Abstract
Analysis of lineage relationships in the naphthalene-injured tracheal epithelium demonstrated that two multipotential keratin 14–expressing cells (K14ECs) function as progenitors for Clara and ciliated cells. These K14EC were distinguished by their self-renewal capacity and were hypothesized to reside at the stem and transit amplifying tiers of a tissue-specific stem cell hierarchy. In this study, we used gene expression and histomorphometric analysis of the steady-state and naphthalene-injured trachea to evaluate the predictions of this model. We found that the steady-state tracheal epithelium is maintained by two progenitor cell pools, secretory and basal cells, and the latter progenitor pool is further divided into two subsets, keratin 14–negative and –positive. After naphthalene-mediated depletion of the secretory and ciliated cell types, the two basal cell pools coordinate to restore the epithelium. Both basal cell types up-regulate keratin 14 and generate a broadly distributed, abundant, and highly mitotic cell pool. Furthermore, basal cell proliferation is associated with generation of differentiated Clara and ciliated cells. The uniform distribution of basal cell progenitors and of their differentiated progeny leads us to propose that the hierarchical organization of tracheal reparative cells be revised to include a facultative basal cell progenitor pool.
Basal cell hyperplasia is a common pathological alteration in chronic lung diseases, including those with an airway bias, asthma, chronic bronchitis, and cystic fibrosis, as well as diseases with lesser airway involvement such as idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease.1 This lesion is identified in part by immunostaining for keratin 14 (see below), although ectopic expression of other type I keratins has been reported.2 Aberrant epithelial regions are also characterized by secretory and ciliated cell hypoplasia. The palisade form of squamous metaplasia has been associated with aberrant Epidermal Growth Factor (EGF) and Epidermal Growth Factor Receptor (EGFR) expression3 or with Wnt signaling.4 In contrast, advanced lesions are associated with aberrant transforming growth factor β signaling and express keratinization markers.5 Although progression of squamous metaplasia to squamous cell carcinoma has been suggested, the cellular origin of these lesions is unknown. Further, the relationship between this pathology and epithelial injury and repair has not been determined. The goal of this study was to shed light on these questions through evaluation of the molecular phenotype and distribution of progenitor cells involved in maintenance and repair of the basal cell–containing compartment of the mouse airway, the tracheal epithelium.
The tracheal compartment of the mouse respiratory tract is analogous to the first six generations of the human airway. It is supported by cartilaginous rings and associated contractile and noncontractile mesenchemyal cells. The epithelium in this region is pseudostratified. Pyramidal basal cells are located adjacent to the basement membrane and anchor the epithelium through hemidesmosomal contacts.6 Clara-like cells are nonciliated secretory cells that are identified morphologically by an apical projection, molecularly by expression of Clara cell secretory protein (CCSP or CC10) and secretoglobins 1A1 and 1A2,7 and biochemically by activity of cytochrome P450–2F2 (CYP-2F2). These cells are similar to bronchiolar Clara cells but exhibit some distinctive ultrastructural, functional, and molecular properties.8 Ciliated cells are distinguished by the presence of motile cilia. This cell type expresses acetylated tubulin in the ciliary bed and the transcription factor FoxJ1. Like secretory cells, ciliated cells exhibit compartment-dependent functional differences.9 Clara-like and ciliated cells interact through adherins and gap junctions and exhibit a progenitor-progeny relationship in the steady state and after nitrogen dioxide exposure.10
The naphthalene injury model has been used to evaluate cell replacement mechanisms for repair of a simple epithelium, the bronchiolar epithelium. Naphthalene is activated to a cytotoxic epoxide by CYP-2F2–mediated metabolism in mice.11 This enzyme is expressed specifically in bronchiolar Clara cells. Parenteral naphthalene treatment results in a dose-, strain-, and sex-dependent depletion of the Clara cell population. For example, exposure of female FVB/n mice to 300 mg/kg naphthalene results in spatially restricted regeneration of the Clara cell population.12 In contrast, treatment of female Swiss-Webster mice with 200 mg/kg naphthalene results in uniform regeneration of the terminal bronchiolar Clara cell population.13 Together the high- and low-dose naphthalene studies identified subsets of Clara cells that were distinguished on the basis of naphthalene sensitivity. The most naphthalene-resistant cell was termed the variant CCSP-expressing cell (vCE).14 Several lines of evidence suggest that this cell may be a tissue-specific stem cell (reviewed in reference15), although this interpretation has been challenged on the basis of the stem cell definition.16 The naphthalene-sensitive Clara cell exhibits differentiated features when in the quiescent state yet has the capacity to proliferate for normal tissue maintenance and in response to injury. These Clara cells have been termed a facultative progenitor cell.15
Keratin proteins are commonly used to identify epithelial cells and to distinguish functional subsets in pseudostratified and stratified epithelia.17 Keratins are the protein subunits that form the 10 nanometer cytoplasmic structures termed intermediate filaments. In epithelia, these filaments interact with desmosomes to form cell–cell contacts and with integrins to allow hemidesmosomal attachment to the extracellular matrix. Thus, intermediate filaments are important for maintenance of cellular structure and form. Intermediate filaments also exhibit cycles of assembly and disassembly in response to alterations in the tissue microenvironment. Keratin proteins are categorized into five subclasses on the basis of isoelectric point and structural differences.18 Type I keratin (including keratins 14, 15, and 18) and type II keratins (including keratins 5 and 8) assemble into heterodimeric protein complexes in a one to one ratio. Immunolocalization of keratins suggested pairing of keratin 5 with 14 in complex epithelia, while keratin 8 is typically paired with 18 in simple epithelia. Keratins 5 and 14 have been used to identify tracheal basal cells in histological sections, while keratins 8 and 18 are used to identify Clara-like, Clara, and ciliated cells. In complex epithelia, keratin 14+/5+ cells are enriched in the mitotic compartment, and these markers are expressed at lower levels as cells exit the mitotic pool.17 Thus, cellular expression of keratins may vary as a specific cell type responds to injury or disease processes.
Analysis of naphthalene-mediated tracheobronchial injury and the subsequent repair process suggested that the basal cell functions as tissue-specific stem cell for this compartment. Lineage tracing identified a multipotential keratin 14–expressing cell (K14EC) that self-renewed and also generated Clara and ciliated cells.19,20 However, interpretation of these studies was confounded by the finding that lineage-traced K14ECs generated two additional colony types. The basal cell-only colony was indicative of a unipotential keratin 14+ cell, while the Clara-like/ciliated colony type was suggestive of a transient bipotential keratin 14+ cell. These data, when considered in light of lineage tracing analysis in human bronchial epithelial cells ex vivo and identification of label retaining basal cells following acid or detergent injury,21 led to organization of keratin 14–expressing mouse basal cells into a classical stem cell hierarchy. Within this hierarchy, the bipotential K14EC was thought to represent the transient amplifying cell tier.
Conducting airway epithelial cell depletion, proliferation, and replacement of injured cell types has been extensively evaluated using quantitative techniques.19,22,23,24 This two-dimensional (2D) method was used to determine the number or representation of various cell types and their mitotic index in anatomically similar regions of the epithelium. The 2D method is valid if nuclear or cellular volume is constant between comparison groups. Stereological methods25 can be used to evaluate this potential bias in a 2D analysis or to avoid this issue entirely.
In this study, we used stereological methods to determine whether nuclear volume differed between control and naphthalene recovery day 6. Although this analysis demonstrated that the 2D method was valid, there are numerous advantages to the stereological approach. First, volume densities (VV) of various cell types can be determined by point counting of tracheal epithelial profiles. The reference space for this type of evaluation is the epithelium. VV is presented in μm3/μm3 but is in reality a unit-less term. Second, the surface area of the basement membrane per reference volume (SV) can be determined by point and intercept counting. The reference volume is the epithelium. Finally, the thickness of a structure or the volume per unit area of basement membrane (Vs) can be calculated by determining the arithmetic mean thickness. This parameter is presented in μm3/μm2. Use of stereological methods permitted a more complete and unbiased evaluation of parameters critical to evaluation of basal cell hyperplasia and phenotypic transitions in the naphthalene injured tracheal epithelium.
In this study, we used gene expression and histomorphometric analysis of the steady-state and naphthalene-injured trachea to evaluate the predictions of the classical and facultative progenitor cell models. We find that the steady-state tracheal epithelium is maintained by two independent progenitor cell pools, the basal and Clara cell types. The basal cell progenitor pool is divided into two subsets, keratin 14–negative and–positive. After naphthalene-mediated depletion of the secretory and ciliated cell populations, the two basal cell pools coordinate to restore the epithelium. Both basal cell subtypes up-regulate keratin 14 and generate a broadly distributed, abundant, and highly mitotic cell pool. Nascent secretory and ciliated cells are detected after generation of this hyperplastic basal cell population. Comparison of the present study with lineage tracing of reparative K14ECs suggests that upregulation of keratin 14 is synonymous with an activated cell type that can differentiate in response to environmental cues. Based on these data we propose that the hierarchical organization of tracheal reparative cells be revised to include both basal and secretory facultative progenitor pools.
Materials and Methods
Animals
All procedures involving animal use were reviewed and approved by the National Jewish Health Institutional Animal Care and Use Committee. Adult (8- to 14-week-old) female FVB/n mice were used for all injury/repair analyses. Neonatal mice aged 1 or 21 days were used for gene expression analysis.
Exposures
Treatments were as previously described19,20 and used 300 mg/kg naphthalene (i.p.) as the standard dose. Naphthalene was dissolved at 30 mg/ml Mazola corn oil immediately before treatment. Mice were injected with 10 μl naphthalene/g body weight (i.p.) between 8:00 and 10:00 A.M. Mice were weighed between 8:00 and 10:00 on a daily basis. Survival to 13 days posttreatment was 95%. Groups of 4 mice were killed on recovery days 3, 6, 9, or 13. For bromodeoxyuridine (BrdU; Sigma, St. Louis, MO) pulse-chase analysis, mice were treated with 0.3 ml 5 mg/ml BrdU (i.p.) on recovery day 3. Groups of 3 mice were killed 2 hours (recovery day 3), 3 days (recovery day 6), or 6 days (recovery day 9) after BrdU exposure.
Histology
Control and naphthalene-treated animals were killed by injection of 17.5 mg of 2,2,2-tribromoethanol in PBS (i.p.) followed by exsanguination. Trachea/esophagus units from each mouse were filled with 10% neutral buffered formalin, immersion fixed for 2 hours at room temperature, and stored in PBS/.01% sodium azide at 4°C. Tissues were dehydrated, embedded in paraffin, and sectioned at 5 μm. Antigens were detected with: Rabbit anti-Keratin 5 (Covance, Princeton, NJ; 1:1000), Mouse IgG3 anti-Keratin 14 (ThermoFisher, Waltham MA; 1:500), Chicken anti-Keratin 15 (Covance, 1:1000), Rat anti-Ki-67 (Dako, Glostrup, Denmark; 1:500), Goat anti-CCSP,26 Mouse IgG2b anti-acetylated tubulin (Sigma, 1:8000), and monoclonal Rat anti-BrdU (Accurate Chemical, Westbury, NY; 1:500). All antigens except CCSP required microwave antigen retrieval with 10 mmol/L Citrate Buffer (pH 6.0). Antigen-antibody complexes were detected with Alexa488 or Alexa594-conjugated secondary antibodies (Molecular Probes, Inc., Eugene, OR): Donkey anti-Rabbit (1:500), Goat anti-Mouse IgG3, Goat anti-Chicken 594 (1:500), Donkey anti-Rat (1:500), or Donkey anti-mouse IgG2b. Slides were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired using Zeiss Imager.Z1 fluorescent microscope (Zeiss) equipped with an Axiocam HRc black and white digital camera.
Morphometry
Quantitative Histopathology
The abundance of tracheal epithelial cell types was analyzed using ×200 high-resolution images and morphometric methods.27,28 Only the cartilaginous region of the trachea was evaluated. Comparisons were made between control and postnaphthalene day 6. A random sampling scheme was used. Volume densities (VV) were determined by point (P) counting using a grid of evenly spaced points. VV was calculated by the formula, VV = PP = PN/PT. PP is the point fraction of PN and is determined by counting the number of test points intersecting the structure of interest divided by the total points intersecting the reverence space (PT). In this analysis, the structures of interest were nuclei defined by DAPI staining, total basal cells defined by keratin 5 expression, and specialized basal cells defined by keratin 14 expression. The reference space was the epithelium and was defined by autofluorescence. VV is presented in μm3/μm3. The surface area of the basement membrane per reference volume (SV) was determined by counting intercepts (I) between the surface and grid of evenly spaced cycloid lines. SV was calculated using the formula, SV = 2Io/Lr. In this formula, I0 is the number of intersections between the cycloid grid and a surface, the basement membrane. Lr is the total length of the test line within the reference volume, the epithelium. The thickness of a structure or the volume per unit area of basement (Vs) membrane was calculated by determining the arithmetic mean thickness: τ = VV/SV. This parameter is presented in μm3/μm2.
Cell Counting Methods
Historically, this group has determined the number of cells or nuclei per unit membrane when evaluating cell density, cellular representation, and mitotic index.19 Stereological methods were used to determine whether this 2D method was confounded by alterations in nuclear volume after naphthalene injury. These data are presented in the Results. Comparison of SV did not detect differences in the surface area of the basement membrane in control and on naphthalene recovery day 6. Comparison of nuclear volume (VV/SV) in control and naphthalene recovery day 6 did not detect significant differences. Thus, the 2D method was valid for analysis of cell density and mitotic index. Cell density was calculated as previously reported with the following modification: basement membrane length was measured in pixels using Scion 4 software and converted into mm using a micrometer. Mitotic indices were calculated as previously reported.19 To maintain positional information in the evaluation of basal cell distribution, tracheas were imaged from the third cartilaginous ring (proximal end) to the hilum (distal end) at a magnification of ×400. Statistical significance was determined by Student’s t test.
Gene Expression Analysis
The trachea or accessory lobe was isolated from the esophagus, larynx, fascia, glands, and fat by blunt dissection at ×12 magnification, incubated overnight at 4°C in RNALater (Ambion, Foster City, CA), and stored in RNALater at −80°C. RNA was prepared using the QuickGene RNA Tissue Kit SII (Fujifilm Life Sciences) and concentration was determined on a NanoDrop (NanoDrop Technologies, Wilmington, DE). cDNA was prepared from 200 ng of RNA as previously described.24 Pools of RNA from trachea or lung were used as a calibrator sample. mRNA abundance was evaluated using Taqman Assays (Applied Biosystems, Foster City, CA) on the Fluidigm high throughput platform (San Francisco, CA). This method makes use of a 14 cycle cDNA pre-amplification step (95°C 15 seconds, 60°C 4 minutes) and Taqman PreAmp Master Mix (Applied Biosystems) in a standard PCR thermocycler. Preamplified cDNA was diluted 1:5 in 10 mmol/L Tris/1 mmol/L EDTA (TE). Diluted cDNA (2.25 μl) was added to 2.5 μl Taqman Universal PCR Master Mix (Applied Biosystems) and 0.25 μl DA Sample Loading Reagent (Fluidigm). In a separate tube, 2.5 μl of 20× Taqman Assay was added to 2.5 μl DA Sample Loading Reagent. The 5 μl cDNA samples were loaded into the sample inlet wells, and 5-μl assay samples were loaded into assay detector inlets. The chip was primed and placed into the NanoFlex IFC controller where 8 nanoliter of cDNA and 1 nanoliter of were mixed. Real time PCR analysis was completed on the BioMark System for Genetic Analysis (Fluidigm). Relative gene expression was calculated using the delta-delta cross-threshold (Ct) method.29 Data are presented as the mean ± SEM (n = 4). Differences between control and treatment groups were determined by Student’s t-test.
An experiment to test the linearity of preamplification was done by preamplifying RNA samples from untreated lung tissue for 0, 5, 10, and 15 cycles and analysis of mRNAs representing high (CCSP), medium (FoxJ1 and βGus), and low abundance (keratin 5) classes (See Supplemental Figure S1A at http://ajp.amjpathol.org). Ct values were plotted against number of preamp cycles, and linear regression was performed. All genes analyzed had an R2 value greater than 0.9774. Data are presented as the mean ± SD, n = 3.
Chip to chip reproducibility was evaluated using a total of 48 samples from 3 untreated control mice and groups of 3 mice that were treated with 300 mg/kg naphthalene and recovered 3, 6, 9, or 13 days. Each sample was converted into cDNA using three separate reverse transcriptase reactions (reverse transcriptase, RT) and 1 no-reverse transcriptase (NRT) control. A lung RNA calibrator (2 RT and 1 NRT) was included on each 48 × 48 chip. Sixteen transcripts were assayed on duplicate chips. Results for duplicate analysis of 3 genes are presented (See Supplemental Figure S1B at http://ajp.amjpathol.org). Data are presented as the mean ΔΔCt ± SD, n = 3. CCSP mRNA abundance was expected to decrease dramatically in response to naphthalene injury. Claudin 10 has been reported to be a Clara cell–specific marker for lower airway secretory cells but showed a disparate pattern in the trachea. Adenomatosis Polyposis Coli (APC) was a transcript of interest but the pattern had not been previously assayed. Analyses on separate chips were indistinguishable. The mean SD for 1813 sets of technical replicates was 0.11 ± 0.004 cycles (Supplemental Figure 1B at http://ajp.amjpathol.org).
Results
Steady-State Phenotype of Tracheal Basal Cells
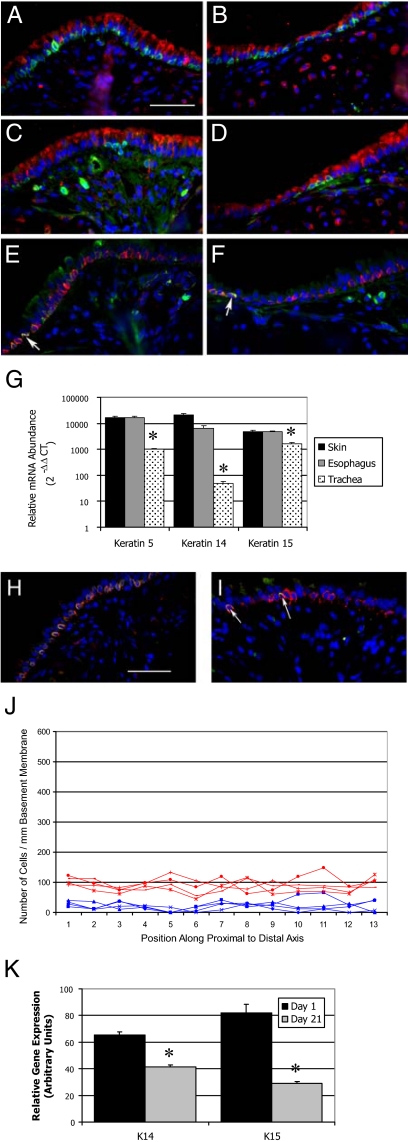
Basal cells that populate the pseudostratified epithelia of glands (mammary, prostate) and cornified epithelia (epidermis, esophagus) coexpress keratins 5 and 14.17 Dual immunofluorescence was used to determine the immunophenotype of steady-state tracheal basal cells. Analysis of keratin 5 and the secretory cell marker CCSP detected keratin 5+ cells located along the basement membrane of both the intercartilagenous (ICR, Figure 1A) and midcartilagenous (MCR, Figure 1B) zones of the steady-state trachea. Similar analysis of keratin 14 detected a subset of basal cells (Figure 1, C and D) in each region of the trachea. Keratin 14–positive cells tended to be clustered in groups of three to five cells. These keratin 14–positive cells coexpressed keratin 5 (Figure 1, E and F). This analysis indicated that tracheal basal cells can be divided into at least two phenotypically-distinct subsets, the keratin 5+/14− and the keratin 5+/14+ subtypes.
Figure 1.
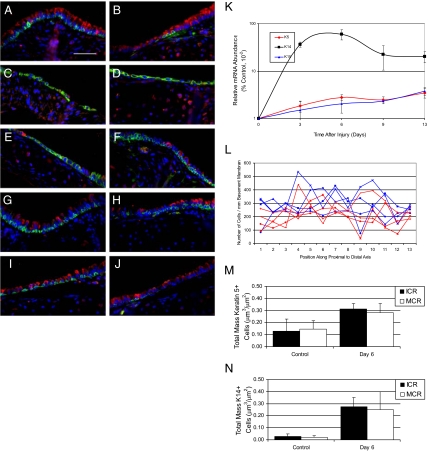
Tracheal basal cell phenotype and distribution. A–F: Dual immunofluorescence analysis of basal cell markers in the intercartilagenous (A, C, E) or midcartilagenous (B, D, F) regions. All cells are counterstained with DAPI (blue). A and B: Red-CCSP, green-keratin 5. C and D: Red-CCSP, green-keratin 14. E and F: Red-keratin 5, green-keratin 14. Arrows in E and F indicate keratin 5+/14+ cells. G: Quantitative RT-PCR analysis of keratin mRNA abundance in basal cell-containing tissues. Note log scale. Mean ± SEM, n = 4. *P < 0.005. H: Dual immunofluorescence analysis of keratin 5 (red) and keratin 15 (green). I: Dual immunofluorescence analysis of keratin 14 (red) and keratin 15 (green). Arrows indicate keratin 14 and 15 dual positive cells. J: Morphometric analysis of keratin 5+ (red) and keratin 14+ (blue) cell distribution as a function of position along the proximal to distal axis of the ventral trachea. Analysis of four mice is shown. K: Quantitative RT-PCR analysis of keratin mRNA abundance as a function of age in days. Mean ± SEM, n = 4. *P < 0.005 relative to day 1. Scale bar in A and H = 50 μm.
Immunofluorescence analysis indicated that the majority of tracheal basal cells were molecularly distinct from the well characterized basal cell populations of glandular and stratified epithelia. To further evaluate this finding, quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was used to evaluate expression of keratins 5 and 14 mRNA in back skin, esophagus, and tracheal RNA (Figure 1G). This analysis detected high-level expression of keratin 5 in all three tissues, with skin and esophagus expressing approximately 10-fold more keratin 5 than trachea (Figure 1G, left). Keratin 14 was also expressed at high levels in skin and esophagus but was 100-fold more abundant in these tissues than in tracheal RNA (Figure 1G, middle). Immunostaining demonstrated that keratin 5+/14+ cells were adjacent to the basement membrane in all three tissues and that the total number of basal cells per unit basement membrane did not vary among skin, esophagus, and trachea (data not shown and Figure 1, A–D). Thus, tissue-specific differences in keratin 5 and 14 mRNA levels could not be accounted for by variation in basal cell number. These data support the conclusion that tracheal basal cells are molecularly distinct from the well-characterized basal cell populations of glandular and stratified epithelia.
Identification of keratin 5+/14− cells raised the question of which type 1 keratin partnered with keratin 5 in the majority of tracheal basal cells. Because keratin 15 has been identified as a type 1 keratin partner of keratin 5 in hair follicle and a potential epidermal stem cell marker,30 keratin 15 mRNA abundance was compared in skin, esophagus, and trachea. This keratin was expressed at similar levels in skin and esophagus and was approximately twofold more abundant in these tissues than in trachea (Figure 1G, right). The cellular distribution of keratins 5, 14, and 15 was evaluated by dual immunofluorescence analysis of the steady-state tracheal epithelium (Figure 1, H and I). This study demonstrated coexpression of keratins 5 and 15 in the majority of steady-state tracheal basal cells located in the ICR (Figure 1H) and MCR (data not shown). Interestingly, keratins 14 and 15 were codetected in basal cells located in the ICR (Figure 1I) and MCR (data not shown) regions. These results were confirmed at the mRNA level using in situ hybridization (data not shown). These data indicate that steady-state basal cells form two molecularly distinct classes, keratin 14-/5+/15+ and keratin 14+/5+/15+.
Distribution of Steady-State Tracheal Basal Cell Subtypes
Previous analysis suggested that the distribution of basal cell–like tissue stem cells might vary as a function of position along the proximal-distal axis of the trachea.21 Consequently, the distribution of keratin 5+ basal cells and the keratin 14+ subset was evaluated (Figure 1J). Keratin 5+ cells were uniformly distributed along the proximal to distal axis of the steady-state mouse trachea (Figure 1J, red lines). Basal cell density was approximately 100 cells per millimeter basement membrane. Slight variation in cell density was noted between ICR and MCR regions but was likely a function of small differences in cell density within these regions. The keratin 14+ subset was also uniformly distributed throughout the proximal to distal axis of the trachea (Figure 1J, blue lines). The keratin 14+ subset was approximately 20% of all basal cells. No proximally-biased localization was detected for this basal cell subset. These data demonstrate that distribution of the two basal cell subsets did not vary along the proximal to distal axis of the trachea.
Differentiation State of Steady-State Tracheal Basal Cells
To determine whether expression of keratins 14 and 15 was indicative of cellular differentiation, expression of these mRNAs was evaluated as a function of age (Figure 1K). This analysis demonstrated that type 1 keratins including keratins 14 and 15 as well as other type 1 keratins and type 2 keratins (data not shown) are down-regulated during the early postnatal period. These data suggest that keratin 14+ cells may be less differentiated than the keratin 14- cell subset.
Contribution of Steady-State Basal Cell Subsets to the Mitotic Pool
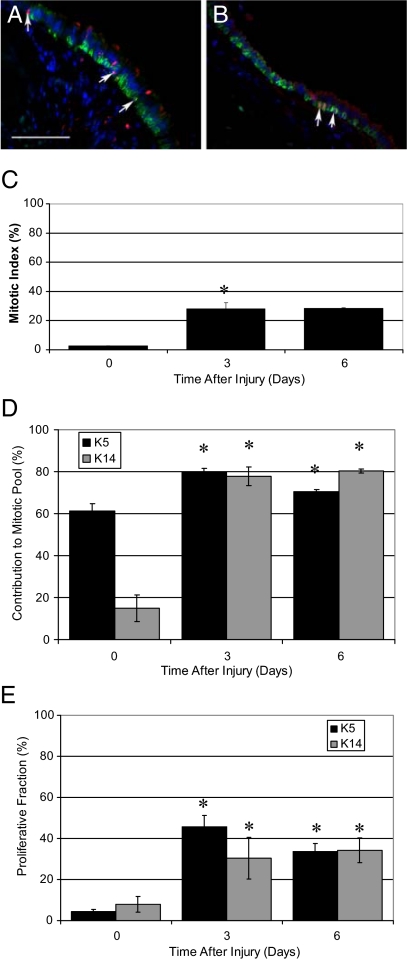
Cellular mechanisms regulating maintenance of the tracheobronchial epithelium were evaluated through analysis of mitotic index, the contribution of basal cell subsets to the mitotic pool, and the cellular mitotic index. Dual immunofluorescence analysis of Ki-67 and keratin 5 or keratin 14 (Figure 2, A and B) was used to identify mitotic cells (Figure 2, C–E). The steady-state tracheal mitotic index was 2.78% ± 0.06% (Figure 2C). In contrast with the bronchial epithelium,19 keratin 5+ basal cells were 61.23%± 3.50% of all Ki-67+ cells while the keratin 14+ subset was 14.99% ± 6.27% of the mitotic pool (Figure 2D, left). These data indicate that basal cells are a major progenitor cell type in the steady-state tracheal epithelium and that both keratin 14- and keratin 14+ cells have the ability to proliferate in the steady state. Within each basal cell class, 4.41% ± 0.98% of all basal cells (keratin 5+) and 8.04% ± 3.86% of keratin 14+ cells were mitotic (Figure 2E). These data indicate that both subsets of basal cells contribute to epithelial maintenance and that differences in their contribution to the mitotic pool reflect their relative abundance.
Figure 2.
Analysis of basal cell proliferation. A and B: Dual immunofluorescence analysis of keratin 5 (green) and Ki-67 (red), and DAPI (blue) in the intercartilagenous region (A) and midcartilagenous region (B). Arrows indicate Ki-67–positive cells. C: Mitotic index in control tissue (0) and as a function of time after injury (days three and six). Mean ± SEM, n = 4. *P < 0.05 relative to control. D: Contribution of basal cells (K5) and the keratin 14+ subset (K14) to the mitotic pool in the steady state (0) and after injury (days three and six). Mean ± SEM, n = 4. *P < 0.005 relative to control. E: Proliferative fraction of basal cells (K5) and the keratin 14+ (K14) subset in control (0) and after injury (days three and six). Mean ± SEM, n = 4. *P < 0.05 relative to control. Scale bar in A = 50 μm.
Naphthalene-Mediated Injury of the Tracheal Epithelium
The size, axial heterogeneity, and noncompartmentalized structure of the trachea prevented simultaneous analysis of cellular and molecular mechanisms in individual mice. Thus, correlations between outcomes that could be analyzed longitudinally (such as body weight change) and those that could be evaluated at single time points in individual mice (histological parameters or gene expression) were established.
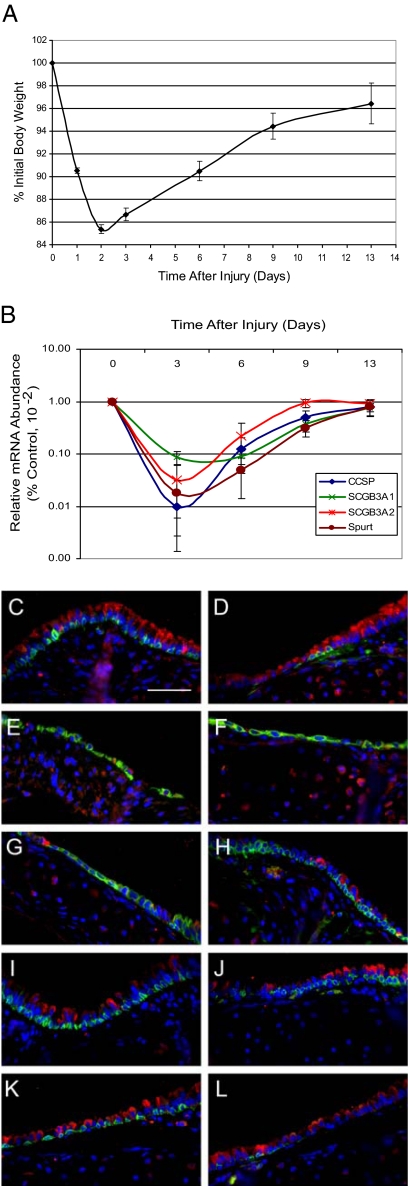
Analysis of body weight demonstrated that exposure of adult female Fvb/n mice to 300 mg/kg naphthalene resulted in body weight decrease to 90.54% ± 0.24% of initial on recovery day 1, 85.38%± 0.40% of initial on recovery day 2, and 86.66% ± 0.56% of initial on recovery day 3. Consistent body weight gain was observed on recovery on days 6 through 13 (Figure 3A).
Figure 3.
Naphthalene-mediated tracheal injury: Clara cell depletion. A: Body weight change as a function of time after naphthalene exposure. Mean ± SEM, n = 4. Values on recovery days two-nine were significantly different from control, P < 0.05. B: mRNA abundance as a function of time after injury. Clara cell secretory markers: CCSP, SCGB3A1 and 3A2, and SPURT. Note the log scale. Values for days three, six, and nine were significantly different from day 0, P < 0.005. Mean ± SEM, n = 4. C–L: Dual immunofluorescence analysis of cell type-specific markers in the intercartilagenous (C, E, G, I, K) and midcartilagenous (D, F, H, J, L) regions. C and D, Control; E and F, Day three; G and H, day six; I and J, day nine; and K and L, day 13. CCSP (red), keratin 5 (green), and nuclei (blue). Scale bar in C = 50 μm.
Analysis of secretory cell marker mRNA abundance demonstrated a rapid decrease in levels of CCSP and other Clara-like cell secretory products including Scgb3A1, Scgb3A2, Spurt (also termed PLUNC) by recovery day 3 (Figure 3B). Secretory cell mRNA levels were 10% of control on recovery day 6 but were ∼80% of control levels by day 13.
Changes in the number of CCSP-expressing Clara-like cells were confirmed by dual-immunofluorescence analysis (Figure 3, C–L). Clara-like cells were located in the ICR and MCR regions in control and were severely depleted by recovery day 3 (Figure 3, E and F). As predicted by body weight change and gene expression analysis, rare Clara cells are detected in the ICR and MCR on recovery day 6 (Figure 3, G and H) and consistently detected in both regions on recovery days 9 (Figure 3, I and J) and 13 (Figure 3, K and L). These studies demonstrated that body weight loss on recovery days 1–3 was predictive of decreases in Clara-like cell-specific mRNA abundance and Clara-like cell number.
Cellular Specificity of Naphthalene Toxicity in the Tracheal Epithelium
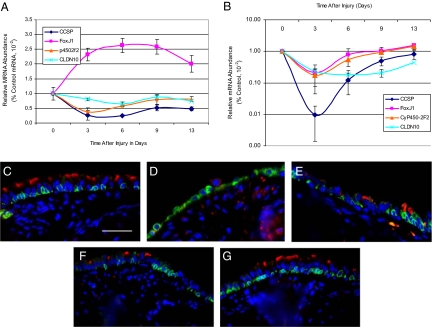
Previous analysis of naphthalene toxicity in the bronchiolar epithelium demonstrated that the Clara cell was the cellular target for this xenobiotic agent and that Clara cell susceptibility was attributable to expression of CyP450-2F2.31 These results were confirmed by gene expression analysis of accessory lobe RNA from mice treated with 300 g/kg naphthalene (Figure 4A). In the lung, Clara cell markers CCSP, SCGB3A2, and CyP450-2F2 decreased to 25% of control levels on recovery day 3 and rebounded to 50% of control on recovery days 9 and 13. Abundance of the ciliated cell marker FoxJ132 was 230% of control on recovery days 3–13. Increased abundance of this transcription factor was likely due to enrichment of ciliated cells within the airway epithelium.33
Figure 4.
Naphthalene-mediated tracheal injury: Ciliated cell depletion. A and B: mRNA abundance in lung (A) and trachea (B) as a function of time after injury. Note the log scale in B. CCSP, SCGB3A2, CyP450-2F2 (p4502F2), and Clauden 10 (CLDN10), FoxJ1. Mean ± SEM, n = 4. Values for days three, six, and nine were significantly different from day 0, P < 0.005. C–G: Dual immunofluorescence analysis of the ciliated cell marker acetylated tubulin (red), keratin 5 (green), and nuclei (blue). C, control; D, day three; E, day six; F, day nine; and G, day 13. Scale bar in C = 50 μm.
In contrast with the bronchiolar epithelium, FoxJ1 mRNA abundance in tracheal RNA decreased in response to naphthalene treatment (Figure 4B). FoxJ1 mRNA values were 20% of control on recovery day 3 and returned to 105 to 170% of control on recovery days 6–13. Depletion of tracheal ciliated cells was confirmed by dual immunofluorescence analysis of acetylated tubulin (ACT), a marker for apical cilia (Figure 4, C–G). In control mice, acetylated tubulin was detected in the ciliary bed of luminal cells located throughout the tracheal epithelium (Figure 4C). On recovery day 3, ACT+ cells were undetectable (Figure 4D) but on recovery days 6–13, ACT+ cells were present (Figure 4, E and F). Decrements in FoxJ1 gene expression and ACT+ cells indicated that tracheal ciliated cells were lost as a consequence of naphthalene exposure.
Alterations in the pattern of ciliated cell gene and marker expression in the trachea suggested that other cell type–specific markers might exhibit distinct expression patterns. Gene expression analysis demonstrated that CyP450-2F2 mRNA abundance followed the FoxJ1 pattern after naphthalene injury (Figure 4B). Gene expression analysis also demonstrated that Claudin 10, a bronchiolar Clara cell–specific marker,34 was expressed in a pattern more similar to that of FoxJ1 than it was to mRNAs encoding secreted proteins (Figure 4, C versus B). These data indicate that the naphthalene injury process differs in the tracheal and bronchiolar compartments.
Changes in Tracheal Epithelial Cell Populations
Our previous analysis of tracheal and bronchial histological parameters was reported as cell number per unit length basement membrane.19,20 However, analysis of the smoke-injured tracheal epithelium35 demonstrated that basal cells change shape over time and that alterations in cell shape and size correlate with posttranslational modifications of type 1 keratins.36 To determine whether naphthalene treatment caused a similar effect, stereological methods27 were used to compare the basement membrane area in control and naphthalene-treated trachea (Figure 5A) and the volume fraction of nuclei as a function of basement membrane area (VV/SV) in these two groups (Figure 5B). Statistically significant differences were not detected for either parameter, indicating that basement membrane area and nuclear size did not vary between control and naphthalene injured trachea.
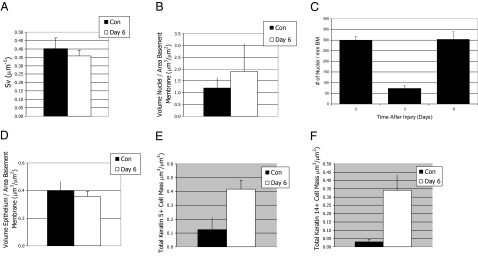
Figure 5.
Stereological comparison of control and repair day six. A and B: The area of the basement membrane (SV) (A) and the volume fraction of nuclei as a function of basement membrane area (VV/SV) (B) of control (black bars) and naphthalene recovery day 6 (white bars) was evaluated using stereological methods. Mean ± SEM, n = 4. Statistically significant differences were not detected for either parameter. C: Cell density change as a function of time after injury. Measurement is the number of nuclei per unit length basement membrane. Mean ± SEM, n = 4. *P < 0.005 relative to control. D: Mass of the epithelium (VS) in control (black bar) and in naphthalene recovery day 6 (white bar). Mean ± SD, n = 4. Differences are not significant. E and F: The mass (VV/SV) of keratin 5+ cells (E) and the keratin 14+ subset (F) was determined in control (black bars) and naphthalene recovery day 6 (white bars). Mean ± SD, n = 4. The mass of keratin 5+ cells increased fourfold and was significantly different from control, *P = 0.02. The mass of the keratin 14+ subset increased 14-fold and was significantly different from control, *P = 0.007.
Analysis of cell number as a function of length of basement membrane (Figure 5C) detected a rapid decrease in total cell density on recovery day 3 (25% of control cell density) and a return to normal cell density on recovery day 6. Total cell density in nuclei per mm basement membrane was 299.23 ± 18.41 in control mice. After injury, cell density decreased to 73.32 ± 13.55 nuclei per mm basement membrane on day 3 and increased to 302.64 ± 35.98 on day 6. Cell density on recovery day 3 was less than control and recovery day 6, P = 0.05. However, cell density on recovery day 6 was not different from control. Stereological methods were used to compare epithelial volume in control and recovery day 6 (Figure 5D) and did not detect a difference in this parameter. This result suggested that injury altered the epithelial cell type without changing epithelial volume.
In contrast with the cell density and epithelial volume analysis, histological analysis suggested that Clara cells (Figure 3, C–L) and ciliated cells (Figure 4, C–G) were hypoplastic on recovery day 6. To determine the cellular mechanism leading to restoration of epithelial cell number and volume without replacement of Clara or ciliated cells, the frequency of keratin 5+ and 14+ cells was evaluated by dual immunofluorescence. This analysis suggested normalization of cell density was due to basal cell hyperplasia (compare Figure 3, C and G or D and H, Figure 4, C and E).
Stereological methods were used to compare the total cell mass of keratin 5+ basal cells (Figure 5E) and the keratin 14+ subset (Figure 5F) in control and on recovery day 6. The mass of keratin 5+ cells increased fourfold after injury and was significantly different from control (P = 0.02). The mass of the keratin 14+ subset increased 14-fold and was also significantly different from control (P = 0.007). This analysis demonstrated the type of cell populating the tracheal epithelium varied between control and recovery day 6 and that normalization of cell density was due to hyperplasia of basal cells.
The Reparative Basal Cell Phenotype
Histological analysis demonstrated that keratin 14+ basal cells were located in both the ICR and MCR (Figure 6, A–J). On recovery day 3 all basal cells were keratin 14+ and exhibited a low cuboidal to squamous morphology (Figure 6, C and D). On recovery day 6, the reparative basal cells assumed a more normal pyramidal morphology (Figure 6, E and F) and this morphology persisted through recovery days 9 and 13 (Figure 6, G–J). The majority of keratin 14+ cells coexpressed keratin 5 although a population of keratin 5-/14+ cells was detected (Figure 6, E and F, arrows). A subset of basal cells exhibited a columnar morphology and was located in pseudostratified subregions of the epithelium. These regions were found primarily within the ICR, and basal cells in these regions were keratin 14+/5+, keratin 14+/5−, or expressed keratin 14 alone. On recovery days 9 and 13, the ratio of keratin 5 and keratin 14 protein increased resulting in the appearance of basal cells that were predominantly keratin 5high and keratin 14low (Figure 6, G–J). However, the number of keratin 14+/5+ positive cells remained elevated relative to control (compare Figure 6, A and I or Figures 6B and 5J).
Figure 6.
Basal cell response to naphthalene-mediated tracheal injury. A–J: Dual immunofluorescence analysis of keratin 5 (red) and keratin 14 (green) in the intercartilagenous (A, C, E, G, I) and midcartilagenous (B, D, F, H, J) regions. Arrows in E and F indicate cells that expressed only keratin 14. K: mRNA abundance as a function of time after injury. Note log scale in K. Keratin (K) 5, 15, and 14. Mean ± SEM, n = 4. All keratin 14 values are significantly different from day 0, P < 0.0005. All keratin 5 and 15 values are significantly different from day 0, P < 0.05. L: Morphometric analysis of keratin 5+ (red) and keratin 14+ (blue) cell distribution as a function of position along the proximal to distal axis of the ventral trachea. Analysis of four mice is shown. M–N: Stereological methods were used to determine the mass of keratin 5+ cells (M) and keratin 14+ cells (N) in the intercartilaginous (ICR) and midcartilaginous (MCR) regions of the control (white bars) and naphthalene recovery day 6 trachea (black bar). Mean ± SD, n = 4. The mass of keratin 5+ cells in the ICR (P = 0.03) and MCR (P = 0.06) was statistically significant. The mass of keratin 14+ cells in the ICR was significant (P = 0.002). Scale bar in A = 50 μm.
Quantitative RT-PCR analysis was used to evaluate alterations in expression of basal cell keratins. Abundance of keratins 5 and 15 increased two- to threefold as a function of recovery time (Figure 6K). In contrast, expression of keratin 14 increased 60-fold by recovery day 3 and persisted at high levels through recovery day 6. A trend toward decreased keratin 14 mRNA abundance was noted on recovery days 9 and 13, but differences were not statistically significant. These data indicated that keratin 14 gene and protein expression was differentially regulated from that of keratins 5 and 15. Parallel results using qRT-PCR and immunofluorescence analysis suggest that naphthalene-injury initiated a change in basal cell phenotype that is most readily recognized by an increase in keratin 14 gene expression and protein abundance.
Spatial Distribution of Reparative Basal Cells
Previous analysis suggested that a subset of keratin 14+ cells were a multipotential stem-like cell20 and that label-retaining basal cells were located within the tracheal gland duct junction in mice.21 To determine whether the reparative basal cells identified in this study were spatially restricted along the proximal to distal axis of the trachea, keratin 5+ and keratin 14+ cell density was determined as a function of position on recovery day 6 (Figure 6L). This analysis demonstrated that reparative keratin 14+ cells were not restricted to the proximal region on recovery day 6.
Because axial differences in basal cell distribution were not identified (Figure 6L), we determined whether keratin 5+ cells and the keratin 14+ subset were localized to the ICR, the subzone of the trachea that was previously defined as stem cell microenvironment.21 Keratin 5+ and keratin 14+ basal cell mass in the ICR and MCR regions was compared in control and on postnaphthalene recovery day 6. The mass of keratin 5+ cells in the ICR increased ∼2-fold after injury and differences were statistically significant (P = 0.03, Figure 6M). Similarly, the mass of keratin 5+ in the MCR increased twofold and differences were statistically significant (P = 0.06, Figure 6M). The mass of keratin 14+ cells increased 10-fold in both the ICR and the MCR (Figure 6N). However, only the differences in the ICR were significant (P = 0.002). This study indicates that basal cell hyperplasia occurred in ICR and the MCR located throughout the trachea.
Mitotic Index of Reparative Basal Cells
Cellular mechanisms leading to basal cell hyperplasia were investigated through analysis of mitotic index. After naphthalene-mediated injury, the overall epithelial mitotic index increased 10-fold, from 2.78% ± 0.06% to 28.05% ± 4.10%, and remained at this high level through recovery day 6 (28.65% ± 0.14%, Figure 2C). On recovery days 3 and 6, basal cells (keratin 5+) and the keratin 14+ subset were approximately 80% of the mitotic pool (Figure 2D), and contribution of these cells to the mitotic pool was indistinguishable (Figure 2E). On recovery days 3 and 6, 30 to 45% of keratin 5+ or keratin 14+ cells were cycling. These data indicated that the increase in mitotic index was the result of basal cell proliferation.
Restitution of Secretory and Ciliated Cells
Previous lineage tracing studies demonstrated a precursor-progeny relationship between tracheal basal cells and the Clara and ciliated cell populations.19,20,37 Dual immunofluorescence was used to compare the distribution of reparative basal cells and nascent secretory or ciliated cells. CCSP immunoreactive secretory cells were first detected on recovery day 6 (Figure 3, G and H). These cells were located in either the ICR or MCR regions but were more prevalent in junctions between these two zones. The number of secretory cells increased dramatically between recovery days 6 and 13 (Figure 3, G–L), resulting in replacement of this cell type throughout the proximal to distal axis of the trachea. The pattern of ciliated cell replacement was similar to that of secretory cells (Figure 4, C–G).
Differentiation of Reparative Basal Cells
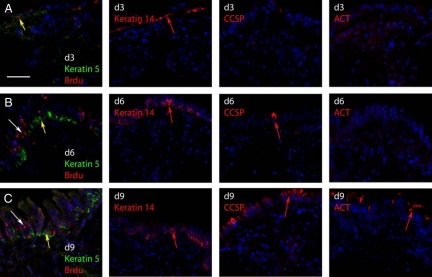
Lineage tracing of mouse tracheal or bronchial keratin 14+ cells demonstrated that cells tagged on postnaphthalene days 2–4 were progenitors for Clara and ciliated cells.19,20 An increase in clone size suggested that tagged cells proliferated as part of the repair process. To determine whether basal cells underwent cytokinesis as part of the Clara-like and ciliated cell replacement process, cells in S-phase of the cell cycle were labeled with BrdU on recovery day 3. At this time point, numerous BrdU+ cells were detected (Figure 7A). These cells were situated on the basement membrane and coexpressed keratin 5. Adjacent serial section analysis indicated that these cells were keratin 14+ (Figure 7A, day 3 keratin 14). No CCSP+ or ACT+ cells were detected at on day 3 (Figure 7A, day 3 CCSP and day 3 ACT). These data confirmed the Ki-67 analysis of proliferation (Figure 2) and demonstrated active DNA synthesis.
Figure 7.
Pulse-chase analysis of basal cell proliferation. A–C: Dual immunofluorescence analysis of BrdU and keratin 5 in naphthalene-injured tracheas that were pulsed with BrdU on recovery day three and killed three hours later (day three) or after a three- (day six) or a six-day chase (day nine). Yellow arrows indicate BrdU+/keratin 5+ dual positive cells. White arrows indicate BrdU+ cells that are keratin 5−. Adjacent serial sections to those presented in A–C are labeled d3, d6, and d9, respectively. These sections were stained for keratin 14, CCSP, or ACT. Examples of antigen-positive cells are indicated by red arrows. Scale bar in A = 50 μm.
A pulse-chase study was used to determine whether cells that synthesized DNA on recovery day 3 generated keratin 5− daughter cells.10 On recovery day 6 (chase day 3) numerous keratin 5+/BrdU+ cells were detected (Figure 7B, yellow arrow). In addition, columnar keratin 5-/BrdU+ cells (Figure 7B, white arrow) were identified. These data indicate that keratin 5+ basal cells labeled on recovery day completed cytokinesis by recovery day 6 and that they passed the BrdU label on to a keratin 5− cell type. Adjacent serial section analysis indicated that the keratin 5−/BrdU+ cells were also keratin 14− (day 6 keratin 14), CCSP− (day 6 CCSP), and ACT− (day 6 ACT). On recovery day 9 (chase day 6), a subset of BrdU+ cells were keratin 5+ and were located along the basement membrane (Figure 7C). In addition, numerous BrdU+ cells were located in the luminal portion of the regenerating epithelium. These cells were keratin 5−. Adjacent serial sections demonstrated that keratin 14+ cells were not located in the luminal region suggesting that the luminal daughter cells were not basal cells. The presence of numerous CCSP+ and ACT+ cells in the luminal region (d9 CCSP and d9 ACT) indicated that the label had been chased into the Clara-like and ciliated cell types. These studies demonstrate that keratin 5+/14+ cells were the precursor to keratin 5−/14− cells that were likely to be Clara-like and ciliated cells.
Discussion
Summary
This study details the cellular mechanisms that regulate tracheal epithelial maintenance and repair. In the steady state, two types of basal cells contribute to epithelial homeostasis: keratin 5+/15+ double positive and keratin 14+/5+/15+ triple positive cells. These two basal cell subtypes are distributed throughout the proximal to distal axis of the trachea and have a similar mitotic index. Both basal cell subsets up-regulate keratin 14 mRNA and protein in response to naphthalene-mediated secretory and ciliated cell depletion. These cells are highly mitotic and are distributed throughout the proximal to distal axis of the trachea. Nascent Clara and ciliated cells are detected on recovery days 6–13. BrdU pulse-chase analysis demonstrated that differentiated cells were derived from basal cells that proliferated on recovery day 3. The distribution of these differentiated cell types follows the unbiased distribution of naphthalene-resistant basal cell population. We conclude that the majority of naphthalene-resistant basal cell progenitors function as a facultative progenitor cell and that this abundant cell is a previously unrecognized member of the tracheal stem cell hierarchy.
Unique Characteristics of Naphthalene Injury in the Trachea
The naphthalene model has been used to evaluate precursor/progeny relationships in the conducting airway epithelium (reviewed in reference15). Within the simple bronchiolar epithelium, naphthalene targets the Clara cell resulting in cell death. Exposure to high doses of naphthalene activates rare naphthalene-resistant variant CCSP expressing cells (vCE) that are spatially restricted to the neuroepithelial body and to the bronchoalveolar duct junction microenvironments. In contrast, we now demonstrate that naphthalene exposure results in depletion of both the tracheal Clara-like cell and tracheal ciliated cells. Ciliated cell depletion is likely due to loss of contacts with Clara-like cells, although naphthalene metabolism in these cells or transfer of toxic epoxies has not been eliminated as a possible mechanism. These differences in the cellular pattern of naphthalene injury in the trachea and bronchiolar epithelium should be considered when using this model to evaluate the presence and function of tissue stem and progenitor cells.
Other agents that are commonly used to evaluate tracheal reparative cells include sulfuric acid and detergent exposure.21,38 These models are relevant to human environmental and occupational exposures. However, damage to the surface epithelium is more extensive than that observed with naphthalene injury. Importantly, published analysis indicates that acid or detergent exposure results in depletion of most basal cells, including the uniformly distributed reparative keratin 14+ cell characterized in this study. Further, the acid and detergent models may deplete or compromise the multipotential basal cell identified by lineage tracing analysis. As a consequence of differences in the extent of injury and the spatial location of surviving progenitor cell pools, great care must be taken in comparing the results of studies using these disparate models.
Tracheal Basal Cell Subsets
Previous analysis of tracheal and bronchial basal cell function used keratins 5 and 14 as interchangeable markers for this cell type.19,20,21,39,40 The assumption that keratin 14 partnered with keratin 5 to form intermediate filaments was based on analysis of basal cells in the mammary gland, prostate, epidermis, and esophagus.17 In glands and cornified epithelia, all basal cell progenitors coexpress keratins 14 and 5. Interestingly, myoepithelial cells found in tracheal submucosal glands share this phenotype (data not shown). However, the present study demonstrates that 80% of steady-state tracheal basal cells, in the FVB/n strain, are keratin 14–negative. Thus, a high proportion of tracheal cells are molecularly distinct from progenitors of stratified epithelia and glands.
In the epidermis, high level keratin 14 expression has been equated with proliferation. As daughter cells differentiate keratin 14 protein levels decrease.41 The present study demonstrated that the mitotic index of steady-state keratin 14− basal cells was equivalent to that of keratin 14+ cells (Figure 2E). These data indicate that expression of keratin 14 is not a prerequisite for proliferation of steady-state tracheal basal cells. However, the mitotic index of the normal trachea (Figure 2C) is very low relative to the epidermis. Demonstration that keratin 14 mRNA and protein levels increase within 3 days of injury (Figure 6, A–K) and that this increase was associated with a 10-fold increase in basal cell proliferation (Figure 2) suggests that upregulation of keratin 14 may play a role in generation of a highly mitotic reparative cell pool. Mechanisms underlying this potential function for keratin 14 are under investigation.
It is currently unknown whether steady-state keratin 14 expression defines independent classes of tracheal basal cells or represents phenotypic transitions of a plastic cell type. However, the finding that most keratin 5+ cells upregulate expression of keratin 14 after injury suggests that the keratin 14+ reparative cell type is a derivative of the steady-state keratin 5+/14− cell. Lineage tracing analysis of steady-state keratin 14+ cells (S.D.R., manuscript in preparation) supports the notion that keratin 14− and keratin 14+ cells are metaplastic variants of a single cell type.
The present study in combination with lineage tracing analysis also suggests that some of these metaplastic cells do not survive the repair process. Lineage tracing identified a bipotential keratin 14+ reparative cell that failed to self-renew but generated secretory/ciliated cell-only clones.19 The present analysis suggests that this bipotential cell was derived from a steady-state keratin 14− cell and that this tagged cell produced two differentiated daughters. Likely explanations for loss of the tagged keratin 14+ cell include metaplasia of one daughter to a Clara cell or a ciliated cell or apoptosis of the initial tagged cell.
Squamous metaplastic lesions of the human bronchial epithelium are characterized by expression of keratin 14 and cornified epithelial markers including keratins 1 and 10, loricrin, and involucrin. In contrast, upregulation of keratin 14 after naphthalene injury preceded expression of Clara-like and ciliated cell–specific genes and replacement of these cell types. Histological analysis of tracheal tissue on post naphthalene injury days 2–13 failed to detect expression of cornified envelope markers (data not shown). These data suggest that the association between expression of keratin 14 and cornified epithelial markers in squamous metaplastic lesions of the human lung reflects a modification of signals that regulate normal repair.5,42,43
Functional Significance of Keratin 14 Upregulation
Posttranslational modification of keratins has been implicated in signaling processes that regulate cell growth and hypertrophy.36 Although this analysis has not included so called “hard keratins” such as keratin 14, increased expression of keratin 14 in tracheal basal cells may indicate a role for this type 1 keratin in regulation of cell shape and size. Variation in tracheal keratin 14 expressing cell morphology was suggested by histological analysis (Figure 6, A–J) and stereological analysis detected differences in basal cell mass on recovery day 6 (Figure 5, E and F). However, increased proliferation between control and recovery day 6 (Figure 2, E–G) was sufficient to account for changes in basal cell mass. Consequently, there is little evidence that the increase in keratin 14 mRNA and protein drives basal cell hypertrophy on recovery day 6.
Keratin 14 containing intermediate filaments are thought to direct asymmetric cell division and establishment of distinct cell subsets that retain or lose contact with the basement membrane.44 Consistent with this notion was the finding that basal cell shape varies as a function of time after injury and that a subset of reparative keratin 14+ cells exhibits a columnar morphology (Figures 6E, arrow). However, most columnar derivatives of BrdU-labeled basal cells were keratin 14− (Figure 7, B and C). This result suggests that the keratin profile shifts rapidly as keratin 14+ reparative cells differentiate into Clara and ciliated cells. Demonstration that cells in contact with the basement membrane retain keratin 14 expression suggests that this intermediate filament protein plays a greater role in basal cell–matrix interactions than in regulation of mitotic spindle orientation.
Previous morphometric analysis demonstrated a positive correlation between basal cell number and epithelial height.45 These studies defined the basal cell as an “anchor” for the pseudostratified epithelium. After injury, tight adhesion of basal cells to the basement membrane may be critical for receipt of signals that initiate the reparative response.46 BrdU pulse-chase analysis demonstrated that BrdU-labeled basal cells produced some daughters that did not express keratin 14 or Clara-like and ciliated cell differentiation markers on recovery day 6 (Figure 7, B and C). Analysis at later time points demonstrated that replacement of Clara-like and ciliated cells involved nascent cells that were the product of basal cell proliferation. However, it is unknown whether the replacement process involves metaplasia of basal cell daughters to Clara-like or ciliated cells or multiple rounds of basal cell proliferation and differentiation. Rapid restoration of ciliated cell gene expression and detection of ACT+ ciliated cells suggests that direct basal to ciliated cell differentiation may occur in the mouse as was previously suggested for human.37 This observation suggests that differentiation of keratin 14+ cells is regulated by short-range signals that impinged on a single cell. Misregulation of these interactions may lead to the basal cell hyperplasia that is characteristic of many chronic lung diseases3,4 and to inappropriate differentiation characterized by squamous metaplasia.5
Tracheal Facultative Progenitor Cells
We demonstrated that naphthalene-mediated depletion of tracheal Clara-like and ciliated cells initiated an epithelial repair process that was driven by an abundant and broadly distributed basal cell population. BrdU pulse-chase analysis demonstrated that basal cells were in S-phase of the cell cycle on naphthalene recovery day 3 and that daughters of these mitotic basal cells contributed to the luminal keratin 14− cell population on recovery days 6 and 9. Thus, keratin 14+ reparative cells can be termed progenitor cells. These progenitor cells were at least 80% of mitotic cells on recovery days 3 and 6. Reparative keratin 14+ cells were highly proliferative with approximately 40% of cells cycling at a given time point (Figure 2).
The unbiased distribution of keratin 14+ reparative cells along the proximal distal axis (Figure 6L) and parallel restitution of the secretory and ciliated cell populations demonstrated that epithelial repair after naphthalene injury was mediated by a broadly distributed population of basal cell progenitors rather than through exclusive activation of a proximally-restricted tissue specific stem cell. Because the reparative keratin 14+ basal cells were uniformly distributed, we suggest that reparative keratin 14+ cells are not transit amplifying cells. Rather, tracheal basal cells exhibit differentiated features when in the quiescent state yet had the capacity to proliferate for normal tissue maintenance and in response to injury. Thus, we propose that majority of tracheal basal cells functions as a facultative progenitor.15
Hierarchical Organization of Tracheal Progenitor Cells
Demonstration that keratin 14 expression is a gain-of-function phenotype and that the majority of keratin 14–expressing cells are facultative progenitor cells may be viewed as being in conflict with studies that identified tracheal and bronchial tissue–specific stem cells. These studies demonstrated spatially-restricted label-retaining stem-like cells in the acid and detergent injury models21 and identified multipotential basal cells using clonal analysis,37 FACS sorting,40,47 and lineage tracing.19,20 Our study does not challenge the concept of a tracheal tissue-specific stem cell. Rather, our study indicates that markers other than keratin 14 are needed to identify the subset of keratin 14+ cells that exhibit tissue specific stem cell characteristics.
Based on this study and previously published thymidine pulse-chase studies we suggest that the tracheal stem cell hierarchy be modified to include two facultative progenitor cell types, the Clara-like cell and the basal cell. Previous analysis demonstrated that the Clara cell facultative progenitor is activated by oxidant gas-induced ciliated cell injury.10 In contrast, this study demonstrates that the basal facultative progenitor is activated by Clara-like and ciliated cell depletion.
The presence of two facultative progenitor cell pools distinguishes the tracheal epithelium from other epithelia. The bronchiolar epithelium is populated by a single facultative progenitor pool, the Clara cell. Similarly, the liver contains a single facultative progenitor cell, the hepatocyte. The distinctive cellular properties of the tracheal hierarchy and the potential for differential activation of its facultative progenitor pools highlight the need to define molecular mechanisms regulating cellular interactions in this region of the respiratory tree in health and disease.
Acknowledgments
We thank Douglas Hicks for animal husbandry and genotyping, Bilan Li for immunostaining and microscropy, Kaili Weil (Carl Zeiss MicroImaging, Inc.) for microscopy training, and Jordan Moore (Fluidigm, Inc.) for high throughput qPCR analyses. Drs. Dennis R. Roop and Maranke Koster (University of Colorado-Denver) provided valuable reagents and advice for analysis of keratinization. Dr. Rubin Tuder provided valuable assistance with the stereological analysis.
Footnotes
Address reprint requests to Susan D. Reynolds, Ph.D., Goodman Building, K1007, 1400 Jackson Street, Denver, CO 80206. E-mail: reynoldss@njhealth.org.
Supported by Award Number R01HL075585 from the National Heart, Lung, and Blood Institute, a Cystic Fibrosis Foundation Research grant, and a Cystic Fibrosis Foundation Pilot Project grant. Some data were generated at the University of Pittsburgh, Department of Environmental and Occupational Health, where K.M.J. was supported by a Summer Undergraduate Research Project (SURP) Fellowship.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current address for B.B.C.: University of Oregon Health Sciences Center, Portland, OR.
References
- Puchelle E, Zahm JM, Tournier JM, Coraux C. Airway epithelial repair, regeneration, and remodeling after injury in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:726–733. doi: 10.1513/pats.200605-126SF. [DOI] [PubMed] [Google Scholar]
- Iyonaga K, Miyajima M, Suga M, Saita N, Ando M. Alterations in cytokeratin expression by the alveolar lining epithelial cells in lung tissues from patients with idiopathic pulmonary fibrosis. J Pathol. 1997;182:217–224. doi: 10.1002/(SICI)1096-9896(199706)182:2<217::AID-PATH833>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Voynow JA, Fischer BM, Roberts BC, Proia AD. Basal-like cells constitute the proliferating cell population in cystic fibrosis airways. Am J Respir Crit Care Med. 2005;172:1013–1018. doi: 10.1164/rccm.200410-1398OC. [DOI] [PubMed] [Google Scholar]
- Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, Cancellieri A, Maestro R, Semenzato G, Doglioni C. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya J, Cambier S, Markovics JA, Wolters P, Jablons D, Hill A, Finkbeiner W, Jones K, Broaddus VC, Sheppard D, Barzcak A, Xiao Y, Erle DJ, Nishimura SL. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest. 2007;117:3551–3562. doi: 10.1172/JCI32526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. Cellular and molecular characteristics of basal cells in airway epithelium. Exp Lung Res. 2001;27:401–415. doi: 10.1080/019021401300317125. [DOI] [PubMed] [Google Scholar]
- Reynolds SD, Reynolds PR, Pryhuber GS, Finder JD, Stripp BR. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am J Respir Crit Care Med. 2002;166:1498–1509. doi: 10.1164/rccm.200204-285OC. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Reynolds SD. Clara Cells. Shapiro S, Laurent G, editors. St. Louis MO,: Elsevier,; 2006:471–477. [Google Scholar]
- Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol. 2007;69:401–422. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Shami SG, Cabral-Anderson LJ, Dekker NP. Role of nonciliated cells in renewal of the bronchial epithelium of rats exposed to NO2. Am J Pathol. 1986;123:126–133. [PMC free article] [PubMed] [Google Scholar]
- Fanucchi MV, Murphy ME, Buckpitt AR, Philpot RM, Plopper CG. Pulmonary cytochrome P450 monooxygenase and Clara cell differentiation in mice. Am J Respir Cell Mol Biol. 1997;17:302–314. doi: 10.1165/ajrcmb.17.3.2774. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Maxson K, Mera R, Singh G. Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am J Physiol. 1995;269:L791–L799. doi: 10.1152/ajplung.1995.269.6.L791. [DOI] [PubMed] [Google Scholar]
- Van Winkle LS, Buckpitt AR, Nishio SJ, Isaac JM, Plopper CG. Cellular response in naphthalene-induced Clara cell injury and bronchiolar epithelial repair in mice. Am J Physiol. 1995;269:L800–L818. doi: 10.1152/ajplung.1995.269.6.L800. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, Stripp BR. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol. 2001;24:671–681. doi: 10.1165/ajrcmb.24.6.4498. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Reynolds SD. Maintenance and repair of the bronchiolar epithelium. Proc Am Thorac Soc. 2008;5:328–333. doi: 10.1513/pats.200711-167DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E. Functional differences between keratins of stratified and simple epithelia. J Cell Biol. 1998;143:487–499. doi: 10.1083/jcb.143.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Coulombe PA. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007;21:1581–1597. doi: 10.1101/gad.1552107. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: Evidence for multipotent and unipotent subpopulations, Am J Physiol Lung Cell Mol Physiol. 2004;286:L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol. 2001;24:662–670. doi: 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol. 2002;161:173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SD, Giangreco A, Power JH, Stripp BR. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol. 2000;156:269–278. doi: 10.1016/S0002-9440(10)64727-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SD, Zemke AC, Giangreco A, Brockway BL, Teisanu RM, Drake JA, Mariani T, Di PY, Taketo MM, Stripp BR. Conditional stabilization of beta-catenin expands the pool of lung stem cells. Stem Cells. 2008;26:1337–1346. doi: 10.1634/stemcells.2008-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde DM, Plopper CG, Kass PH, Alley JL. Estimation of cell numbers and volumes of bronchiolar epithelium during rabbit lung maturation. Am J Anat. 1983;167:359–370. doi: 10.1002/aja.1001670306. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Reynolds SD, Boe IM, Lund J, Power JH, Coppens JT, Wong V, Reynolds PR, Plopper CG. Clara cell secretory protein deficiency alters clara cell secretory apparatus and the protein composition of airway lining fluid. Am J Respir Cell Mol Biol. 2002;27:170–178. doi: 10.1165/ajrcmb.27.2.200200270c. [DOI] [PubMed] [Google Scholar]
- Hyde DM, Tyler NK, Plopper CG. Morphometry of the respiratory tract: avoiding the sampling, size, orientation, and reference traps. Toxicol Pathol. 2007;35:41–48. doi: 10.1080/01926230601059977. [DOI] [PubMed] [Google Scholar]
- Van Winkle LS, Fanucchi MV, Miller LA, Baker GL, Gershwin LJ, Schelegle ES, Hyde DM, Evans MJ, Plopper CG. Epithelial cell distribution and abundance in rhesus monkey airways during postnatal lung growth and development, J Appl Physiol. 2004;97:2355–2363. doi: 10.1152/japplphysiol.00470.2004. discussion 2354. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Buckpitt A, Chang AM, Weir A, Van Winkle L, Duan X, Philpot R, Plopper C. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. IV Metabolism of naphthalene and naphthalene oxide in microdissected airways from mice, rats, and hamsters. Mol Pharmacol. 1995;47:74–81. [PubMed] [Google Scholar]
- You Y, Huang T, Richer EJ, Schmidt JE, Zabner J, Borok Z, Brody SL. Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2004;286:L650–L657. doi: 10.1152/ajplung.00170.2003. [DOI] [PubMed] [Google Scholar]
- Zemke AC, Teisanu RM, Giangreco A, Drake JA, Brockway BL, Reynolds SD, Stripp BR. {beta}-Catenin is not necessary for maintenance or repair of the bronchiolar epithelium. Am J Respir Cell Mol Biol. 2009;41:535–543. doi: 10.1165/rcmb.2008-0407OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemke AC, Snyder JC, Brockway BL, Drake JA, Reynolds SD, Kaminski N, Stripp BR. Molecular staging of epithlieial maturation using secretory cell-specific genes as markers. Am J Respir Cell Mol Biol. 2008;40:340–348. doi: 10.1165/rcmb.2007-0380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CZ, Evans MJ, Cox RA, Burke AS, Zhu Q, Herndon DN, Barrow RE. Morphologic changes in basal cells during repair of tracheal epithelium. Am J Pathol. 1992;141:753–759. [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Schlossberg H, Yankaskas JR, Dudus L. Progenitor cells of the adult human airway involved in submucosal gland development. Development. 1995;121:2031–2046. doi: 10.1242/dev.121.7.2031. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Hogan BL. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am J Physiol Lung Cell Mol Physiol. 2008;295:L231–L234. doi: 10.1152/ajplung.90209.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch KG, Lori A, Burns KA, Eldred T, Olsen JC, Randell SH. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol. 2004;286:L631–L642. doi: 10.1152/ajplung.00112.2003. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium, Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- Liu M, Yang SC, Sharma S, Luo J, Cui X, Peebles KA, Huang M, Sato M, Ramirez RD, Shay JW, Minna JD, Dubinett SM. EGFR signaling is required for TGF-beta 1 mediated COX-2 induction in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2007;37:578–588. doi: 10.1165/rcmb.2007-0100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MB, Ramirez RD, Wright WE, Minna JD, Shay JW. A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006;74:141–148. doi: 10.1111/j.1432-0436.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. p63: Revving up epithelial stem-cell potential. Nat Cell Biol. 2007;9:731–733. doi: 10.1038/ncb0707-731. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Cox RA, Shami SG, Wilson B, Plopper CG. The role of basal cells in attachment of columnar cells to the basal lamina of the trachea. Am J Respir Cell Mol Biol. 1989;1:463–469. doi: 10.1165/ajrcmb/1.6.463. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Van Winkle LS, Fanucchi MV, Baker GL, Murphy AE, Nishio SJ, Schelegle ES, Gershwin LJ, Sannes PL, Plopper CG. Fibroblast growth factor-2 in remodeling of the developing basement membrane zone in the trachea of infant rhesus monkeys sensitized and challenged with allergen. Lab Invest. 2002;82:1747–1754. doi: 10.1097/01.lab.0000043911.94235.f3. [DOI] [PubMed] [Google Scholar]
- Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25:139–148. doi: 10.1634/stemcells.2006-0288. [DOI] [PubMed] [Google Scholar]