.
Immunoblot Analysis
Following PDGF treatment, pBSMC were washed twice with PBS and lysed in 1 X lysis buffer (10 mmol/L Tris-Cl, pH 7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton X-100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, and 1 μg/ml leupeptin) for 15 minutes on ice. Cell lysates were centrifuged at ∼16,000 × g for 10 minutes at 4°C to pellet insoluble material. Supernatants were transferred to fresh tubes, and protein was quantified using the MicroBCA Protein assay kit (Pierce Chemical Co., Rockford, IL). Bladder tissue was snap frozen in liquid nitrogen following distension, ground to a fine powder using a pestle and mortar and solubilized in 1× lysis buffer. Equal amounts of cell lysates were resolved by SDS-PAGE and electrotransferred to nitrocellulose membranes. Following transfer, membranes were stained with Ponceau S to confirm equal loading where applicable. Membranes were blocked with PBS/0.1% Tween 20/10% nonfat dried milk, rinsed with PBS-T and incubated with protein-specific primary antibodies overnight at 4°C. After washing, membranes were incubated with species-specific horseradish peroxidase-conjugated secondary antibodies, and proteins were visualized following incubation with SuperSignal WestPico chemiluminescence reagent (Pierce Chemical Co.) and exposure of membranes to X-ray film. Quantification of band intensity was performed using Adobe Photoshop software.
Indirect Immunofluorescence Imaging of pBSMC
For imaging experiments, pBSMC were seeded on coverslips at 105 cells/well in 6-well tissue culture dishes. Cells were serum depleted in DMEM/0.5% FBS for 24 hours before stimulation with PDGF for 8 hours. To inhibit the PDGFR, cells were preincubated with 10 μmol/L PDGFR inhibitor (Calbiochem) for 30 minutes before growth factor treatment. Following incubation, cells were fixed in 4% paraformaldehyde in PBS for 10 minutes, followed by 2 × 5-minute washes with PBS. Nonspecific binding sites were blocked in PBS/5% BSA/0.2% Triton X-100 for 1 hour at room temperature. Coverslips were incubated with TM monoclonal antibody (D3 (sc-13164); Santa Cruz Biotechnology) at a dilution of 1/50 in PBS/5% BSA/0.2% Triton X-100 overnight at 4°C. Coverslips were washed four times with PBS for 10 minutes before addition of Cy3-conjugated goat anti-mouse secondary antibody (1/250) for 1 hour at room temperature. Coverslips were washed four times with PBS for 10 minutes and then mounted in Vectashield medium containing DAPI (Vector Laboratories, Burlingame, CA). Cells were visualized using an Axioplan-2 microscope (Carl Zeiss MicroImaging, Thornwood, NY).
Ex Vivo Rat Bladder Distension
An ex vivo model of bladder injury was used as described previously.10,13,16 Briefly, 45-day-old-female rats were anesthetized with isoflurane inhalation. A low midline incision was made to expose the bladder. The urethra was catheterized, and the ureters were ligated before bladder distension with serum-free DMEM to 40- or 70-cm water pressure using a gravity manometer. The bladder neck was isolated, tightened with a 4–0 silk suture, and the organ excised and placed in a 50-ml tube containing serum-free DMEM. Specimens were maintained in culture for different times at 37°C in a humidified 5% CO2/95% air atmosphere incubator. Catheterized, nondistended bladders (ie, sham-operated specimens) were harvested and incubated in parallel with the distended specimens as controls. All animal experiments were performed in compliance with the Animal Care and Use Committee at Children’s Hospital Boston.
Immunostaining of Rat Bladder Tissue Sections
Sham and distended rat bladders were fixed in 10% neutral buffered formalin at room temperature for 48 hours, rinsed with PBS, dehydrated in ethanol, and embedded in paraffin. Sections of 8-μm thickness were obtained with microtome and mounted on glass slides. For immunohistochemical staining, tissue sections were deparaffinized in xylene, rehydrated through graded ethanols and equilibrated in PBS for 10 minutes. Quenching of endogenous peroxidase activity was achieved by incubation for 30 minutes at room temperature in peroxidase block (DakoCytomation, Carpinteria, CA). Sections were washed and blocked in 1% bovine serum albumin (BSA) for 30 minutes. TM antibody (at 1/50 dilution) was prepared in 1% BSA and added to sections for 90 minutes at room temperature in a moist chamber. Slides were washed four times with PBS for 6 minutes before addition of horseradish peroxidase-conjugated anti-mouse secondary antibody (1/500) for an additional 30 minutes at room temperature in a moist chamber. Slides were washed four times with PBS for 6 minutes and signals developed with a NovaRed kit (Vector Laboratories, Burlingame, CA). Sections were counterstained with hematoxylin, dehydrated, and mounted using Permount. Sections were visualized using a Zeiss Axioplan-2 fluorescence microscope (Carl Zeiss MicroImaging).
Wound Healing Assay
Confluent monolayers of pBSMC nucleofected with either 1 μmol/L nontargeting control siRNA or siRNAs to Fra1 or TM were “wounded” by creating a scratch across the cell layer with a sterile pipette tip, following serum depletion for 24 to 36 hours. Migration of cells into the wound in response to vehicle or 30 ng/ml PDGF was monitored visually over 8 hours.
Migration Assay
Serum-depleted pBSMC (1 × 106) were nucleofected with 1 μmol/L nontargeting siRNA (Dharmacon) or TM-targeting siRNA (Santa Cruz Biotechnology) and 1 μg of pmaxGFP (Amaxa) to allow for visualization of cells. Approximately 1.6 × 105 cells were seeded in each of four Transwell FluoroBlok inserts containing 200 μl of DMEM/0.5% FBS, and Transwells were placed in the corresponding wells of a companion plate containing 1 ml/well DMEM/0.5% FBS with or without 30 ng/ml PDGF. The remaining cells were seeded in 2 wells of a 6-well plate for confirmation of transfection efficiency and knockdown. Twenty-four hours after seeding, Transwell inserts were rinsed three times with PBS for 5 minutes each and then transferred to a glass-bottomed 24-well black plate (Greiner, Monroe, NC) for measurement. Green fluorescent protein (GFP) fluorescence signal was measured using a FLUOstar Omega microplate reader (BMG LabTech, Durham, NC) using the bottom optic, with excitation and emission wavelengths of 485 and 520 nm respectively.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT Express kit from Active Motif. Briefly, pBSMC from three confluent 15-cm dishes (∼5 × 106 cells/dish) treated for 2 hours with vehicle or 30 ng/ml PDGF were cross-linked with formaldehyde. Chromatin was harvested, sheared by sonication into fragment sizes from 0.2 to 1 kb, and immunoprecipitated overnight at 4°C with protein G magnetic beads and 4 μg each of phospho-c-jun antibody (KM-1, sc-822-X; Santa Cruz Biotechnology) or control IgG (The Jackson Laboratory, Bar Harbor, ME). The beads were washed, chromatin was eluted, cross-links were reversed, and DNA was purified according to the manufacturer’s instructions. Enrichment of transcription factor binding on PDGF treatment was assessed by real-time PCR, using primers that amplified a 144-bp region in the TM promoter spanning the AP-1 binding site. Binding to a 175-bp region in the TM coding region was used as a control. Fold changes (PDGF treatment over vehicle) are depicted relative to signal obtained from immunoprecipitation with control IgG under identical conditions. Primer sequences are as follows: promoter region, 5′-GGCTGTGCACCGGAGTCACG-3′ (forward); 5′-ACACAGCTGCGCCTCCCCTG-3′ (reverse); and coding region, 5′-GCTCCTCTGCCACCTGCGCA-3′ (forward); 5′-AGGAGGCACAGGCTCCTGGA-3′ (reverse).
Statistical Analysis
Where appropriate, comparisons between experimental groups were performed using Student’s t-test. P values are indicated in figure legends.
Results
Activation of the PDGFR ex Vivo and in Vitro Induces TM Expression in Bladder SMC
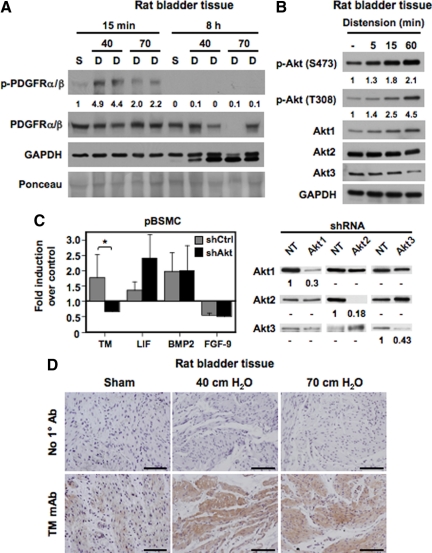
Our previous work demonstrated activation of similar intracellular signaling cascades in pBSMC exposed to cyclic stretch relaxation or PDGF treatment.12 In addition, stretch-sensitive genes in pBSMC were also found to be PDGF responsive.13 Published reports have suggested that mechanical stimuli can activate the PDGFR in vascular SMC in vitro. However, direct activation of the PDGFR has not been reported in intact tissue. To determine whether the PDGFR signaling axis was activated in the bladder wall, we used a rat model of bladder injury, as described previously.10,13,16 Briefly, bladders were distended ex vivo to 40- or 70-cm water pressure for different times, and extracts were immunoblotted with anti-phospho-PDGFRα(Tyr849)/PDGFRβ(Tyr857) antibody; phosphorylation at these sites in the α and β PDGFRs is known to be required for full kinase activation.17 As shown in Figure 1A, distension led to a rapid increase (within 15 minutes) in PDGFR phosphorylation in bladder tissue compared with control (sham), which declined to baseline by 8 hours, consistent with direct activation of the PDGFR by mechanical stimulation. In addition, we also demonstrated an increase in Akt phosphorylation in bladder tissue exposed to distension (Figure 1B), consistent with previous data from us showing that both cyclic stretch and ligand-mediated PDGFR activation activate the PI3K/Akt pathway.12 To determine the consequences of Akt signaling in bladder smooth muscle, we next determined the effect of perturbing this pathway on expression of a subset of PDGF-sensitive genes in pBSMC as described previously.13 Knockdown of all three Akt isoforms in pBSMC using a short hairpin RNA approach abrogated the PDGF-stimulated increase in TM mRNA (*P < 0.01) (Figure 1C) but did not alter expression of other PDGF-responsive genes such as leukemia-inhibitory factor (LIF), bone morphogenetic protein-2 (BMP2), or fibroblast growth factor-9. In view of our in vitro observations in pBSMC, we then determined whether TM was also induced following PDGFR activation in tissue and demonstrated that ex vivo bladder distension led to robust induction of TM expression in the detrusor smooth muscle compared with sham-operated controls (Figure 1D). Taken together, these observations suggest that wall stretch in the intact bladder can directly stimulate signaling through the PDGFR and that PDGFR activation leads to increased TM expression in tissue and in pBSMC.
Figure 1.
Activation of the PDGFR ex vivo and in vitro induces TM expression in bladder SMC. A: Rat bladders were hydrodistended to 40- or 70-cm water pressure for the indicated times to promote injury. Nondistended bladders from sham-operated rats served as controls. Lysates from nondistended sham bladders (S) and distended organs (D) were immunoblotted with antibodies to total and phosphorylated (activated) PDGFRα/β or glyceraldehyde-3-phosphate dehydrogenase. Tissues showed a rapid and transient phosphorylation of the PDGFR by 15 minutes following distension (compare 40- and 70-cm distension conditions at 15 minutes with nondistended controls). PDGFR activation disappeared by eight hours. No corresponding change in total PDGFR levels was observed with time. The values represent quantification of the increase in PDGFR phosphorylation. B: Lysates from rat bladders hydrodistended as in A for the indicated times were immunoblotted with antibodies to Akt1, Akt2, Akt3, Akt phosphorylated at Ser473 or Thr308, and glyceraldehyde-3-phosphate dehydrogenase. The data indicate a time-dependent increase in Akt phosphorylation with bladder distension. The values represent quantification of the increase in Akt phosphorylation at S473 and T308. C: Populations of pBSMC infected with retroviruses expressing shRNAs against Akt1, Akt2, and Akt3 (black bars) or control nontargeting shRNA (gray bars) were exposed to PDGF treatment for four hours and changes in gene expression assessed by real-time RT-PCR analysis. The graph shows fold change in expression of the indicated genes with PDGF relative to vehicle control. Data represent the mean ± SD of two independent trials, in which samples were assayed in duplicate. Knockdown of Akt ablated PDGF-induced TM expression (P < 0.01), whereas expression of other genes such as LIF, BMP2, and fibroblast growth factor-9 (FGF-9) was not significantly changed by Akt knockdown. The panel on the right shows immunoblot analysis confirming specificity of Akt knockdown in stable populations of pBSMC following infection with retroviruses individually targeting the three Akt isoforms. The extent of knockdown is presented as signal remaining for each Akt isoform relative to nontargeting control. D: Sections from rat bladders distended ex vivo for eight hours to 40- or 70-cm water pressure were stained sequentially with anti-TM primary antibody and horseradish peroxidase-conjugated species-specific secondary antibody. Increased TM staining was evident in the detrusor smooth muscle of distended specimens but not of nondistended, sham-operated control bladders. Data are representative of three independent experiments. Scale bar = 100 μm.
PDGF-Stimulated Akt Activation Mediates TM Expression in pBSMC
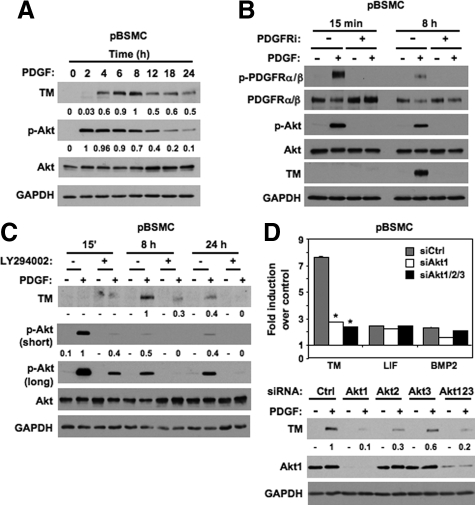
To further investigate PDGF-stimulated TM expression in vitro, we first performed a kinetic experiment, in which pBSMC exposed to PDGF were harvested for immunoblot analysis. As shown in Figure 2A, TM expression was evident within 4 hours after growth factor treatment, reached a peak of expression at 8 hours and declined to ∼50% of peak level by 24 hours. Consistent with previous observations, Akt was robustly phosphorylated following exposure to PDGF, peaking at 2 hours and decreasing in a time-dependent manner up to 24 hours. Pretreatment of pBSMC with a pharmacological inhibitor of intrinsic PDGFR kinase activity abolished both the PDGF-stimulated increase in TM protein levels as well as PDGF-induced Akt phosphorylation (Figure 2B and Supplemental Figure 1, see http://ajp.amjpathol.org). We also performed additional experiments to confirm the role of Akt in TM regulation in pBSMC. Treatment of cells with LY294002, the well-characterized inhibitor of PI3K-dependent Akt activation, attenuated PDGF-stimulated TM protein levels in pBSMC (Figure 2C). In addition, to determine which Akt isoform was the dominant regulator of TM expression, pBSMC nucleofected with nontargeting control siRNA or siRNAs to Akt1, Akt2, Akt3 or all three together were treated without or with PDGF for 8 hours and assayed for both mRNA and protein expression. As shown in Figure 2D, silencing of either Akt1 or all three Akts completely suppressed PDGF-stimulated TM mRNA expression in pBSMC (*, **P < 0.01; upper panel) but did not significantly diminish PDGF-stimulated LIF or BMP2 mRNA levels, consistent with the data in Figure 1C. Immunoblot analysis confirmed the role of Akt1 as the principal isoform regulating TM expression (Figure 2D, lower panel). Thus, in both cases, knockdown of Akt1 alone was sufficient to significantly attenuate PDGF-induced TM mRNA and protein expression in pBSMC. Taken together, these observations identify Akt1 as a dominant regulator of PDGF-stimulated TM expression in pBSMC.
Figure 2.
Expression of TM in pBSMC is regulated by a PDGFR/Akt pathway. A: Lysates of quiescent pBSMC stimulated with PDGF for the indicated times were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. The values represent quantification of PDGF-induced TM levels and of pAkt levels normalized to total Akt at the indicated times. In each case the maximum signal was set to a value of one, and the intensity of other signals was determined relative to this reference. B: pBSMC pretreated with vehicle (dimethyl sulfoxide) or 10 μmol/L PDGFR inhibitor (PDGFRi) for 30 minutes were stimulated without (−) or with (+) 30 ng/ml PDGF for the indicated times. Lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Rapid activation (phosphorylation) of the PDGFR and Akt was observed at 15 minutes, with activation attenuated by eight hours. TM protein expression was detected at 8 hours. The PDGF-induced increase in TM levels, as well as PDGFR and Akt activation, was abolished by PDGFR inhibition but not with vehicle alone (dimethyl sulfoxide). C: Lysates of pBSMC treated without (−) or with (+) 30 ng/ml PDGF for the indicated times following 30 minutes pretreatment with vehicle (dimethyl sulfoxide) or 10 μmol/L LY294002, were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. PDGF-induced phosphorylation of Akt and induction of TM protein expression was attenuated at all time-points by LY294002, whereas total Akt levels were unchanged. The values represent quantification of PDGF-induced TM levels at eight and 24 hours, and of pAkt levels (normalized to total Akt) at 15 minutes, 8 hours, and 24 hours in the absence and presence of LY294002. As for A, the maximum signal was set to a value of 1, and the intensity of other signals was determined relative to this reference. A long exposure for phospho-Akt is included to illustrate presence of signal at the 8- and 24-hour time points in cells treated with PDGF, consistent with the data in Figure 2, A and B. D: Upper panel, Total RNA isolated from pBSMC nucleofected with nontargeting siRNA duplexes or duplexes against Akt1, Akt2, Akt3, or all three Akt isoforms together, and treated without or with PDGF for eight hours was reverse transcribed to cDNA and amplified with primers for TM, LIF, BMP2, or glyceraldehyde-3-phosphate dehydrogenase. Data are presented as mean fold change in gene expression with PDGF treatment compared with nontreated controls. Only the reduction in TM expression was statistically significant with Akt knockdown (*P < 0.01 relative to siCtrl), with LIF and BMP2 showing little or no change with Akt silencing. Lower panel: Lysates from pBSMC nucleofected with siRNAs to Akt1, Akt2, Akt3 individually or combined, or nontargeting control siRNA were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. The values represent quantification of the change in PDGF-induced TM levels following Akt isoform knockdown.
PDGF Selectively Activates Fra Components of the AP-1 Transcription Factor Family
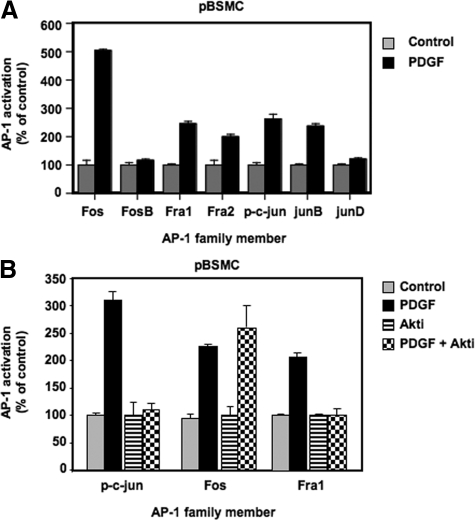
Our previous microarray analysis of pBSMC revealed an overrepresentation of putative binding sites for the AP-1 transcription factor in the promoters of stretch- and PDGF-stimulated genes.13 AP-1 is a dimer made up of members of the Jun (c-jun, JunB, and JunD) and Fos families (c-fos, FosB, ΔFosB, Fra1, and Fra2) and mediates a range of cellular activities such as growth, differentiation, and apoptosis in a context-dependent manner.18 To determine the extent of activation of AP-1 in pBSMC downstream of PDGFR activation, cells were exposed to PDGF treatment for 2 hours, and AP-1 DNA binding activity was assessed using a transcription factor ELISA. As shown in Figure 3A, AP-1 was robustly induced by PDGF, with increased binding of c-jun, JunB, c-fos, Fra1, and Fra2. Having demonstrated that PDGF-induced TM expression was sensitive to both pharmacological and genetic silencing of Akt, we proceeded to verify whether PDGF-induced AP-1 activation was also regulated by Akt. Quiescent pBSMC were pretreated for 30 minutes with 10 μmol/L triciribine or vehicle (dimethyl sulfoxide), followed by stimulation with PDGF for 2 hours. The DNA-binding activity of c-jun, Fos, and Fra1 was determined by AP-1 ELISA. Interestingly, although the DNA-binding activity of c-fos in response to PDGF was unaltered in Akt-inhibited cells, c-jun and Fra1 activation was abolished (Figure 3B). Similar findings were observed in cells treated with the PI3K inhibitor LY294002 (data not shown). These observations suggest that Akt regulates AP-1 through selective induction of c-jun and Fra1.
Figure 3.
PDGF activates select members of the AP-1 family of transcription factors. A: Nuclear extracts from pBSMC treated with 30 ng/ml PDGF or vehicle for two hours were assayed using a transcription factor ELISA to determine DNA-binding activity of AP-1 proteins. The graph shows AP-1 binding expressed as a percentage of control for each of 7 AP-1 subunits and data are representative of at least two independent experiments, each performed in duplicate. Increased binding of c-jun, JunB, Fos, Fra1, and Fra2 proteins to a consensus AP-1 oligonucleotide was observed in PDGF-treated cells (black bar) relative to controls (gray bar). JunD and FosB binding was unaffected by PDGF stimulation. B: Nuclear extracts from pBSMC pretreated for 30 minutes without (gray, black bar) or with (stippled, striped bar) 10 μmol/L triciribine followed by vehicle (gray, striped bar) or PDGF (black bar, stippled) for two hours were assayed as in (A). The graph shows AP-1 binding expressed as a percentage of control (vehicle or triciribine alone control). Data are representative of two independent trials. Inhibition of Akt ablated PDGF-induced DNA binding of c-jun and Fra1 but not Fos (compare stippled and black bar).
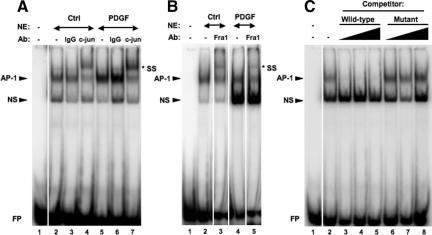
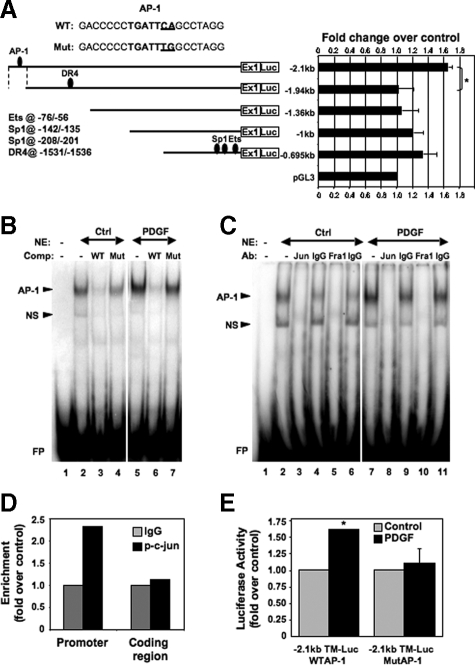
To verify the ELISA data and confirm the role of AP-1 in PDGF-mediated cell signaling, we performed EMSAs in conjunction with supershift analyses to evaluate AP-1 binding using an oligonucleotide probe containing a consensus AP-1 site. Consistent with our observations from the AP-1 ELISA, we confirmed the presence of phospho-c-jun and Fra1 within the AP-1-DNA complex (Figures 4, A and B) by antibody supershift analyses. In competition analyses, the AP-1-DNA complex observed in nuclear extracts from PDGF-treated pBSMC was competed out with increasing amounts of unlabeled wt consensus AP-1 oligos but not with a mutated version of the oligo (Figure 4C), thereby confirming specificity of the interaction.
Figure 4.
EMSAs detailing AP-1 activation in pBSMC on treatment with PDGF. Nuclear extracts prepared from control or PDGF-treated pBSMC were used in an EMSA and assessed for binding to a radiolabeled oligonucleotide carrying a consensus AP-1 binding sequence motif. Increased AP-1-DNA complex formation was detected in PDGF-treated pBSMC (A, compare lanes 2 and 5), and the complex was supershifted (SS) in the presence of antibody to (A) phospho-c-jun (lanes 4 and 7) or (B) Fra1 (lanes 3 and 5). An isotype-matched IgG control (A, lanes 3 and 6) did not affect mobility of the AP-1-containing complex. C: The PDGF-stimulated AP-1-DNA complex in pBSMC nuclear extracts (lane 2) was specifically competed out by increasing concentrations (25-, 50 -,and 100-fold molar excess) of unlabeled wt AP-1 consensus oligonucleotide (lanes 3–5) but not by an oligonucleotide carrying a mutation in the AP-1 binding motif (lanes 6–8). NS, nonspecific; FP, free (unbound) probe.
PDGF Up-Regulates TM Gene Expression via AP-1 Binding to the TM Promoter
We further investigated the mechanism of PDGF-mediated TM gene activation by evaluating a series of promoter-deletion luciferase reporter constructs for responsiveness to PDGF. Sequential deletions of the TM promoter, from 695- to ∼2.1-kb upstream, relative to the translational start site, were used to evaluate sensitivity of the promoter to PDGF in pBSMC. The −2.1-kb promoter region was found to show the greatest PDGF-mediated increase in reporter gene expression versus untreated control (Figure 5A). Furthermore, deletion of a short (∼160 bp) region significantly reduced PDGF-stimulated promoter activity (*P < 0.02), consistent with the presence of PDGF-responsive elements within this stretch of sequence. Although we observed an increase in PDGF-induced luciferase activity with progressive deletion, the activation was less than that observed with the −2.1-kb promoter construct and did not differ significantly between successive deletions. Informatics analysis of the ∼160-bp region between the −1.94- and −2.1-kb promoter sequences to identify transcription factor binding sites revealed a putative AP-1 binding element at −2010/−2004 (Figure 5A). Having shown earlier that PDGF activates AP-1—specifically c-jun and Fra1—we proceeded to explore the potential role of these components in binding to and activating the TM promoter. We used oligonucleotide probes encompassing the AP-1 binding site at −2010/−2004 (wt) in EMSAs and observed a specific complex that was competed out by an excess of homologous unlabeled competitor (wt) but not by an oligonucleotide carrying a point mutation in the AP-1 motif (Mut) (Figure 5B). Addition of either phospho-c-jun or Fra1 antibody to the binding reaction substantially immunodepleted AP-1-DNA complex formation (Figure 5C), consistent with the presence of c-jun and Fra1 in the complex. We went on to show binding of AP-1 to this site in vivo by carrying out a ChIP assay. Recruitment of phospho-c-Jun to the AP-1 motif on the TM promoter was enhanced on PDGF treatment, relative to control IgG binding (Figure 5D). No corresponding increase in DNA binding was observed at a site ∼4 kb downstream, within the coding region of the gene. We also determined the functional significance of the AP-1 motif at −2010/−2004 by site-directed mutagenesis. As shown in Figure 5E, although PDGF significantly increased activity of the wt TM 2.1kb promoter-reporter construct (P < 0.01) point mutation of the AP-1 site abolished PDGF-responsiveness of the construct, thus establishing the requirement for AP-1 DNA binding to elicit the increase in TM gene expression by PDGF. Together these findings implicate c-jun and Fra1 as the principal components of AP-1 associated with the DNA binding motif at −2010/−2004 in the TM promoter.
Figure 5.
An AP-1 binding site in the TM gene promoter mediates its activation by PDGF. A: Promoter-deletion luciferase constructs, comprising up to 2.1-kb sequence upstream of the TM gene, were transiently expressed in pBSMC and evaluated for their response to PDGF. The graph shows luciferase activity in extracts from PDGF-treated cells expressed as fold change relative to vehicle-treated controls. Data are representative of at least two independent experiments. Deletion of a region from −2.1 to −1.94 kb upstream of the translational start site caused a significant reduction in reporter gene expression following PDGF treatment (*P < 0.02), whereas further promoter deletion did not alter activity significantly. Binding sites for other transcription factors (Ets, Sp1, and DR4) that have been implicated previously in TM gene regulation are also represented. A putative AP-1 binding site present in the −2.1/−1.94 kb region is indicated. The sequence of the AP-1 site (Wt) present in the PDGF-responsive region of the TM promoter, along with mutation of this sequence (Mut), used in EMSAs and reporter gene analyses, is also shown. B: Nuclear extracts prepared from control or PDGF-treated pBSMC were used in an EMSA and assessed for binding to a radiolabeled oligonucleotide carrying the putative AP-1 binding motif at −2010/−2004 in the TM promoter. An increase in intensity of the AP-1 complex was detected on PDGF treatment relative to control (compare lanes 2 and 5). The complex was specifically competed out by a 25-fold molar excess of the unlabeled wt oligonucleotide (WT; lanes 3 and 6) but not by the mutant oligonucleotide (Mut; lanes 4 and 7). C: c-jun and Fra1 bind to the TM AP-1 site. The EMSA showed that the AP-1 DNA-protein complex in control (lane 2) or PDGF-treated nuclear extracts (lane 7) was specifically immunodepleted using antibodies against c-jun (lanes 3 and 8) and Fra1 (lanes 5 and 10) but not by normal IgG controls (lanes 4, 6, 9, and 11). NS, non-specific; FP, free (unbound) probe. D: AP-1 binds to the TM gene promoter in vivo. ChIP assay followed by real-time PCR shows binding of phospho-c-Jun to the AP-1 site present in the −2.1/−1.94-kb region (promoter) or to the coding region (gene) ∼4 kb downstream of the AP-1 site. Binding efficiency is depicted relative to control IgG binding. Values are expressed as fold change of PDGF-treated samples relative to untreated controls. E: TM-luc reporter constructs with the −2.1 kb upstream promoter region carrying either the wt or mutated (MutAP-1) AP-1 site were transiently expressed in pBSMC, which were subsequently treated without (gray bar) or with (black bar) 30 ng/ml PDGF. The graph shows fold change in reporter activity with PDGF relative to control and the values represent the mean ± SD of three independent experiments. The WT construct showed a significant increase in luciferase expression on PDGF treatment relative to control (*P < 0.01) that was abolished on mutation of the AP-1 site (P = 0.703).
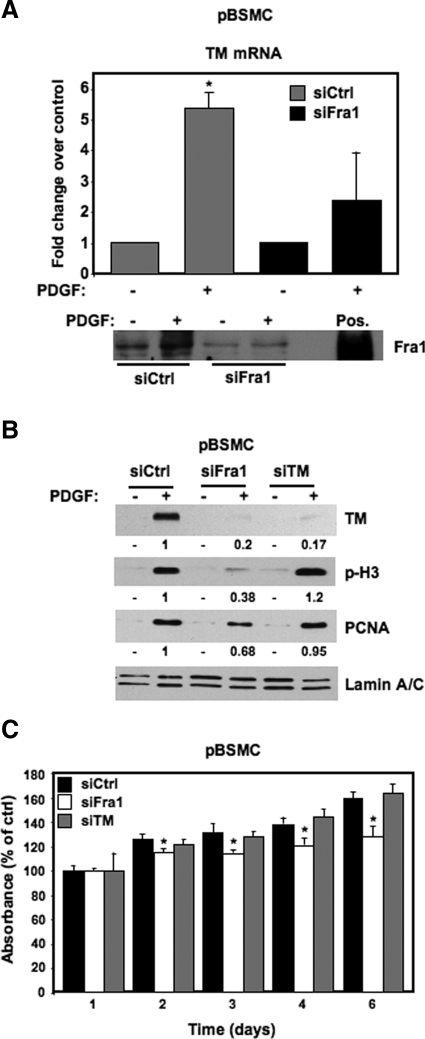
Fra1 Mediates PDGF-Induced Cell Growth and TM Expression
Our results so far demonstrate an increase in c-jun and Fra1 activity as well as TM protein expression in response to PDGF treatment in pBSMC. TF ELISA (Figure 3), measurement of AP-1 DNA-binding activity by EMSA (Figures 4 and 5B), ChIP assay (Figure 5D), and promoter-reporter analyses (Figure 5, A and E) implicated c-jun and Fra1 as the major AP-1 components involved in TM gene regulation. However, to directly test the requirement for AP-1 in PDGF-mediated TM gene regulation, we targeted Fra1 protein expression using a knockdown approach and evaluated its effect on TM mRNA and protein induction in response to PDGF treatment. We chose to target Fra1 as opposed to c-jun in these experiments for the following reason. We were intrigued by the activation of Fra1 by PDGF because, unlike c-jun and c-fos, it was not activated on treatment of pBSMC with another activator of PI3K/Akt, namely cyclic stretch relaxation (A. Ramachandran and R.M. Adam, unpublished observations). Thus, in contrast to c-jun and c-fos, the increase in Fra1 activity appeared selective for PDGF and suggested that activation of Fra1 was required for PDGF-induced TM expression. Quiescent pBSMC were transfected with Fra1 siRNA or nontargeting control (Ctrl) siRNA, treated with 30 ng/ml PDGF or vehicle, and evaluated for induction of TM mRNA and protein, and other proteins. As shown in Figure 6A, in cells nucleofected with control siRNA, PDGF significantly induced TM mRNA (P < 0.01), whereas Fra1 knockdown significantly attenuated the induction of TM mRNA by PDGF, strongly suggesting that Fra1 induction by PDGF is required for subsequent TM gene activation. PDGF is a classic SMC mitogen: thus, we reasoned that the activation of Fra1 and TM by PDGF may play a role in PDGF-induced DNA synthesis and cell division. To this end, we examined known markers of cell proliferation in PDGF-stimulated pBSMC that were transfected with siRNAs targeting Fra1 or TM. Consistent with the effect on TM mRNA levels, Fra1 knockdown abolished TM protein expression and also led to a decrease in proliferating cell nuclear antigen (PCNA, a marker of DNA replication) and histone H3 phosphorylation (a marker of chromosome segregation during mitosis) (Figure 6B). However, TM knockdown did not affect PDGF-induced up-regulation of PCNA or phospho-histone H3 (p-H3) proteins (Figure 6B), suggesting that TM does not regulate cell cycle transit in pBSMC. To confirm the effects of TM and Fra1 knockdown on proliferation, we assessed pBSMC biomass for up to 6 days following nucleofection with control, TM, or Fra1 siRNAs. As shown in Figure 6C, knockdown of Fra1, but not TM, led to a statistically significant reduction in cell number at days 2 to 6 (P < 0.05), in agreement with the immunoblot data in Figure 6B. Fra1 silencing, but not TM knockdown, also attenuated the PDGF-stimulated increase in pBSMC biomass (data not shown).
Figure 6.
Fra1 mediates the PDGF-induced activation of TM. A: Real-time RT-PCR of TM mRNA in pBSMC following nucleofection with either nontargeting control siRNA (Ctrl, gray bar) or siRNA targeting Fra1 (black bar), and treatment of serum-depleted cells with vehicle (−) or 30 ng/ml PDGF (+) for 2 hours. The graph shows fold increase in TM mRNA with PDGF treatment relative to control; data are the average of two independent experiments assayed in duplicate. TM mRNA was increased ∼5-fold on PDGF treatment (+) for two hours relative to untreated cells (−) in the Ctrl siRNA condition (P = 0.009). Knockdown of Fra1 decreased the PDGF-mediated induction of TM mRNA levels, such that the effect of PDGF was no longer significant (P = 0.28). An immunoblot showing Fra1 protein levels in pBSMC 48 hours after nucleofection of control or Fra1 siRNA, and subsequent treatment with 30 ng/ml PDGF for two hours is shown below the graph. Lysate from HEK293 cells transfected with a Fra1 expression construct was used as a positive control (Pos.) to verify the position of the Fra1 protein on the blot. B: Fra1 knockdown in pBSMC ablates the PDGF-mediated increase in TM protein as well as markers of cell proliferation. Twenty-four hours after nucleofection of pBSMC with control, Fra1- or TM-targeting siRNAs, cells were serum depleted for 24 hours, and subsequently treated with 30 ng/ml PDGF or vehicle for 48 hours. PDGF caused a robust increase in TM, p-H3, and PCNA levels in the control siRNA-transfected cells. The PDGF-stimulated increase in TM expression was significantly reduced by both Fra1 and TM knockdown. Fra1 knockdown also attenuated the PDGF-induced increase in p-H3 and PCNA levels, whereas cells in which TM was knocked down showed no appreciable change in p-H3 or PCNA up-regulation in response to PDGF. The values represent quantification of the change in PDGF-induced TM, p-H3, and PCNA levels with Fra1 or TM silencing relative to the nontargeted control. C: The effect of Fra1 or TM knockdown on pBSMC proliferation was determined over time using the crystal violet biomass assay. pBSMC nucleofected with siRNA duplexes against Fra1 or TM or with nontargeting siRNA were assayed for up to six days. Data are presented as absorbance expressed as a percentage of control (absorbance at the start of the experiment) and represent mean ± SD of six replicates. Fra1 knockdown led to reduced biomass at days 2–6 (*P < 0.05, siFra1 relative to siCtrl).
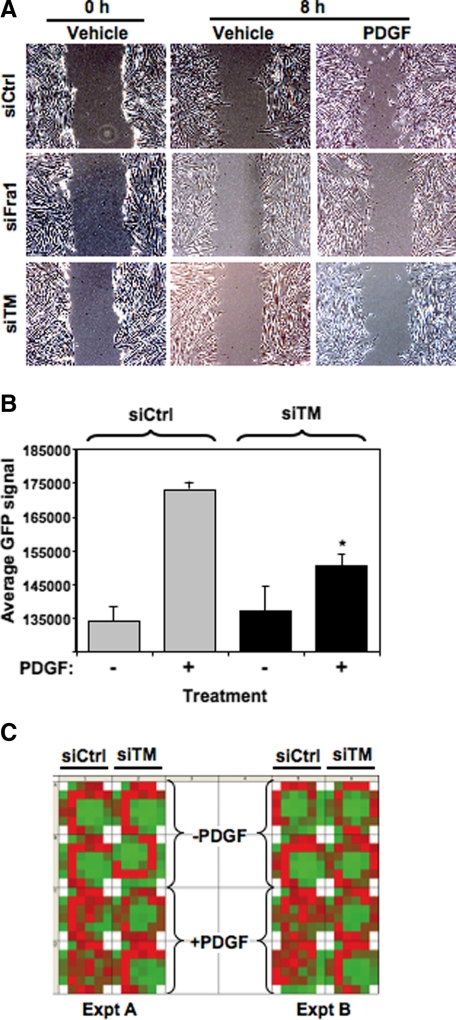
The apparent dichotomy between Fra1 and TM in regulating growth in cells exposed to PDGF suggested that TM participates in PDGF-induced cell behaviors distinct from cell growth. Because PDGF is known to stimulate cell migration in addition to cell division, we investigated the role of TM in influencing PDGF-induced migratory activity of SMC using two independent approaches. First, confluent monolayers of pBSMC transiently transfected with either nontargeting control siRNA or siRNAs to Fra1 or TM were “wounded” by creating a scratch across the cell layer, and the ability of cells to fill the wound in response to vehicle or PDGF was monitored over time. As shown in Figure 7A, PDGF treatment promoted cell migration into the wound in cells transfected with control siRNA but not Fra1 or TM siRNA. To verify the effect of TM on migration observed in the scratch assay, pBSMC transiently nucleofected with a GFP expression vector and either TM siRNA or nontargeting control siRNA were seeded in Transwell chambers and placed in a 24-well culture plate of a modified Boyden chamber migration assay system. PDGF or vehicle was added to the medium in the bottom wells to induce migration that was subsequently detected by assaying for GFP fluorescence. A Fluoroblok insert in the membrane blocked GFP signal from nonmigrated cells present on the upper side of the membrane. After 24 hours of PDGF treatment, we observed a distinct increase in migration compared with nontreated controls (P < 0.05). Significantly, we observed that cell migration in response to PDGF treatment was significantly attenuated in cells with diminished TM levels (*P < 0.05) (Figure 7B). A heat map of two independent experiments is shown (Figure 7C), visually illustrating the reduction in PDGF-stimulated migration in cells transfected with TM siRNA. To our knowledge, this is the first direct demonstration of a role for endogenous TM in influencing cell migration.
Figure 7.
TM regulates PDGF-induced cell migration. A: pBSMC were transfected with control nontargeting, Fra1, or TM siRNAs and allowed to grow to confluence. The cells were serum depleted for 24 hours and a scratch was made in the confluent monolayer, followed by addition of 30 ng/ml PDGF (or vehicle alone) for 8 hours. Migration of cells into the wound caused by the scratch was monitored by phase contrast imaging. The data are representative of three independent trials. B: pBSMC cotransfected with a GFP-expressing plasmid and either control or TM siRNA were used in a modified Boyden Transwell chamber migration assay, as described in Materials and Methods. The GFP signal from cells that migrated through to the bottom of the Transwell membrane was evaluated under conditions of control or PDGF treatment. The graph shows the average GFP signal of migrated cells in control siRNA (gray bar) or TM siRNA (black bar) conditions, in the absence (−) or presence (+) of PDGF. Data are representative of four independent experiments. The PDGF-stimulated increase in pBSMC migration was significantly attenuated in cells in which TM was silenced (*P = 0.024, siTM + PDGF relative to siCtrl + PDGF). C: Heat map of a 24-well plate shows an increase in GFP signal from migrated cells (red) in response to PDGF in pBSMC nucleofected with control but not with TM siRNA. Two independent experiments, each performed in duplicate, are shown.
Discussion
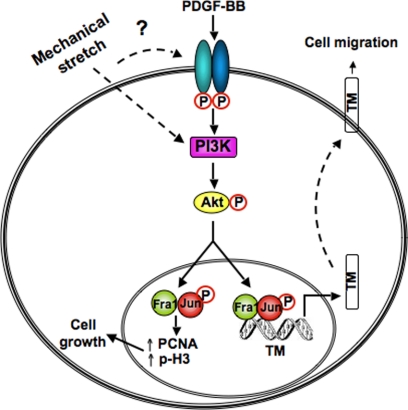
In this study, we describe the mapping of a novel PDGF-dependent signaling mechanism in bladder SMC that results in enhanced expression of TM via an Akt- and AP-1-dependent pathway. We also provide evidence to support a functional role for TM in smooth muscle remodeling. The evidence supporting our conclusions is as follows: i) ex vivo bladder distension induced activation of the PDGFR and TM expression in the detrusor smooth muscle; ii) knockdown of Akt in pBSMC by RNA interference or pharmacological inhibition attenuated PDGF-stimulated TM expression, with Akt1 identified as the dominant TM-regulating Akt isoform; iii) PDGF treatment of pBSMC activates a select subset of AP-1 family proteins, of which Fra1 is essential for PDGF-induced up-regulation of TM; iv) PDGF regulates TM transcription through binding of c-jun and Fra1 to a novel AP-1 motif at −2010/−2004 in the TM promoter; and v) knockdown of TM in pBSMC attenuates PDGF-induced migration, a hallmark of pathological remodeling. These findings are summarized in schematic form in Figure 8.
Figure 8.
A model of regulation of TM by the PDGFR axis in bladder smooth muscle. Exposure to PDGF in vitro or bladder wall distension in vivo causes phosphorylation and activation of the PDGFR, initiating downstream signaling cascades such as the PI3K/Akt pathway. PDGF-stimulated, PI3K/Akt-dependent signals activate specific AP-1 components (c-jun and Fra1), which bind to the TM promoter to increase its expression and thereby promote cell migration. PDGF also stimulates cell cycle transit in a Fra1-dependent manner.
Although numerous studies have described the downstream targets of PDGF that effect phenotypic modulation of smooth muscle in vascular smooth muscle (reviewed in Ref. 19), there is a dearth of knowledge regarding PDGF-stimulated mechanisms of remodeling in visceral smooth muscle, such as that which makes up the bladder wall. Previous reports have described increases in PDGF ligand and receptor levels in mechanically stimulated tissues, as well as increased PDGFR phosphorylation in SMC exposed to mechanical stimuli in vitro. Wilson et al20 reported an increase in PDGFs AA and BB production by neonatal rat vascular SMC subjected to mechanical strain in vitro and proceeded to demonstrate autocrine stimulation of DNA synthesis by secreted PDGF. In contrast, Hu et al9 observed rapid phosphorylation of the PDGFR in VSMC subjected to cyclic stretch in vitro that could not be blocked by PDGF-neutralizing antibody and concluded that PDGFR activation occurred in a ligand-independent manner. Increased PDGFR mRNA levels were observed in the arteries of rats with arterial distension as a result of pulmonary hypertension,21 whereas an increase in both PDGF ligand and receptor levels was evident in human bladder fibroblasts exposed to hydrostatic pressure in the physiological range.8 A modest increase in PDGFR level in SMC was also observed, although in all cases, up-regulation of ligand and receptor expression was observed over 1 to 2 days,8 consistent with a role for paracrine effects of PDGF ligand on bladder wall remodeling following its release into the microenvironment. Despite these observations linking up-regulation of the PDGFR signaling axis to physical forces, direct activation of the PDGFR has not been reported in intact tissue. Our data, in contrast, suggest a rapid (within minutes) induction of PDGFR phosphorylation within the bladder wall in response to distension, ie, in a time frame that cannot be explained by de novo synthesis of PDGF. To our knowledge, this is the first demonstration of PDGFR phosphorylation in an intact organ or tissue and implicates PDGFR-dependent signaling in the response of bladder smooth muscle to pathological stimuli, such as overdistension.
TM has been widely studied in the context of endothelial cell biology and coagulation. TM is a single membrane-spanning glycoprotein with an N-terminal lectin-like domain, six EGF-like motifs, a serine/threonine-rich juxtamembrane region, and a short cytoplasmic tail (reviewed in22). The anticlotting activity of TM derives primarily from its ability to bind thrombin, thereby preventing thrombin from catalyzing the conversion of fibrinogen to fibrin, the penultimate step of the coagulation cascade. However, its function in smooth muscle, and particularly visceral smooth muscle, has not been explored in detail. In vascular smooth muscle, TM has been shown to have both growth stimulatory and growth inhibitory effects. Treatment of aortic SMC in vitro with a recombinant TM peptide, comprising the six EGF-like domains but lacking the native N-terminal region and the Ser/Thr-rich region, increased DNA synthesis in a dose-dependent manner.23 In contrast, exposure of carotid artery SMC and balloon-injured vessels to recombinant TM comprising the entire ectodomain had no effect on DNA synthesis when used alone but inhibited thrombin-induced cell cycle transit in vitro24 and neointimal hyperplasia in vivo.25 Thus, the activity of recombinant TM in vascular SMC appears to be highly dependent on TM conformation and target cell. To address the function of TM in bladder smooth muscle, we used RNA interference to target the endogenous molecule. Knockdown of TM in pBSMC had no appreciable effect on either basal or PDGF-stimulated cell cycle transit, whereas down-regulation of Fra1 by siRNA markedly inhibited PDGF-induced growth, consistent with previous studies linking Fra1 to SMC mitogenesis.26 In addition to its robust mitogenic activity, PDGF is also a potent inducer of cell motility and migration.27 Because SMC migration is an important component of pathological smooth muscle remodeling, we investigated the potential function of TM in migratory behavior and found that in contrast to its effects on growth, down-regulation of TM by RNA interference reduced PDGF-induced pBSMC migration by >50%. These observations provide the first demonstration of a role for endogenous TM in migration of any cell type and suggest that expression of TM in pBSMC in vitro and in the intact bladder downstream of PDGFR and Akt activation may be a mediator and/or marker of bladder SM remodeling.
The mechanism whereby TM elicits effects on migration is unknown but may be related to its ability to regulate cell adhesion. Published findings have demonstrated that deletion of the N-terminal lectin-like domain in TM prevented intercellular association of melanoma cells.28 In that study, inhibition of the lectin-like domain with function-blocking antibody was also found to inhibit clustering of keratinocytes.28 Conversely, forced expression of TM in an insulinoma cell line increased cell aggregration, an effect believed to result, in part, from TM interacting with neural cell adhesion molecule (N-CAM).29 The authors also speculated in that study that TM overexpression may increase the function of β1 integrin, a critical regulator of migration (reviewed in Ref. 30). Other reports have suggested that exogenous recombinant TM could stimulate expression of mediators of matrix proteolysis in endothelial cells, including matrix metalloproteinases and plasminogen activators.31 Taken together, these observations suggest that TM may regulate migration through its ability to alter cell association with the microenvironment, including cell-cell and cell-matrix interactions.
Our data show that the effect of PDGF on TM expression is mediated at the transcriptional level through Akt-dependent effects on the AP-1 transcriptional complex. Deletion analysis identified a novel PDGF-responsive AP-1 binding site at −2010/−2004 in the human TM promoter. Activity of the TM promoter is also known to be regulated by Ets1,32 Sp1, and DR433 transcription factors. Indeed, Lo et al34 recently implicated Akt and Ets1 as mediators of PDGF-induced TM expression in vascular SMC. In that study, however, the function of TM was not described. An important aspect of our ongoing studies is the demonstration that discrete hyperplastic stimuli for pBSMC, such as PDGF or mechanical stretch, elicit distinct patterns of AP-1 activation. In particular, PDGF stimulated the DNA-binding activity of c-jun, c-fos, Fra1, Fra2, and JunB in pBSMC. In contrast, cyclic stretch showed greater selectivity, activating only c-jun, c-fos, and FosB to a significant extent (A. Ramachandran and R.M. Adam, unpublished observations). Furthermore, of the AP-1 subunits tested, only c-jun and Fra-1 emerged as functionally relevant mediators of PDGF- and Akt-induced TM expression in pBSMC. These observations lend support to the concept that AP-1 dimer composition is tightly regulated depending on the initiating stimulus and is likely to be a key determinant of AP-1 function.35,36
In summary, we have described a novel PDGFR-dependent pathway that regulates expression of TM in bladder smooth muscle. We identify AP-1, specifically c-jun and Fra-1, as specific regulators of TM gene expression and provide the first evidence to support a role for endogenous TM in regulating SMC migration. Last, our demonstration that TM is robustly induced in a model of bladder injury suggests that TM may be an early marker of pathological smooth muscle remodeling.
Supplementary Material
Acknowledgments
We thank Drs. Carlos Estrada, Dolores Di Vizio, Keith Solomon, and Michael Freeman for critical reading of the manuscript and members of the Adam, Estrada, and Freeman laboratories for helpful discussions. We also acknowledge the technical contributions from Samuel Eaton in the early part of the study.
Footnotes
Address reprint requests to Rosalyn M. Adam, Ph.D., Urological Diseases Research Center, John F. Enders Research Laboratories, Room 1077, Children’s Hospital Boston, 300 Longwood Ave., Boston, MA 02115. E-mail: rosalyn.adam@childrens.harvard.edu.
Supported by National Institutes of Health grants R01 DK077195 (R.M.A.) and R21 DK66412 (R.M.A.), the Edwin Beer Fellowship from the New York Academy of Medicine (R.M.A.), and the Children’s Urological Foundation. This work was also supported in part by grants from the AUA Foundation Research Scholar Program and Boehringer Ingelheim (A.R.) and by the Children’s Hospital Boston Office of Fellowship Training (A.R.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Hershenson MB, Brown M, Camoretti-Mercado B, Solway J. Airway smooth muscle in asthma. Annu Rev Pathol. 2008;3:523–555. doi: 10.1146/annurev.pathmechdis.1.110304.100213. [DOI] [PubMed] [Google Scholar]
- Orlandi A, Bochaton-Piallat ML, Gabbiani G, Spagnoli LG. Aging, smooth muscle cells and vascular pathobiology: implications for atherosclerosis. Atherosclerosis. 2006;188:221–230. doi: 10.1016/j.atherosclerosis.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Adam RM. Recent insights into the cell biology of bladder smooth muscle. Nephron Exp Nephrol. 2005;102:e1–e7. doi: 10.1159/000088310. [DOI] [PubMed] [Google Scholar]
- Halayko AJ, Solway J. Molecular mechanisms of phenotypic plasticity in smooth muscle cells. J Appl Physiol. 2001;90:358–368. doi: 10.1152/jappl.2001.90.1.358. [DOI] [PubMed] [Google Scholar]
- Levin RM, Monson FC, Haugaard N, Buttyan R, Hudson A, Roelofs M, Sartore S, Wein AJ. Genetic and cellular characteristics of bladder outlet obstruction. Urol Clin N Am. 1995;22:263–283. [PubMed] [Google Scholar]
- Park JM, Borer JG, Freeman MR, Peters CA. Stretch activates heparin-binding EGF-like growth factor expression in bladder smooth muscle cells. Am J Physiol Cell Physiol. 1998;275:C1247–C1254. doi: 10.1152/ajpcell.1998.275.5.C1247. [DOI] [PubMed] [Google Scholar]
- Borer JG, Park JM, Atala A, Nguyen HT, Adam RM, Retik AB, Freeman MR. Heparin-binding EGF-like growth factor expression increases selectively in bladder smooth muscle in response to lower urinary tract obstruction. Lab Invest. 1999;79:1335–1345. [PubMed] [Google Scholar]
- Akbal C, Lee SD, Jung C, Rink R, Kaefer M. Up-regulation of both PDGF-BB and PDGF-BB receptor in human bladder fibroblasts in response to physiologic hydrostatic pressure. J Pediatr Urol. 2006;2:402–408. doi: 10.1016/j.jpurol.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Hu Y, Bock G, Wick G, Xu Q. Activation of PDGF receptor α in vascular smooth muscle cells by mechanical stress. FASEB J. 1998;12:1135–1142. doi: 10.1096/fasebj.12.12.1135. [DOI] [PubMed] [Google Scholar]
- Estrada CR, Adam RM, Eaton SH, Bagli DJ, Freeman MR. Inhibition of EGFR signaling abrogates smooth muscle proliferation resulting from sustained distension of the urinary bladder. Lab Invest. 2006;86:1293–1302. doi: 10.1038/labinvest.3700483. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Adam RM, Bride SH, Park JM, Peters CA, Freeman MR. Cyclic stretch activates p38 SAPK2-, ErbB2-, and AT1-dependent signaling in bladder smooth muscle cells. Am J Physiol Cell Physiol. 2000;279:C1155–C1167. doi: 10.1152/ajpcell.2000.279.4.C1155. [DOI] [PubMed] [Google Scholar]
- Adam RM, Roth JA, Cheng HL, Rice DC, Khoury J, Bauer SB, Peters CA, Freeman MR. Signaling through PI3K/Akt mediates stretch and PDGF-BB-dependent DNA synthesis in bladder smooth muscle cells. J Urol. 2003;169:2388–2393. doi: 10.1097/01.ju.0000063980.99368.35. [DOI] [PubMed] [Google Scholar]
- Adam RM, Eaton SH, Estrada C, Nimgaonkar A, Shih SC, Smith LE, Kohane IS, Bagli D, Freeman MR. Mechanical stretch is a highly selective regulator of gene expression in human bladder smooth muscle cells. Physiol Genomics. 2004;20:36–44. doi: 10.1152/physiolgenomics.00181.2004. [DOI] [PubMed] [Google Scholar]
- Kanematsu A, Ramachandran A, Adam RM. GATA-6 mediates human bladder smooth muscle differentiation: involvement of a novel enhancer element in regulating α-smooth muscle actin gene expression. Am J Physiol Cell Physiol. 2007;293:C1093–C1102. doi: 10.1152/ajpcell.00225.2007. [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capolicchio G, Aitken KJ, Gu JX, Reddy P, Bagli DJ. Extracellular matrix gene responses in a novel ex vivo model of bladder stretch injury. J Urol. 2001;165:2235–2240. doi: 10.1016/S0022-5347(05)66173-3. [DOI] [PubMed] [Google Scholar]
- Baxter RM, Secrist JP, Vaillancourt RR, Kazlauskas A. Full activation of the platelet-derived growth factor β receptor kinase involves multiple events. J Biol Chem. 1998;273:17050–17055. doi: 10.1074/jbc.273.27.17050. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Owens GK. Molecular control of vascular smooth muscle cell differentiation and phenotypic plasticity. Novartis Found Symp. 2007;283:174–191. doi: 10.1002/9780470319413.ch14. discussion 191–173, 238–141. [DOI] [PubMed] [Google Scholar]
- Wilson E, Mai Q, Sudhir K, Weiss RH, Ives HE. Mechanical strain induces growth of vascular smooth muscle cells via autocrine action of PDGF. J Cell Biol. 1993;123:741–747. doi: 10.1083/jcb.123.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Saito M, Ueno A, Nakamura M, Takeishi K, Nakayama K. Mechanical stretch augments PDGF receptor β expression and protein tyrosine phosphorylation in pulmonary artery tissue and smooth muscle cells. Mol Cell Biochem. 2000;215:103–113. doi: 10.1023/a:1026506801659. [DOI] [PubMed] [Google Scholar]
- Weiler H, Isermann BH. Thrombomodulin. J Thromb Haemost. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- Tohda G, Oida K, Okada Y, Kosaka S, Okada E, Takahashi S, Ishii H, Miyamori I. Expression of thrombomodulin in atherosclerotic lesions and mitogenic activity of recombinant thrombomodulin in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 1998;18:1861–1869. doi: 10.1161/01.atv.18.12.1861. [DOI] [PubMed] [Google Scholar]
- Li J, Garnette CS, Cahn M, Claytor RB, Rohrer MJ, Dobson JG, Jr, Gerlitz B, Cutler BS. Recombinant thrombomodulin inhibits arterial smooth muscle cell proliferation induced by thrombin. J Vasc Surg. 2000;32:804–813. doi: 10.1067/mva.2000.107992. [DOI] [PubMed] [Google Scholar]
- Li JM, Singh MJ, Itani M, Vasiliu C, Hendricks G, Baker SP, Hale JE, Rohrer MJ, Cutler BS, Nelson PR. Recombinant human thrombomodulin inhibits arterial neointimal hyperplasia after balloon injury. J Vasc Surg. 2004;39:1074–1083. doi: 10.1016/j.jvs.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Cao H, Dronadula N, Rao GN. Thrombin induces expression of FGF-2 via activation of PI3K-Akt-Fra-1 signaling axis leading to DNA synthesis and motility in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C172–C182. doi: 10.1152/ajpcell.00284.2005. [DOI] [PubMed] [Google Scholar]
- Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev. 2004;15:255–273. doi: 10.1016/j.cytogfr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Huang HC, Shi GY, Jiang SJ, Shi CS, Wu CM, Yang HY, Wu HL. Thrombomodulin-mediated cell adhesion: involvement of its lectin-like domain. J Biol Chem. 2003;278:46750–46759. doi: 10.1074/jbc.M305216200. [DOI] [PubMed] [Google Scholar]
- Iino S, Abeyama K, Kawahara K, Yamakuchi M, Hashiguchi T, Matsukita S, Yonezawa S, Taniguchi S, Nakata M, Takao S, Aikou T, Maruyama I. The antimetastatic role of thrombomodulin expression in islet cell-derived tumors and its diagnostic value. Clin Cancer Res. 2004;10:6179–6188. doi: 10.1158/1078-0432.CCR-03-0750. [DOI] [PubMed] [Google Scholar]
- Al-Jamal R, Harrison DJ. Beta1 integrin in tissue remodelling and repair: from phenomena to concepts. Pharmacol Ther. 2008;120:81–101. doi: 10.1016/j.pharmthera.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Shi CS, Shi GY, Chang YS, Han HS, Kuo CH, Liu C, Huang HC, Chang YJ, Chen PS, Wu HL. Evidence of human thrombomodulin domain as a novel angiogenic factor. Circulation. 2005;111:1627–1636. doi: 10.1161/01.CIR.0000160364.05405.B5. [DOI] [PubMed] [Google Scholar]
- von der Ahe D, Nischan C, Kunz C, Otte J, Knies U, Oderwald H, Wasylyk B. Ets transcription factor binding site is required for positive and TNF-α-induced negative promoter regulation. Nucleic Acids Res. 1993;21:5636–5643. doi: 10.1093/nar/21.24.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S, Ishii H, Matsumoto F, Kusano M, Kizaki K, Matsuda J, Kazama M. Acceleration of thrombomodulin gene transcription by retinoic acid: retinoic acid receptors and Sp1 regulate the promoter activity through interactions with two different sequences in the 5′-flanking region of human gene. J Biol Chem. 2001;276:2440–2450. doi: 10.1074/jbc.M004942200. [DOI] [PubMed] [Google Scholar]
- Lo IC, Lin TM, Chou LH, Liu SL, Wu LW, Shi GY, Wu HL, Jiang MJ. Ets-1 mediates platelet-derived growth factor-BB-induced thrombomodulin expression in human vascular smooth muscle cells. Cardiovasc Res. 2009;81:771–779. doi: 10.1093/cvr/cvn351. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrzynska B, Mosieniak G, Kaminska B. Changes of the trans-activating potential of AP-1 transcription factor during cyclosporin A-induced apoptosis of glioma cells are mediated by phosphorylation and alterations of AP-1 composition. J Neurochem. 2000;74:42–51. doi: 10.1046/j.1471-4159.2000.0740042.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.