Abstract
Most non-small cell lung cancer (NSCLC) patients harboring activating epidermal growth factor receptor (EGFR) mutations respond to tyrosine kinase inhibitor (TKI) therapy. However, about 30% exhibit primary resistance to EGFR TKI therapy. Here we report that Met protein expression and phosphorylation were associated with primary resistance to EGFR TKI therapy in NSCLC patients harboring EGFR mutations, implicating Met as a de novo mechanism of resistance. In a separate patient cohort, Met expression and phosphorylation were also associated with development of NSCLC brain metastasis and were selectively enriched in brain metastases relative to paired primary lung tumors. A similar metastasis-specific activation of Met occurred in vitro in the isogenous cell lines H2073 and H1993, which are derived from the primary lung tumor and a metastasis, respectively, from the same patient. We conclude that Met activation is found in NSCLC before EGFR-targeted therapy and is associated with both primary resistance to EGFR inhibitor therapy and with the development of metastases. If confirmed in larger cohorts, our analysis suggests that patient tumors harboring both Met activation and EGFR mutation could potentially benefit from early intervention with a combination of EGFR and Met inhibitors.
Activating mutations in EGFR (primarily del19 EGFR and L858R) are associated with sensitivity to epidermal growth factor receptor (EGFR) small molecule tyrosine kinase inhibitor therapy, but patients uniformly develop disease recurrence.1,2,3,4,5,6 In addition, about 30% of patients with EGFR sensitizing mutations show primary resistance to EGFR inhibitor therapy.1,2,3,4,5,6,7 While disease recurrence in formerly responsive patients has been associated with an EGFRT790M mutation,8 MET amplification,9,10 or hepatocyte growth factor (HGF) expression,11 the factors involved in de novo resistance remain unidentified. Sequist et al reported primary resistance to EGFR tyrosine kinase inhibitor (TKI) in a patient harboring Met copy gain, but analysis of larger cohorts has not been reported and Met protein expression and activation were not tested.12 We examined a cohort of EGFR mutant non-small cell lung cancer (NSCLC) patients before treatment with EGFR TKI (designated cohort 1), and we report here that Met protein expression and phosphorylation were found in a subset of tumors before EGFR TKI therapy. Importantly, the subset harboring Met expression and phosphorylation was associated with poor response to subsequent EGFR TKI therapy despite the presence of EGFR inhibitor sensitizing mutations.
In addition to primary resistance, poor outcome to EGFR targeted therapy in NSCLC can result from development of metastases, especially to the central nervous system. Since HGF/Met signaling is uniquely positioned to be a key factor in cell migration and tumor dissemination,13,14 we compared Met status in a separate cohort of NSCLC patients (designated cohort 2) with paired brain metastases. Met expression has been correlated with both development of metastases and poor prognosis in some tumor types,13 but no studies have demonstrated Met activation in metastatic lung cancer. In this study we found that Met expression and phosphorylation in primary NSCLC tumors were strongly associated with subsequent development of brain metastases. Furthermore, we showed an enrichment of cells positive for Met expression and phosphorylation in brain lesions compared with matched primary lung tumors.
Materials and Methods
Cell Lines and Reagents
Cell lines were from American Type Culture Collection (ATCC) except: EBC-1 (Health Science Research Resources Bank, Japan Health Sciences Foundation) and H1993 and H2073 (from J. Minna and A. Gazdar, Hamon Cancer Center, University of Texas Southwestern). Cells were maintained in RPMI plus 10% fetal calf serum and 100 μg/ml Pennicillin/Streptavidin (Sigma, St. Louis, MO). SU1127415 was from Sigma.
Clinical Samples
Cohort 1: EGFR Kinase Inhibitor–Treated NSCLC Cohort
Sixty-nine patients with a diagnosis of lung adenocarcinoma were identified who had undergone EGFR mutation analysis between 2003 and 2007 and who had archived formalin-fixed paraffin-embedded (FFPE) samples at Brigham and Women’s Hospital Department of Pathology. Of these, 46 were excluded because they either did not receive TKI therapy or did not have available follow-up data. Of the remaining 23, all were women, 11 were nonsmokers, and 10 were smokers; smoking status was not available for two patients. Patient response was scored according to Response Evaluation Criteria in Solid Tumors (RECIST).16
Cohort 2: Primary NSCLC and Paired Brain Metastases Cohort
FFPE tumor samples were from a subset of a previously described collection of primary NSCLC diagnosed between 1989 and 2003.17 Patients with adenocarcinoma or squamous cell carcinoma with metastases to the brain were matched for clinicopathological features with a control group of NSCLC patients who did not develop brain metastasis during six years of follow-up or until death. We excluded from the Control Group patients who developed metastases in a distant site different from brain, to have a homogeneous control group that did not develop metastases. We also excluded neuroendocrine tumors. We analyzed 40 patients and a total of 58 specimens, including 18 primary tumors (10 adenocarcinoma and 8 squamous cell carcinoma), with paired 18 brain metastases and 22 nonmetastatic tumors (14 adenocarcinoma and 8 squamous cell carcinoma).
Fluorescence in Situ Hybridization
The MET probe (BAC clone RP11-95i20,CHORI; http://bacpac.chori.org) was labeled with SpectrumOrange dUTP (Abbott Molecular Inc., Des Plaines, IL) and hybridized to full tissue sections along with the centromere probe (CEP7/D7Z1, SpectrumGreen, Abbott Molecular Inc., Des Plaines, IL), using standard conditions.18 A minimum of 100 cells were analyzed. MET amplification was defined as MET:CEP7 ratio greater than 2.2.19 Samples having MET:CEP7 ratio between 1 and 2.2 were further defined as either low copy gain (MET copy number from 3 to 5) or high copy gain (MET copy number >5).20,21 A score analogous to the H score for immunohistochemistry (IHC) was derived by multiplying the percentage of cells harboring MET abnormalities by the level of copy gain (low copy gain multiplied by 1, high copy gain multiplied by 2).
Immunohistochemistry on FFPE Sections
The following antibodies were used: anti-c-Met (Zymed-Invitrogen, clone3D4, diluted 1:100); anti-Y1234/35 Met (3077, Cell Signaling Technology, 1:50); anti-EGFR (Novocastra, clone EGFR.25, 1:50). Negative controls for the Y1234/35 Met antibody included multiple FFPE embedded cell lines with various well-described kinase amplification or activation as indicated in Supplemental Table S2. Negative control for Met was LNCAP cells.22 Positive controls for Met and Y1234/35 Met were Met-amplified GTL16 and nonamplified A549 cells ± HGF. The negative control for EGFR staining was MDAMB453 cells (negative for EGFR23), while HCC827 cells were a positive control.
Epitope retrieval used boiling 0.01 M citrate buffer, pH 6.0 for 15 minutes in microwave (anti-c-Met,) or in pressure cooker (anti-EGFR). For Y1234/35 Met, Ventana Benchmark autostainer and Rabbit Ultra-HRP were used. Detection was with ChromoMap kit (Ventana Molecular Discovery Systems, Tucson, AZ). Results of the IHC were reviewed independently by two pathologists (M.F. and L.R.C.), who were blinded as to the outcome of the tumors. Membrane stain intensity was scored from 0 to 3+ for total Met and EGFR: 0, absent or faint staining in less than 5% of cells; 1+, ≥5% tumor cells faint stain; 2+, tumor cells moderate stain; 3+, tumor cells strong staining. For Y1234/35 Met phosphospecific antibody membrane stain intensity was scored from 0 to 2+ as follows: 0, absent or faint staining in less than 5% of cells; 1+, tumor cells with weak staining; 2+, tumor cells with moderate-strong staining. The results of the two reviewers were compared, and any tumors for which there was not agreement were reviewed together by both pathologists and a consensus was reached. The H score for each sample was calculated by multiplying the stain intensity value by the percentage of positive cells. Images were acquired using a Leitz Diaplan microscope bright field and a CRI Nuance spectral analyzer (CRI Inc., Woburn, MA) and collected at 5-nm wavelength intervals from 450 to 700 nm.
DNA Extraction and Amplification
DNA was extracted from five 0.8-mm cores of each FFPE specimen with Qiagen BioRobot (Qiagen, Hilden, Germany). Whole genome amplification (WGA) was performed with GenomePlex (Sigma) as described.24
Somatic Mutation Profiling (Oncomap)
The set of somatic EGFR kinase domain activating mutations along with 15 MET mutations (E168D, L229F, R988C, T1010I, H1112Y, H1112R, N1118Y, L1213V, Y1248H, Y1248C, Y1253D, M1268T, splicing site IVS13del22, exon 14 splice site mutation, splicing site ex14del28)25 were tested as described.24
Cell Line Genotyping
DNA typing was done with Gene Print Multiple PowerPlex 2.1 (Promega). Cell identity typing (Wellcome Trust Sanger Institute, Cambridge, UK) is listed at http://www.sanger.ac.uk/genetics/CGP/Genotyping/synlinestable.shtml.
Migration Assays
Wounding migration assays used 1-mm-thick scotch tape (Bristol) placed in the center of 24-well plates. Cells were plated at confluency, and tape was removed after to create a clear zone. Cells that migrated into the cleared region were photographed for counting at 72 hours later. A separate migration assay format used 96-well Fluoroblock insert wells (BD Biosciences). H1993 or H2073 cells (n = 10,000) were plated and 24 hours later treated with compounds for 12 hours before HGF addition to both the top and bottom wells. After 12 hours, calcein AM (Invitrogen) at 2.5 μg/ml was added to the bottom chamber for 1 hour. Signal was measured on a Wallace 1420 Multilabel Victor V counter (Perkin Elmer) with bottom reading capacity.
shRNA and Cell Growth
Met and control shRNA were as described.26 Growth factor–mediated rescue of gefitinib growth inhibition in HCC827 cells used 4000 cells per well in 96-well plates. Gefitinib (LC labs, 1 μmol/L) was added, followed by addition of growth factors (R&D Systems) at 50 ng/ml. Cell growth was measured by Vialight 4 days later.
Western Blotting
Lysis buffer containing 30 mmol/L Tris-HCL, pH 7.5, 50 mmol/L NaCl, 5 mmol/L EDTA, 50 mmol/L NaF, 30 mmol/L NaPPi, 1% Triton, 0.5% IGEPAL, 10% Glycerol, 1 mmol/L Sodium Vanadate, 1 mmol/L bpPhen (Calbiochem), and protease inhibitor cocktail (Roche) was added with shaking for 10 minutes at 4°C. Lysates were clarified by centrifugation at 20,000g for 5 minutes at 4°C. SDS-PAGE on 40 μg of cell lysates and Western blotting followed standard procedures. Antibody to Met (AF276) was from R&D Systems, GAPDH was from Fitzgerald industries, and antibodies from Cell Signaling Technology included Y1234/35 Met (3077), Met (3127), Ser473 Akt (4058), Akt (4691), T202/Y204 Erk (4370), S235/236 S6 (4857), cleaved PARP (9544), Y1173 EGFR (4407), and EGFR (4267).
Statistical Analysis
Wilcoxon rank-sum test was used to compare the distribution of phosphorylation H scores between independent tumor groups defined by Met expression and MET copy number. The Signed rank test was applied to the analysis of paired difference in H scores between primary lung tumors and brain metastases. When H scores were dichotomized as positive versus negative, the Fisher exact test was used to compare the frequencies of expression and phosphorylation between independent patient groups, while the McNemar test was used in the analysis of paired binary data. Exact P values were computed for the log rank test to compare the time of progression between patient groups, stratifying by stage to control for the underlying prognosis. Analysis was performed using SAS 9.1 (SAS Institute, Cary, NC) and StatXact 6.1 (Cytel Software Corp, Cambridge, MA). All P values are based on a two-sided hypothesis test.
Results
Met Phosphorylation in NSCLC Patients not exposed to EGFR Inhibitors Is Associated with Poor Response to EGFR Inhibitor Therapy
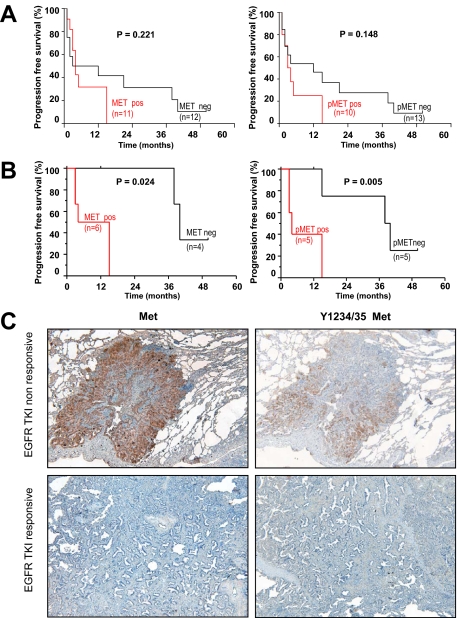
To investigate the role of Met in de novo resistance to EGFR TKI therapy, we correlated Met expression, phosphorylation, and gene copy gain with patient response. We analyzed 23 NSCLC samples, including ten with activating EGFR mutations (L858R, L861Q, or exon19 deletion) and 13 with wild-type EGFR. Importantly, our analysis was conducted on patient tumors resected before EGFR inhibitor therapy (cohort 1, see supplemental Table S1, available at http://ajp.amjpathol.org). Met phosphorylation was tested using a recently developed antibody specific for Y1234/35 activation loop phosphorylation (Ab 3077, Cell Signaling Technology). Importantly, this antibody did not cross-react by Western blot or IHC with cell lines harboring other amplified or activated receptor tyrosine kinases (including EGFR family kinases, PDGFR, FGFR, insulin receptor, and Ron kinases), providing a specificity not available with prior reagents (see supplemental Table S2, available at http://ajp.amjpathol.org). We determined whether progression free survival (PFS) was different in tumors that expressed Y1234/35 Met compared with tumors that did not express Y1234/35 Met. In a stage-matched analysis, Met expression and phosphorylation were more common in tumors that progressed during EGFR TKI therapy, but this trend was not significant (P = 0.221 and P = 0.148, respectively; Figure 1A). However, when considering only patients harboring EGFR mutation, both Met phosphorylation and expression were strongly associated with shorter time to progression (P = 0.024 and P = 0.005, respectively; Figure 1B and C). Furthermore, two patients with EGFR– activating mutations exhibited primary resistance to EGFR TKI therapy, displaying progressive disease (PD) three months after treatment initiation (Table 1, patients 1 and 8). Importantly, these resistant tumors exhibited extensive Met expression and activation (in 70% of tumor, P = 0.022).
Figure 1.
Met expression and phosphorylation in EGFR mutation containing NSCLC tumors are associated with poor response to EGFR TKI therapy. Kaplan–Meier curves of progression-free survival (PFS) for Met and Y1234/35 Met in 23 NSCLC patients before EGFR inhibitor therapy (A), and the subset of patients (n = 10) harboring EGFR mutations (B). C: Top panels (EGFR TKI nonresponsive) indicate elevated Met and Y1234/35 Met in the tumor specimen of a nonresponsive patient (Patient eight, Table one, PFS of three months); bottom panels (EGFR TKI responsive) indicate undetectable Met and Y1234/35 Met in the tumor from a responsive patient tumor (PFS = 40 months, patient five, Table one). FISH analysis revealed low Met copy gain in both patients (Table 1).
Table 1.
NSCLC Patient Outcome on EGFR TKI Therapy in Relation to Met Expression, Phosphorylation, and Gene Copy Number Gain
| Pt | RECIST | PFS (months) | Met
|
Y1234/35 Met
|
EGFR mutation
|
Site | EGFR TKI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Pos cells | Intensity | H score | FISH | % Pos cells | Intensity | H score | Type | % Pos cells | |||||
| 1 | PD | 3 | 70 | 3+ | 210 | H | 70 | 2+ | 140 | ex19del | 90 | local LN | Er |
| 2 | PR | 15 | 10 | 1+ | 10 | H | 30 | 2+ | 60 | ex19del | 100 | PWR | Er |
| 3 | PR | 15 | 40, 10 | 1+, 2+ | 60 | L | 10 | 1+ | 10 | ex19del | 90 | PWR | Er |
| 4 | SD | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ex19del | 20 | local LN | Ge |
| 5 | SD | 40 | 3 | 1+ | 3 | L | 0 | 0 | 0 | ex19del | 70 | PWR | Ge |
| 6 | PR | 4 | 20, 5 | 1+, 2+ | 30 | H | 30 | 2+ | 60 | ex19del | 90 | PWR | Er |
| 7 | PR | 38 | 0 | 0 | 0 | na | 0 | 0 | 0 | ex19del | na | PWR | Er |
| 8 | PD | 3 | 70 | 2+ | 140 | L | 70 | 2+ | 140 | L861Q | na | PWR | Er |
| 9 | PR | 15 | 40, 10 | 1+, 2+ | 60 | H | 0 | 0 | 0 | ex19del | 40 | local LN | Er |
| 10 | SD | 12 | 0 | 0 | 0 | L | 0 | 0 | 0 | L858R | 90 | lobe | Er |
All tumors harbor EGFR-activating mutations and were resected before EGFR TKI therapy. RECIST indicates patient response according to RECIST criteria; PFS, progression-free survival during EGFR inhibitors treatment. IHC: when multiple intensity values are present in the same sample, the respective percentages of positive cells are separated by a comma. FISH for Met is scored as described in Materials and Methods: D indicates disomic; L, low copy gain; H, high copy gain; na, not assessable; Local LN, loco-regional lymph node; PWR, pulmonary wedge resection; Er, erlotinib; Ge, gefitinib.
We also conducted fluorescence in situ hybridization (FISH) analysis and found MET copy gain resulting from chromosome 7 polysomy but not MET focal amplification (MET/CEP7 ratio >2.2). However MET copy gain was not associated with time to progression (P = 0.198, data not shown), similar to a previous report.27 We conclude that Met activation (in the absence of gene amplification) was correlated with de novo resistance to EGFR tyrosine kinase inhibitors.
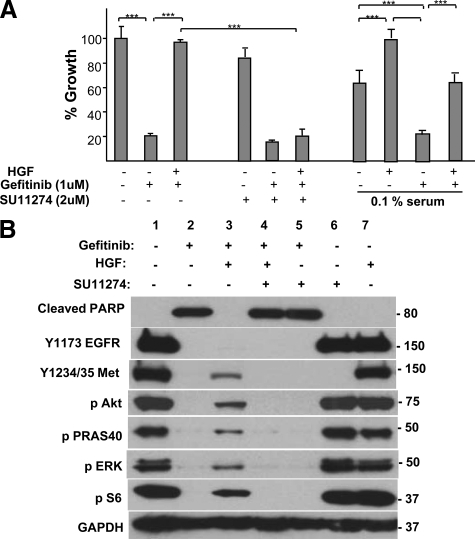
We next used the EGFR mutation containing HCC827 cell line to define a potential mechanism for the poor outcome of EGFR-mutated tumors harboring Met activation. We found that Met activation by HGF in HCC827 cells caused a striking resistance to Gefitinib (Figure 2A). In fact, HGF was the only ligand in a panel of 12 growth factors (HGF, EGF, Amphiregulin, Epiregulin, neuregulin, TGFα, FGF1, PDGFBB, Kit ligand, insulin, IGF1, IGF2) able to maintain HCC827 cell growth in the presence of Gefitinib (data not shown). HGF-mediated rescue also occurred in 0.1% serum, revealing that Met activation did not require cooperation with serum factors (Figure 2A). Importantly, the HGF-mediated rescue was blocked by an anti-HGF antibody or the Met-specific inhibitor SU11274 (Figure 2A). HGF also rescued Gefitinib growth inhibition in the EGFR mutation-containing cell lines HCC4006 and H2935 (see supplemental Figure S1, available at http://ajp.amjpathol.org). These in vitro results provide a mechanism for our observation that Met activation, independent of MET amplification, is associated with de novo resistance and poor disease control with EGFR targeted therapy.
Figure 2.
A: Gefitinib-induced growth inhibition of HCC827 is rescued by HGF. Gefitinib (1 μmol/L) addition was followed by addition of 50 ng/ml HGF. SU11274 (2 μmol/L) addition blocked HGF-mediated rescue. HGF rescue was maintained in 0.1% serum as indicated. Cell growth was measured after four days of treatment and is shown relative to untreated cells. Data represent mean ± SEM. ***P < 0.001. B: HGF restores signal transduction pathways in Gefitinib-treated cells. Gefitinib (1 μmol/L) was added to HCC827 cells, followed by addition of 50 ng/ml HGF. Seventy-two hours later, lysates were prepared and analyzed by SDS-PAGE and Western blotted for the indicated phospho proteins.
We next identified the signal transduction pathways necessary for Met to maintain cell growth and survival in HCC827 cells. Gefitinib inhibited EGFR, Erk, S6 ribosomal protein, Akt, and PRAS40 phosphorylation (Figure 2B, column 2). Elevated basal Y1234/35 Met phosphorylation in HCC827 cells was also dependent on activated EGFR, as also reported in Guo et al.28 Addition of HGF to Gefitinib-treated cells caused a durable (72 hours post HGF addition) reactivation of Met and Erk, S6, and Akt phosphorylation (Figure 2B, column 3). Activation of the Akt target PRAS40 confirmed functional reactivation of Akt. Interestingly, we found that reactivation of Erbb3 was not required for HGF-mediated rescue of growth and signal transduction pathways. Strikingly, HGF completely blocked Gefitinib-mediated PARP cleavage (Figure 2B, top panel). SU11274 reversed HGF-mediated reactivation of signaling and survival pathways (Figure 2B column 4), consistent with SU11274-mediated reversal of HGF-stimulated proliferation (above, Figure 2A). We conclude that HGF blocks Gefitinib-mediated growth arrest and cell death by reactivating major signal transduction pathways.
EGFR Mutations and Met Activation Occur in the Same Tumor Cells
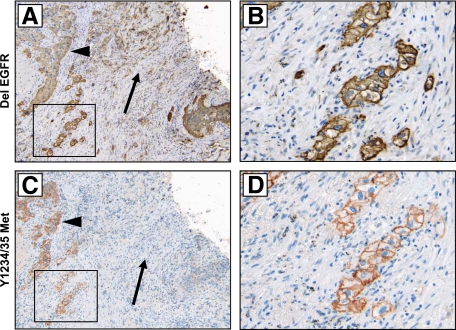
Having shown EGFR mutation and Met activation were commonly found in the same tumors, we next determined whether both occur in the same tumor cells. Tumors harboring either L858R or exon 19 deletion (del 19) EGFR mutation were tested for the expression of mutated EGFR by IHC analysis with mutation-specific antibodies.29 All samples stained positively, and 75% (6 of 8) of these cases showed widespread reactivity, suggesting that the majority of tumor cells (70 to 100%) harbored EGFR mutation (Table 1). Staining of consecutive sections revealed Met activation in the same cells harboring EGFR mutation (Figure 3, A–D). Because the majority of cells stained positively for EGFR mutation, Met phosphorylation was not found in cells independently of EGFR mutation. However, we commonly found EGFR mutant–positive cells that were negative for Met expression and phosphorylation (Figure 3). Our in vitro results strongly suggest that a combination of Met and EGFR-targeted therapy will be required for cell death in these cancer cells harboring both Met activation and EGFR mutation.
Figure 3.
Colocalization of Met phosphorylation and EGFR mutation. A and B: Exon 19 EGFR deletion, detected with a delEGFR-specific antibody. C and D: Met Y1234/35 phosphorylation. IHC images on consecutive sections (patient 2, Table 1) reveal colocalization of delEGFR and Y1234/35 Met. Arrowheads indicate regions of positivity for both antibodies; arrows indicate regions harboring EGFR deletion but not Met activation. B and D are magnification (×400) of A and C (×100), respectively.
Met Expression and Activation Are Associated with the Development of Brain Metastases in NSCLC
Met expression is associated with metastasis development in a wide range of cancers.13,14 We determined whether Met expression or activation were associated with the development of brain metastases in NSCLC by comparing the primary tumors from a group of 18 NSCLC patients that later developed brain metastases to a group of 22 tumors from NSCLC patients that did not develop brain metastases17 (cohort 2). Importantly these groups were otherwise matched for similar clinicopathological characteristics (see supplemental Table S3, available at http://ajp.amjpathol.org). The number of samples positive for Met expression and phosphorylation was much higher in the tumors from patients that developed brain metastases (P = 0.011 and P = 0.031, respectively; see supplemental Table S3, available at http://ajp.amjpathol.org). Thus, both Met expression and phosphorylation in patient primary tumors were associated with a higher risk of developing brain metastases.
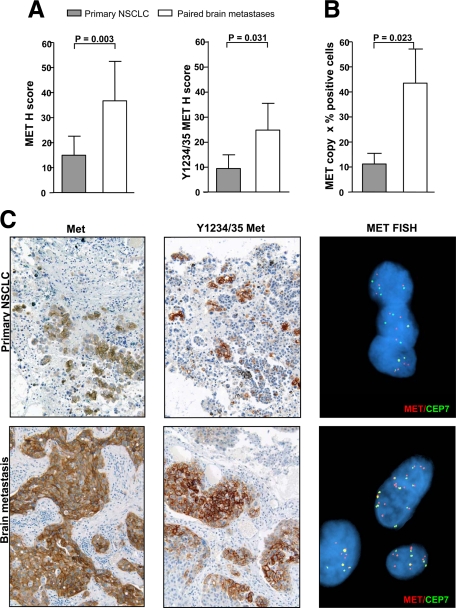
Met Activation Is Increased in NSCLC Metastases
Having seen that Met phosphorylation and expression in primary NSCLC was correlated with the development of brain metastases, we considered that Met may in fact be upregulated in metastatic lesions. We assessed Met activation, protein expression, gene copy number, and mutation in an annotated cohort of 18 primary NSCLC and paired brain metastases.17 Three primary NSCLC tumors were negative for Ki-67 staining and were therefore excluded from IHC analysis (Table 2). IHC revealed an enrichment of cells positive for Met and Y1234/35 Met (P = 0.003 and P = 0.031, respectively) in brain metastases (Figure 4A, and Supplemental Tables S3 and S4, available at http://ajp.amjpathol.org). Figure 4C reveals increased Met expression and phosphorylation in a brain metastasis relative to the paired primary tumor (from patient 10, Table 2). By contrast, there was not an enrichment of EGFR-expressing cells in the metastatic lesions (P = 0.793, Supplemental Table S4, available at http://ajp.amjpathol.org).
Table 2.
Profile of Met Expression, Y1234/35 Phosphorylation, Gene Copy Number, and EGFR Expression in NSCLCs and Their Paired Brain Metastases
| Pt | Type | Primary NSCLC
|
Paired brain metastasis
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Met IHC | Y1234-5 Met IHC | Met FISH
|
EGFR IHC H score | Met IHC | Y1234–5 Met IHC | Met FISH
|
EGFR IHC H score | ||||||||||||
| % Pos cells | Intensity | H score | % Pos cells | Intensity | H score | Score | % Cells w copy gain | % Pos cells | Intensity | H score | % Pos cells | Intensity | H score | Score | % Cells w copy gain | ||||
| 1 | A | 0 | 0 | 0 | 0 | 0 | 0 | D | 0 | 0 | 15, 5 | 1+, 2+ | 25 | 0 | 0 | 0 | L | 15 | 150 |
| 2 | A | 15 | 1+ | 15 | 15 | 2+ | 30 | H | 25 | 105 | 15, 5 | 1+, 2+ | 25 | 20 | 2+ | 40 | H | 40 | 12 |
| 3 | S | na | na | na | na | na | na | L | 10 | na | 20, 10, 2 | 1+, 2+, 3+ | 46 | 20 | 2+ | 40 | D | 0 | 25 |
| 4 | S | 0 | 0 | 0 | 0 | 0 | 0 | D | 0 | 215 | 10 | 1+ | 10 | 0 | 0 | 0 | L | 20 | 200 |
| 5 | A | 3 | 1+ | 3 | 0 | 0 | 0 | D | 0 | 10 | 5 | 2+ | 10 | 0 | 0 | 0 | L | 70 | 30 |
| 6 | S | 0 | 0 | 0 | 0 | 0 | 0 | L | 10 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | na | na | 0 |
| 7 | A | 15, 20, 5 | 1+, 2+, 3+ | 70 | 5 | 2+ | 10 | D | 0 | 30 | 40, 35 | 2+, 3+ | 185 | 30 | 2+ | 60 | L | 10 | 0 |
| 8 | S | 5 | 1+ | 5 | 5 | 1+ | 5 | na | na | 90 | 60 | 1+ | 60 | 10 | 1+ | 10 | L | 30 | 190 |
| 9 | S | 5 | 2+ | 10 | 0 | 0 | 0 | H | 80 | 50 | 10 | 1+ | 10 | 20 | 2+ | 40 | na | na | 20 |
| 10 | A | 5, 2 | 2+, 3+ | 16 | 5 | 2+ | 10 | L | 15 | 70 | 20 | 1+ | 20 | 40 | 2+ | 80 | H | 80 | 140 |
| 11 | S | 3 | 1+ | 3 | 0 | 0 | 0 | D | 0 | 164 | 0 | 0 | 0 | 0 | 0 | 0 | D | 0 | 20 |
| 12 | A | 0 | 0 | 0 | 0 | 0 | 0 | D | 0 | 85 | 5 | 1+ | 5 | 0 | 0 | 0 | H | 40 | 0 |
| 13 | S | na | na | na | na | na | na | L | 20 | na | 0 | 0 | 0 | 0 | 0 | 0 | L | 40 | 70 |
| 14 | A | 0 | 0 | 0 | 0 | 0 | 0 | D | 0 | 0 | 10 | 2+ | 20 | 0 | 0 | 0 | D | 0 | 15 |
| 15 | A | 60, 2 | 1+, 2+ | 100 | 40 | 2+ | 80 | na | na | 30 | 10, 40, 30 | 1+, 2+, 3+ | 180 | 80 | 2+ | 140 | H | 30 | 10 |
| 16 | A | na | na | na | na | na | na | D | 0 | na | 0 | 0 | 0 | 0 | 0 | 0 | na | na | 0 |
| 17 | S | 0 | 0 | 0 | 0 | 0 | 0 | L | 30 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | H | 0 | 60 |
| 18 | A | 0 | 0 | 0 | 0 | 0 | 0 | L | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | L | 90 | 50 |
A indicates adenocarcinoma; S, squamous cell carcinoma. IHC: when multiple intensity values are present in the same sample, the respective percentages of positive cells are separated by a comma. na indicates not available because of Ki67-negative staining. Met FISH is scored as described in Materials and Methods: D indicates disomy, L, low copy gain, H, high copy gain; na, not assessable. Met and EGFR H score: 0–300; Y1234/35 Met H score: 0–200.
Figure 4.
Met expression, phosphorylation, and gene copy gain are enriched in brain metastases relative to their paired NSCLC. A: Met and Y1234/35 Met were assessed by immunohistochemistry (H score) in primary NSCLCs and paired brain metastases. Both Met expression (P = 0.003) and Y1234/35 Met phosphorylation (P = 0.031) are significantly upregulated in metastases. Data represent mean ± SEM. B: Prevalence of MET copy gain (factoring MET copy number gain and % positive cells as in Materials and Methods) is higher in the metastatic lesions relative to the paired primary NSCLCs (P = 0.024). C: Met IHC (magnification ×200) and FISH analysis on a primary NSCLC and its paired brain metastasis (patient 10, Table 1). Met and Y1234/35 Met staining was heterogeneous/focal in the primary cancer but more widespread in the paired brain metastasis. MET FISH reveals low copy gain (MET copies, n = 3 to 5) in the primary lung tumor, and high copy gain (MET copies, n = 6 to 10) in the metastatic lesion.
We also found that high MET copy number gain as assessed by FISH occurred more commonly in metastastic samples (23%) relative to matched primary tumors (8%), but this was not significant (P = 0.500, see supplemental Table S4, available at http://ajp.amjpathol.org). However, the prevalence of MET copy gain (determined similarly to H score by factoring MET copy number gain and percentage of MET FISH-positive cells) was significantly higher in brain lesions relative to the paired primary lung tumor (P = 0.023, Figure 4B). We conclude that MET copy number gain as well as protein expression and phosphorylation were enriched in brain metastases relative to primary NSCLC tumors. We expected that increased MET copy gain would result in overexpression and activation of Met, and we confirmed that Met and Y1234/35 Met phosphorylation in cohort 2 were increased in samples with high MET copy gain compared with samples with lower MET copy gain (≤5 copies of MET, P = 0.002 and P = 0.007, respectively; see supplemental Figure S2, available at http://ajp.amjpathol.org).
Met Activation Is Not a Result of Gene Mutations
We analyzed both cohorts for 15 MET mutations, some of which were previously reported to be selectively upregulated in metastases.30,31 Forty-six samples (32 primary NSCLCs and 14 brain metastases) were informative. Activating MET mutations were found in two NSCLC samples (both from the group of primary tumors that did not develop metastases), but curiously these tumors were negative for Met protein expression. No mutations were found in the primary tumors that had paired brain lesions. We also analyzed the same cases for EGFR kinase domain mutations and found EGFR exon 19 deletion mutations (del19 EGFR) in two primary tumors, but not in the paired brain metastases. Others have reported discordance in the EGFR mutation status of primary NSCLC tumors and matched metastases.32
Met Activation Occurs in a Metastasis-Derived Cell Line but not in the Isogenous Cell Line Derived from the Primary Lung Tumor
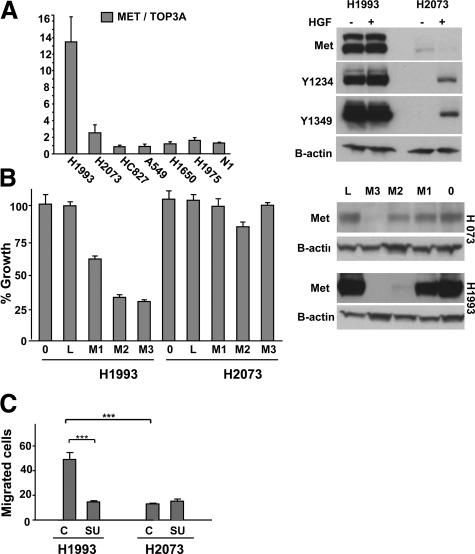
The MET-amplified H1993 lung cancer cell line was isolated from a lymph node metastasis while the H2073 cell line was isolated from the primary lung tumor from the same patient. The common origin of these lines was confirmed by DNA fingerprinting (data not shown) and cell line identity typing (Wellcome Trust Sanger Institute, Cambridge, UK). While H1993 cells harbor MET amplification and highly activated kinase, as reported previously,26 H2073 cells do not have MET amplification and have very low basal Met expression and phosphorylation (Figure 5A). Also in contrast to H1993 cells, H2073 cells were not growth inhibited by MET shRNA (Figure 5B). Finally, Met-specific small molecule inhibitor SU1127415 also did not inhibit H2073 cell growth (IC50 > 10 μmol/L for H2073 compared with IC50 = 460 ± 70 nmol/L for H1993 cells, data not shown).
Figure 5.
MET is amplified and constitutively activated in the metastasis-derived H1993 cell line but not in the paired primary lung tumor–derived H2073 cell line. A: Quantitative PCR for MET copy number. MET is not amplified in H2073 cells but is amplified in H1993 cells. Data represent mean ± SEM. Immunoblot reveals elevated Met expression and Y1234/35 and Y1349 phosphorylation in H1993 cells but not in H2073 cells. B-actin is the loading control. B: Growth of H2073 cells is not inhibited by Met shRNA, whereas H1993 cell growth is inhibited. Data represent mean ± SEM. Immunoblot revealing efficient Met knockdown in H2073 and H1993 cells treated with Met shRNA M3 and less efficient knockdown with Met shRNA M2 and M1. L indicates Luciferase shRNA; 0, no shRNA. C: Migration assay was performed in the indicated cell lines as described in Methods. 0, initial time point; 72, 72 hours later; SU, addition of 2.5 μmol/L SU11274 Met inhibitor. Data represent mean ± SEM. *P < 0.5, ***P < 0.001. Results are representative of three independent experiments.
We compared the metastasis-derived H1993 cells to H2073 cells in in vitro migration assays. Using a “wound healing” migration assay, we found that H1993 cells had a much higher capacity for migration relative to H2073 cells (Figure 5C, supplemental Figure S3, A and B, available at http://ajp.amjpathol.org; P < 0.001). SU11274 Met inhibitor blocked cell migration only in H1993 cells (Figure 5C; P < 0.001), revealing that Met activation is required for migration of this MET-amplified cell line. H2073 cells also migrated poorly relative to H1993 cells in a transwell migration format (see supplemental Figure 3D, available at http://ajp.amjpathol.org). As a control for compound specificity, SU11274 (2.5 μmol/L) did not inhibit the robust migration of A549 cells in which Met is not activated26 (see supplemental Figure 3C, available at http://ajp.amjpathol.org). These results demonstrate that the elevated Met phosphorylation in H1993 cells is required for migration, and the enhanced capacity for migration in H1993 cells is consistent with our finding that cells harboring Met activation were enriched in NSCLC brain metastases.
Discussion
MET amplification has been considered a key parameter for eligibility for Met inhibitor therapy33,34 and an indicator of acquired resistance to EGFR TKI therapy.9,10 In our cohort of EGFR TKI therapy-naïve NSCLC (cohort 1), we observed increased MET copy number resulting from polysomy of chromosome 7, rather than focal MET amplification, but it was not associated with poor response to EGFR targeted therapy, similar to previous results in NSCLC and colorectal cancer.21,27 We instead show that Met expression and activation (before EGFR TKI treatment) predicted poor response to subsequent EGFR inhibitor treatment, despite the presence of EGFR TKI sensitizing mutations. We therefore demonstrate that Met expression and phosphorylation are relevant parameters in stratifying EGFR mutation-containing NSCLC patients for potential Met inhibitor treatment. We further provide a rationale for this poor response to EGFR TKI in patients harboring Met activation by showing in vitro that Met activation in HCC827, HCC4006, and H2935 EGFR mutant cell lines mediates potent resistance to treatment with EGFR TKI.
Taken together, our cell line and clinical data suggest a model whereby tumors with extensive Met activation before EGFR TKI therapy can exhibit primary resistance to subsequent treatment with EGFR TKI (see supplemental Figure S4C, available at http://ajp.amjpathol.org). By contrast, initial disease control (partial response or stable disease) is predicted in tumors with low levels of Met activation (see supplemental Figure S4B, available at http://ajp.amjpathol.org). However, because these Met-activated tumor cells can presumably proliferate despite EGFR TKI therapy, initial disease control is then followed by a relatively short PFS compared with tumors without Met activation (see supplemental Figure S4, A and B, available at http://ajp.amjpathol.org). Interestingly, both Met and T790M mutation12 may be factors in primary resistance as well as acquired resistance. Whether a patient exhibits primary versus acquired resistance may then be related to the percentage of resistant cells (containing either Met activation or T790M mutation12) before EGFR TKI therapy (see supplemental Figure S4, available at http://ajp.amjpathol.org). However larger studies will be required to confirm these relationships.
Although Met expression and activation can be heterogeneous in NSCLC tumors, our IHC analysis with antibodies specific for L858R EGFR and del19 EGFR reveal that EGFR mutations are primarily homogenous within tumors. Therefore poor response to EGFR TKI therapy was not related to a low percentage of EGFR mutant cells within the tumor but was instead associated with Met activation. If confirmed in larger cohorts, our results also suggest that the EGFR mutation-specific and Y1234/35 Met antibodies could identify EGFR mutation-containing patients harboring Met activation who could potentially benefit from early intervention with a combination of EGFR and Met inhibitor therapy.
Finally, we found that the heterogeneous Met expression, activation, and gene copy gain in primary NSCLC is significantly enriched in paired brain metastases. An isogenous pair of cell lines derived from a primary lung tumor and a paired metastatic lesion also revealed that Met activation was required for in vitro migration. These results suggest that the enrichment of Met-activated lung tumor cells in brain metastases may result from an increased capacity for Met activated primary tumor cells to migrate and establish metastases.
Acknowledgments
We thank Alessandro Fornari and Carmen Priolo for discussions and Catarine Campbell and Laura MacConaill for assistance with Oncomap analysis.
Footnotes
Address reprint requests to Bart Lutterbach, Ph.D., Merck Research Laboratories, Boston, MA 02115. E-mail: Bart_lutterbach@merck.com.
Supported by Lung Cancer Specialized Program of Research Excellence (SPORE) 2P50 CA090578–06 (to M.L., L.C., B.Y.Y.) and SPORE P50CA70907 (to J.D.M. and A.G.). M.L. is a Novartis Investigator. E.B. is supported by a Ph.D. Program fellowship in Molecular Medicine, University of Milan, Italy.
L.R.C., B.L., and M.L. contributed equally to this work as principal investigators.
A guest editor acted as editor-in-chief for this manuscript. No person at Thomas Jefferson University was involved in the peer review process or final disposition for this article.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Asahina H, Yamazaki K, Kinoshita I, Sukoh N, Harada M, Yokouchi H, Ishida T, Ogura S, Kojima T, Okamoto Y, Fujita Y, Dosaka-Akita H, Isobe H, Nishimura M. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AY. The role of gefitinib in the management of Asian patients with non-small cell lung cancer. Expert Opin Investig Drugs. 2008;17:401–411. doi: 10.1517/13543784.17.3.401. [DOI] [PubMed] [Google Scholar]
- Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Yeap BY, Lindeman NI, Fidias P, Rabin MS, Temel J, Skarin AT, Meyerson M, Holmes AJ, Borras AM, Freidlin B, Ostler PA, Lucca J, Lynch TJ, Johnson BE, Janne PA. Phase II clinical trial of chemotherapy-naive patients > or = 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol. 2007;25:760–766. doi: 10.1200/JCO.2006.07.5754. [DOI] [PubMed] [Google Scholar]
- Sunaga N, Tomizawa Y, Yanagitani N, Iijima H, Kaira K, Shimizu K, Tanaka S, Suga T, Hisada T, Ishizuka T, Saito R, Dobashi K, Mori M. Phase II prospective study of the efficacy of gefitinib for the treatment of stage III/IV non-small cell lung cancer with EGFR mutations, irrespective of previous chemotherapy. Lung Cancer. 2007;56:383–389. doi: 10.1016/j.lungcan.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Yatabe Y, Park JY, Shimizu J, Horio Y, Matsuo K, Kosaka T, Mitsudomi T, Hida T. Prospective validation for prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer. J Thorac Oncol. 2007;2:22–28. [PubMed] [Google Scholar]
- Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Janne PA, Lynch T, Johnson BE, Miller VA. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer, J Clin Oncol. 28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka Y, Uehara H, Mitsudomi T, Yatabe Y, Nakamura T, Sone S. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, Kuhlmann GL, Han M, Goldberg JS, Settleman J, Iafrate AJ, Engelman JA, Haber DA, Johnson BE, Lynch TJ. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- Sattler M, Pride YB, Ma P, Gramlich JL, Chu SC, Quinnan LA, Shirazian S, Liang C, Podar K, Christensen JG, Salgia R. A novel small molecule met inhibitor induces apoptosis in cells transformed by the oncogenic TPR-MET tyrosine kinase. Cancer Res. 2003;63:5462–5469. [PubMed] [Google Scholar]
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- Saad AG, Yeap BY, Thunnissen FB, Pinkus GS, Pinkus JL, Loda M, Sugarbaker DJ, Johnson BE, Chirieac LR. Immunohistochemical markers associated with brain metastases in patients with nonsmall cell lung carcinoma. Cancer. 2008;113:2129–2138. doi: 10.1002/cncr.23826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;1:19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG, Buttitta F, Incarbone M, Toschi L, Finocchiaro G, Destro A, Terracciano L, Roncalli M, Alloisio M, Santoro A, Varella-Garcia M. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;10:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Varella-Garcia M, Finocchiaro G, Skokan M, Gajapathy S, Carnaghi C, Rimassa L, Rossi E, Ligorio C, Di Tommaso L, Holmes AJ, Toschi L, Tallini G, Destro A, Roncalli M, Santoro A, Janne PA. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–89. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey PA, Zhu X, Zarnegar R, Swanson PE, Ratliff TL, Vollmer RT, Day ML. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am J Pathol. 1995;147:386–396. [PMC free article] [PubMed] [Google Scholar]
- Moasser MM, Basso A, Averbuch SD, Rosen N. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 2001;61:7184–7188. [PubMed] [Google Scholar]
- Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L, Severin C, Rangell L, Schwall R, Amler L, Wickramasinghe D, Yauch R. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Janne PA, Skokan M, Finocchiaro G, Rossi E, Ligorio C, Zucali PA, Terracciano L, Toschi L, Roncalli M, Destro A, Incarbone M, Alloisio M, Santoro A, Varella-Garcia M. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Kane S, Wu J, Benedettini E, Li D, Reeves C, Innocenti G, Wetzel R, Crosby K, Becker A, Ferrante M, Cheung WC, Hong X, Chirieac LR, Sholl LM, Haack H, Smith BL, Polakiewicz RD, Tan Y, Gu TL, Loda M, Zhou X, Comb MJ. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res. 2009;15:3023–3028. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- Di Renzo MF, Olivero M, Martone T, Maffe A, Maggiora P, Stefani AD, Valente G, Giordano S, Cortesina G, Comoglio PM. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene. 2000;19:1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- Lorenzato A, Olivero M, Patane S, Rosso E, Oliaro A, Comoglio PM, Di Renzo MF. Novel somatic mutations of the MET oncogene in human carcinoma metastases activating cell motility and invasion. Cancer Res. 2002;62:7025–7030. [PubMed] [Google Scholar]
- Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, Mavroudis D, Voutsina A. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, Christensen JG, Settleman J, Haber DA. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci USA. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi L, Janne PA. Single-agent and combination therapeutic strategies to inhibit hepatocyte growth factor/MET signaling in cancer. Clin Cancer Res. 2008;14:5941–5946. doi: 10.1158/1078-0432.CCR-08-0071. [DOI] [PubMed] [Google Scholar]