Abstract
The effect of functionalized nickel (Ni) nanoparticles capped with positively charged tetraheptylammonium on cellular uptake of drug quercetin into hepatocellular carcinoma cells (SMMC-7721) has been explored in this study via microscopy and electrochemical characterization as well as MTT assay. Meanwhile, the influence of Ni nanoparticles and/or quercetin on cell proliferation has been further evaluated by the real-time cell electronic sensing (RT-CES) study. Our observations indicate that Ni nanoparticles could efficiently improve the permeability of cancer cell membrane, and remarkably enhance the accumulation of quercetin in SMMC-7721 cells, suggesting that Ni nanoparticles and quercetin would facilitate the synergistic effect on inhibiting proliferation of cancer cells.
Keywords: Nickel nanoparticles, Real-time cell electronic sensing (RT-CES) study, MTT assay, Electrochemical analysis, Hepatocellular carcinoma cell line
Introduction
With the development of nanotechnology, nanomaterials are now widely produced and applied in biomedical and biologic engineering [1-4]. Due to their unique characteristics, nanomaterials and nanotechnologies are changing many basic scientific concepts in a great variety of fields, and are receiving intensively increasing interest in the relative research and industrial applications.
It is well known that the efficiency of many conventional pharmaceutical therapies can be significantly improved through the drug delivery system (DDS). DDS could be designed to alter the pharmacokinetics and biodistribution of their associated drugs and/or to function as drug reservoirs [5]. Some biocompatible nanoparticles, such as gold nanoparticles, iron oxide nanoparticles, have been used in DDS because of their feasibility to produce, characterize, and specifically tailor their functional properties [6-9].
Flavonoids are plant metabolites that are dietary antioxidants and exert significant antiallergic and antiviral effects. Quercetin is one of the most abundant flavonoids in the human diet and has been associated with a large number of biologic activities, many of which may contribute to the prevention of human diseases due to their effects of antihypertensive, antiinflammatory, and anticardiovascular disease [10-13]. Recently, an increasing number of reports have shown that quercetin has multiple effects on cancer cells, which can induce the apoptosis of cancer cells to exert the antitumor effect [14-16]. Additionally, the electrochemical assays for quercetin have been extensively studied due to its sensitive electroactive property [17-20]. Based on these observations, in this study we have utilized electrochemical strategy in the quantitative analysis of quercetin in the cellular systems; meanwhile, the relevant effects of functionalized nickel (Ni) nanoparticles on cellular uptake of drug quercetin into SMMC-7721 cancer cells have also been explored by means of atomic force microscopy (AFM), fluorescence microscopy and electrochemical assay. The result of our studies has afforded the first evidence that the functionalized Ni nanoparticles capped with positively charged tetraheptylammonium could improve the permeability of hepatocellular carcinoma cell membrane, and remarkably enhance the accumulation of quercetin in SMMC-7721 cells.
Meanwhile, the real-time cell electronic sensing (RT-CES) assay also provides the dynamic information that could be used to identify the interaction between cells and chemicals. The RT-CES array has proven valuable, sensitive and reliable for real time monitoring of dynamic changes induced by cell–chemical interaction [21-23]. During the RT-CES assay, the electrode sensor array was specially designed and integrated onto the bottom of a standard microtiter plate and cells directly grow on the sensor surface. The basic principle of the RT-CES system is to monitor the changes in electrode impedance induced by the interaction between testing cells and electrodes, where the presence of the cells will lead to an increase in the electrode impedance. The more cells attached to the sensor, the higher the impedance that could be monitored with RT-CES. Because the test is labeling free and quantitative, the RT-CES assay allows real-time, automatically and continually monitoring cellular status changes during the whole process of the cell–chemical interaction. Accordingly, in this study we have combined the MTT (3-(4,5-dimethylthiazol-2-yl)2,5-diphenyl-tetrazolium bromide) assay with RT-CES study [24]. Initially, we have explored the in vitro effect of quercetin in the absence and presence of Ni nanoparticles on SMMC-7721 cancer cells (a hepatocellular carcinoma cell line) by MTT assay, while the dynamic response of cancer cells exposure to Ni nanoparticles and/or quercetin has been determined by the RT-CES system. Our results demonstrate that Ni nanoparticles can readily facilitate the cellular drug uptake of quercetin into cancer cells and enhance the cytotoxicity suppression of quercetin on the proliferation of cancer cells, indicating their great potential in clinical and biomedical applications.
Experimental Section
Preparation and Characterization of Ni Nanoparticles
The fabrication of the Ni nanoparticles capped with positively charged tetraheptylammonium and the transmission electron microscopy image are similar to that we previously reported in the literature [25]. Briefly, the Ni nanoparticles capped with tetraheptylammonium were produced by electrochemical deposition, where the electrolysis processes were carried out in a 0.1 M tetraheptylammonium 2-propanol solution using an anode of high-purity Ni-sheets and a cathode of glassy carbon. For the electrolysis, a current density of 10–40 mA cm−2 was applied. The deposited clusters were capped with positively charged tetraheptylammonium. The functionalized Ni nanoparticles were characterized by TEM, and the average size was about 30 nm, as shown in Fig. 1.
Figure 1.
Typical TEM image of the functionalized Ni nanoparticle. The average size of Ni nanoparticles was about 30 nm.Bar100 nm
Cell Culture
SMMC-7721 cancer cells (purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) were maintained in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (Sigma, USA), 100 U/ml penicillin (Sigma, USA), and 100 μg/ml streptomycin (Sigma, USA), and grown at 37 °C in a 5% CO2humidified environment.
Morphological Assay of Cells Treated with Ni Nanoparticles
The SMMC-7721 cells were plated on coverlips in 6-well plates (105cells/well) and treated with different concentration of Ni nanoparticles. The concentrations of Ni nanoparticles cocultured with cells for optical microscopy assay were 3.12, 12.5 and 50 μg/mL, respectively, and cultured in the incubator for 72 h at 37 °C with 5% CO2; while the concentration of Ni nanoparticles cocultured with cells for AFM assay was 12.5 μg/mL and incubated for 6 h at 37 °C with 5% CO2. After treatment, the cells were observed by optical microscopy (Olympus BX 51, Japan) and atomic force microscopy (AFM, SPI 3800N, Japan).
Olympus IX71 Inverted Fluorescence Microscopy
The experiment was performed as described in the literature [26]. The SMMC-7721 cells were seeded on the coverlips in 6-well plates (105 cells/well) and cultured for 24 h at 37 °C with 5% CO2, then both quercetin and Ni nanoparticles were injected to cells and their concentrations were 5.0 and 2.0 μg/mL, respectively. Meanwhile, the cells treated with the same concentration of quercetin were taken as control experiments. All specimens were subsequently incubated for 1 h at 37 °C with 5% CO2, quickly washed with PBS, and then was followed by fixation with 4% formaldehyde for 5 min. Finally, specimens were observed by inverted fluorescence microscopy (Olympus IX71, Japan).
Electrochemical Study
SMMC-7721 cell suspensions (8 × 105cells/mL) containing 50 μmol/L of quercetin were cultured in the absence and presence of 2.0 μg/mL of Ni nanoparticles at room temperature (22 ± 2 °C) for 2 and 6 h, respectively. All samples were diluted with sterile phosphate buffer saline (PBS, 100 mmol/L, pH 7.2). The electrochemical signal was determined with differential pulse voltammetry (DPV) assay for each sample by CHI660C electrochemical analyzer. All measurements were carried out in a three-component electrochemical cell consisting of a glassy carbon electrode as the working electrode, a Pt wire as the counter electrode and a saturated calomel electrode as the reference electrode.
MTT Assay
The effect of different quercetin concentrations on SMMC-7721 cancer cells in the absence and presence of Ni nanoparticles was carried out by MTT assay. The final concentrations of quercetin and Ni nanoparticles were 25 (or 50) and 2.0 μg/mL, respectively. Initially, 1 × 104cells were seeded into each well containing 200 μL cell culture medium in 96-well plates and incubated for 24 h, then the relevant chemicals were added and incubated at 37 °C with 5% CO2for 72 h. Controls were cultivated under the same condition without addition of quercetin and/or the Ni nanoparticles. The relevant experiments were repeated thrice independently. The inhibition efficiency (%) was expressed as follows: (1−[A]test/[A]control) × 100, where [A]testand [A]controlrepresent the optical density at 540 nm for the test and control experiments, respectively.
In vitro RT-CES Cytotoxicity Assay
The cell culture condition, the starting cell number and cell culture medium volume used for the 16× sensor device were similar to that of MTT assay. About 50 μmol/L of quercetin in the absence and presence of 2.0 μg/mL of Ni nanoparticles were seeded in the plate. The relevant controls were also seeded in the same plate simultaneously. Once the cells were added to the sensor wells, the sensor devices were placed into the incubator and the real-time cell index (CI) data acquisition was initiated by the RT-CES analyzer (ACEA Bioscience. Inc., USA).
Statistics
Data were expressed as the means ± SD (standard deviation) from at least three independent experiments. One-tailed unpaired Student’st-test was used for significance testing, andp < 0.05 is considered significant.
Results and Discussion
The Cellular Microscopical Morphology
Initially, the effect of the functionalized Ni nanoparticles on SMMC-7721 cells has been studied by optical microscopy and atomic force microscopy (AFM). In comparison with the general morphology of SMMC-7721 cells in the absence of Ni nanoparticles (Fig. 2a), the majority of SMMC-7721 cells still grew well after an incubation of 72 h with 3.13 μg/mL Ni nanoparticles (Fig. 2b) and little influence was observed for the cellular microscopical morphology. However, when the concentration of Ni nanoparticles increased to 12.5 μg/mL, only a portion of SMMC-7721 cells could survive while their morphologies had remarkable changes (Fig. 2c). It is noted that 50 μg/mL of Ni nanoparticles could inhibit almost all SMMC-7721 cell proliferation (Fig. 2d). These results suggest that the functionalized Ni nanoparticles could induce cell-cycle arrest and increase apoptosis and/or necrosis to inhibit cell proliferation at higher doses of nanoparticles, whereas it has little effects on target cancer cells at lower doses.
Figure 2.
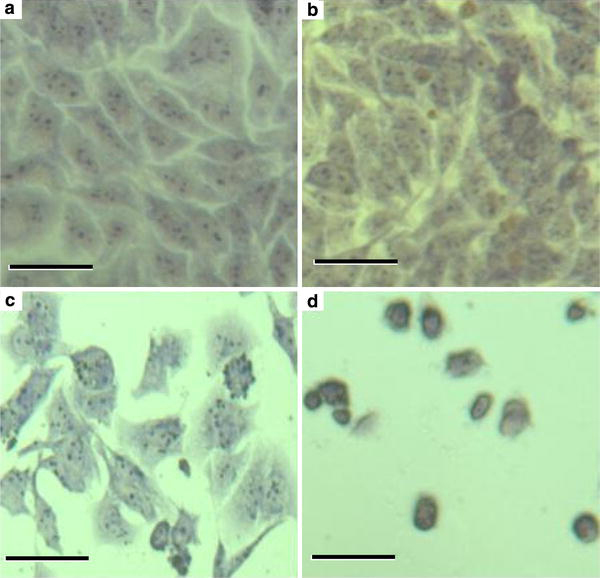
Morphological images of SMMC-7721 cells treated with or without Ni nanoparticles. The SMMC-7721 cells grown on coverlips were treated with different concentrations of Ni nanoparticles for 72 h, respectively. The cancer cells were washed with PBS, fixed in methanol, stained with Wright’s solution and then photographed (original magnification, 200).aSMMC-7721 cells without Ni nanoparticles (control experiment);bSMMC-7721 cells treated with 3.13 μg/mL of Ni nanoparticles;cSMMC-7721 cells treated with 12.5 μg/mL of Ni nanoparticles, anddSMMC-7721 cells treated with 50 μg/mL of Ni nanoparticles.Bar10 μm
AFM Assay
AFM is a surface analytical technique, which can image the nanoscale topography by scanning across the surface. As shown in Fig. 3, compared with the control experiments without Ni nanoparticles (Fig. 3a), the AFM image captured after incubation for 6 h with 12.5 μg/mL of Ni nanoparticles showed that cellular uptake of Ni nanoparticles would lead to some holes generated on SMMC-7721 cells, just as the dark spots arrowed in Fig. 3b. The formation of holes on the cell membrane induced by Ni nanoparticles can alter the permeability of the respective cell membrane and thus facilitate the relevant drug uptake into cancer cells and increase the concentration of drug in cancer cells and thus could greatly inhibit the proliferation of cancer cells.
Figure 3.
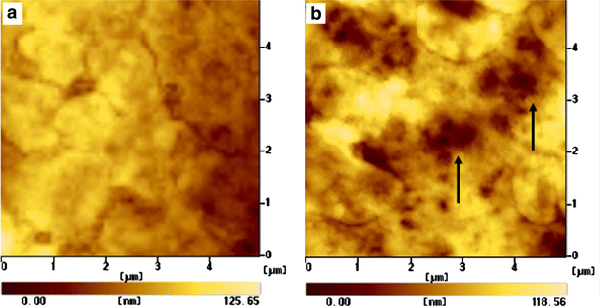
AFM images for cell uptake of Ni nanoparticles.aSMMC-7721 cell without Ni nanoparticles (control experiment); andbthe cell surface after incubation with 12.5 μg/mL of Ni nanoparticles for 6 h, which illustrates the apparent uptake of Ni nanoparticles during the process of endocytosis that led to the change of the cell membrane and formation of holes (arrows) on the surface of the relevant cell
Hence, these observations indicate that upon application of these Ni nanoparticles at higher concentrations, the relevant effect of Ni nanoparticles can efficiently produce some holes and/or result in the release of the cytosol out of the cancer cell and thus make the cell death, whereas at the lower concentrations of Ni nanoparticles, the relevant Ni nanoparticles have little effect on the cell morphology. The induced holes in the cell membrane in the presence of Ni nanoparticles could lead to the alteration of permeability of cell membrane and facilitate the drug uptake of quercetin into the cancer cells, which will further induce the apoptosis and/or necrosis of cancer cells. Moreover, when the combination with the anticancer drug quercetin, the lipid–lipid affinity of the lipid-soluble quercetin and the long alkane groups of functionalized Ni nanoparticles may lead to the formation of the relevant nanocomposites and make more drug molecules readily enter into the cancer cell. Thus, it is evident that in the presence of Ni nanoparticles, quercetin could diffuse through the cell membrane holes created by Ni nanoparticles or the Ni nanoparticles carrying a significant amount of quercetin into cells due to the surface concentrating effect. As shown in Scheme 1, the functionalized Ni nanoparticles have positive charges and hydrophobic groups, which could readily carry more drug molecule into cancer cells. It has been reported that some polymers with the greatest density of positively charged groups on the chains show the most dramatic increase in membrane thinning and membrane permeability, which could induce the formation of holes on both supported lipid bilayers and cellular membranes [27-29]. In our study, the effect of Ni nanoparticles capped with positively charged tetraheptylammonium on the cell membrane is also in favor of the formation of holes (arrows, Fig. 3b), which will facilitate the entry of drug into cell and increase the intracellular accumulation of drug molecules in cancer cells.
Scheme 1.
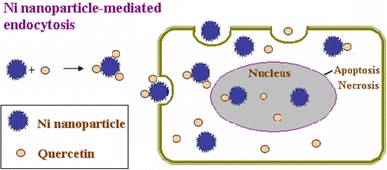
Possible mechanism for quercetin uptake into SMMC-7721 cells via Ni nanoparticle-mediated endocytosis
Enhanced uptake of quercetin by Ni nanoparticles—Fluorescence and electrochemical study
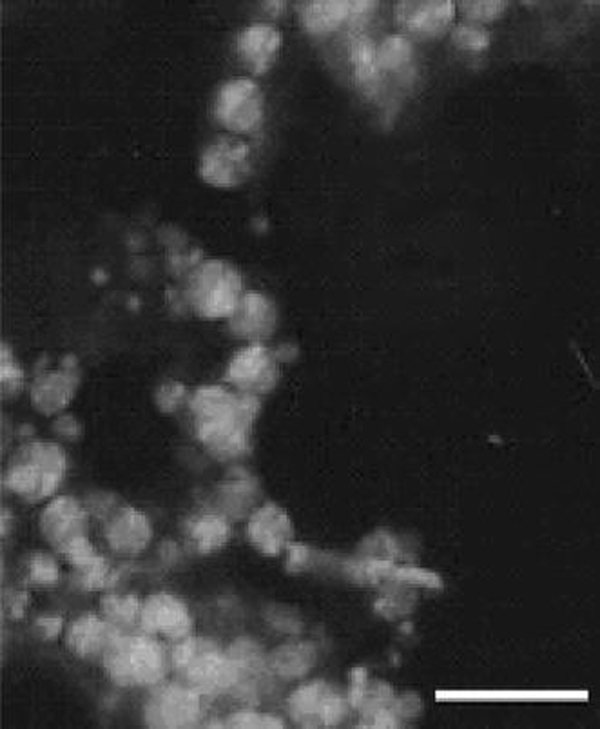
Thus, based on the above studies, we have further investigated the effective effect of the functionalized Ni nanoparticles on the quercetin uptake into cancer cells by using the inverted fluorescence microscopy. It is noted that upon binding and chemical crosslinking within unknown protein molecules in cells, the fluorescence of quercetin could be observed and detected [26]. As shown in Fig. 4, the apparent differences of drug accumulation in cancer cells were observed in the absence and presence of Ni nanoparticles. The cellular fluorescence in the Ni nanoparticles-treated system (Fig. 4b) is much higher than that with free of Ni nanoparticles (Fig. 4a). Hence, these Ni nanoparticles could remarkably facilitate the relative drug uptake and accumulation into SMMC-7721 cancer cells and could thus act as an efficient agent to enhance drug delivery.
Figure 4.
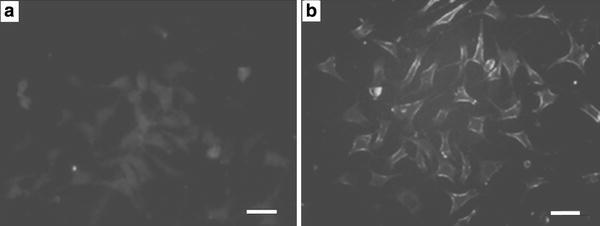
Inverted fluorescence micrographs of SMMC-7721 cells after incubation witha5.0 μmol/L of quercetin, andb5.0 μmol/L of quercetin in the presence of Ni nanoparticles (2.0 μg/mL) at 37 °C for 1 h. Thescale barrepresents 10 μm
Meanwhile, our electrochemical studies also provide the fresh evidence for the remarkable enhancement effect of the Ni nanoparticles in the drug uptake of quercetin in cancer cells. It is already known that quercetin (i.e., 3,3′,4′,5-7-pentahydroxyflavone), a chemical cousin of the glycoside rutin, is a unique flavonoid that has been extensively studied for its multiple effects as anticancer drug. In the present study, we have utilized the differential pulse voltammetry (DPV) to explore the effect of quercetin alone and quercetin in the presence of Ni nanoparticles on the drug uptake of relevant cancer cells. It is observed that with the anticancer drug quercetin as the electrochemical probe, the drug uptake efficiency for different cancer cells could be probed by the differential pulse voltammetry (DPV) technique. As we know, when different cells treated with quercetin, the unadsorbed drug molecules are still in the environmental solution, and the electrochemical response of this part of the molecules can be readily detected. Thus, the quercetin residue (unadsorbed quercetin) can be adopted as the referential value of the cellular uptake efficiency.
As shown in Fig. 5, the results of electrochemical study of the amount of quercetin residues outside SMMC-7721 cells in the absence and presence of Ni nanoparticles at room temperature illustrate that the DPV peak currents of quercetin residue outside SMMC-7721 cells show an evident decrease after treating the SMMC-7721 cells with quercetin for 2 and 6 h. Compared with that of the original concentration of quercetin (50 μM), the peak current of quercetin in the culture media decreased by 25% after a 2 h culture, whereas it decreased by 48% when exposed together with Ni nanoparticles. These results indicated that the quercetin residue outside the SMMC-7721 cells in the presence of Ni nanoparticles decreased more apparently than that in the absence of Ni nanoparticles. In addition, the decrease percentages of the peak current increased with the increasing incubation time. So it could be deduced that after coculture with cancer cells, the quercetin in the culture media could be admitted in cancer cells by endocytosis, which led to the remarkable decrease of the peak current.
Figure 5.
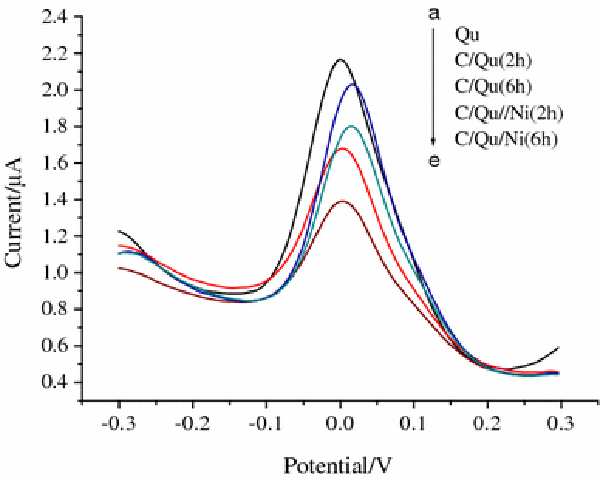
DPV study of quercetin residue outside SMMC-7721 cells in the absence and presence of Ni nanoparticles.a50 μmol/L of quercetin solution;b50 μmol/L of quercetin solution and cells incubated for 2 h;c50 μmol/L of quercetin solution and cells incubated for 6 h;d50 μmol/L of quercetin solution and cells exposed to Ni nanoparticles cocultured for 2 h ande50 μmol/L of quercetin solution and cells exposed to Ni nanoparticles incubated for 6 h. Here, C stands for SMMC-7721 cells, Qu for quercetin, and Ni for Ni nanoparticles. Pulse amplitude: 0.05 V. Pulse width: 0.05 s. Pulse period: 0.1 s
After adding the Ni nanoparticles, apparently more considerable changes of the electrochemical response of quercetin residue outside SMMC-7721 cells were observed than that in the absence of Ni nanoparticles, suggesting that much more quercetin molecules could be diffused into the relative cancer cells in the presence of Ni nanoparticles, which demonstrated that Ni nanoparticles can efficiently enhance the permeation and uptake of quercetin into SMMC-7721 cancer cells.
Cytotoxicity Evaluation
On the basis of these observations, the cytotoxicity suppression effect of quercetin on SMMC-7721 cells in the absence and presence of Ni nanoparticles has been further explored by MTT assay. As shown in Fig. 6, our studies indicate that inhibition efficiency of cancer cell proliferation in vitro could be remarkably improved in the presence of quercetin together with Ni nanoparticles when compared with either relevant quercetin control or Ni nanoparticles alone, where the inhibition rate of Ni nanoparticles alone to cancer cells is only 9.8%, but the relevant inhibition efficiency was improved from 13.8% (25 μmol/L of quercetin) to 36.7% (25 μmol/L of quercetin in the presence of Ni nanoparticles), and from 28.3% (50 μmol/L of quercetin) to 46.9% (50 μmol/L of quercetin in the presence of Ni nanoparticles). Thet-test results suggest that it has great statistical difference when compared to the controls (p < 0.05). These results show that quercetin together with Ni nanoparticles could inhibit cancer cell proliferation more efficiently than that caused by the single addition of quercetin and Ni nanoparticles, which indicates that Ni nanoparticles and quercetin have synergic effect on inhibiting the proliferation of cancer cells.
Figure 6.
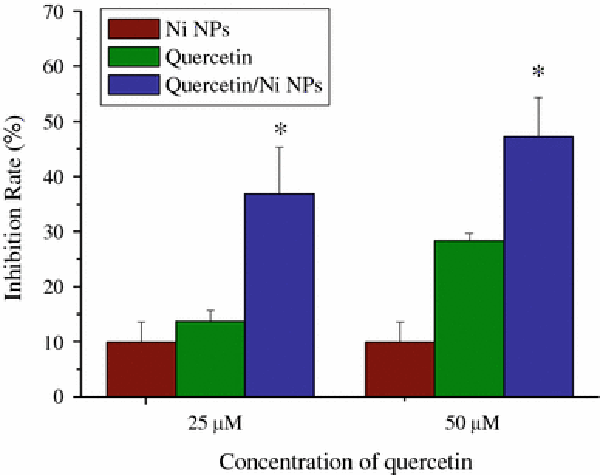
MTT assay on the inhibition rate of SMMC-7721 cells after treatment with quercetin together with Ni nanoparticles, which indicates significant inhibition effect of quercetin in the presence of Ni nanoparticles on SMMC-7721 cells in comparison with either quercetin or Ni nanoparticles, where * representsp < 0.05.Error barsindicate standard deviation
The rational behind this may be attributed to that the functionalized Ni nanoparticles could efficiently improve the penetration of the drug quercetin into the cell membrane, i.e., appropriate concentration of Ni nanoparticles can effectively facilitate the permeation and uptake of quercetin and increase the accumulation of quercetin in cancer cells. Hence, it appears that when SMMC-7721 cells incubated with quercetin exposed to Ni nanoparticles, it shows greatly efficient quercetin uptake via introduction by functionalized Ni nanoparticles.
RT-CES Assay
Unlike traditional end-point assays the RT-CES readout of impedance is non-invasive and could continuously and quantitatively provide a dynamic measurement of the cytotoxic activity. The time resolution in the assay processes provides high content information regarding the extent of the cytotoxicity in addition to the exact time, while the cytotoxicity takes place at different effect to ratio. Cell Index (CI) is used to represent cell status based on the measured electrical impedance [22]. The calculation of frequency dependent electrode impedance with or without cells present in the wells and corresponding CI value has been described in the literature [22]. In general, under the same physiologic conditions, CI value depends on the number of cells attached to the electrodes: If no cells are present on the electrodes, CI value is 0. The more cells attached to the electrodes (e.g., an increase in cell adhesion or cell spread), the higher the CI value obtained. All the factors that affect the number of cells will result in the change in CI value, e.g., cell proliferation will induce more cells to attach to the sensors and lead to a higher CI value. On the other hand, cell death or toxicity induced cell detachment will result in a lower CI value.
Based on these considerations, the effect of cellular interaction with Ni nanoparticles and/or quercetin have been further explored with RT-CES assay, and our results are consistent with those of our microscopy and electrochemical studies as well as MTT assay. As shown in Fig. 7, little effect was observed for 2.0 μg/mL of Ni nanoparticles on SMMC-7721 cancer cells (curve B) compared to the control cells alone (curve A). However, 50 μmol/L of quercetin (curve C) can greatly inhibit the cell proliferation in vitro. When 50 μmol/L of quercetin and 2.0 μg/mL of Ni nanoparticles were coapplied to the system, the cytotoxicity suppression of quercetin on cancer cells was further enhanced (curve D), which was well in agreement with that determined by MTT assay. These observations demonstrate that the synergistic effect on cell proliferation suppression between Ni nanoparticles and quercetin could take place, i.e., Ni nanoparticles could efficiently facilitate the cellular drug uptake into the cancer cells to increase the drug accumulation and thus inhibit the cell proliferation.
Figure 7.
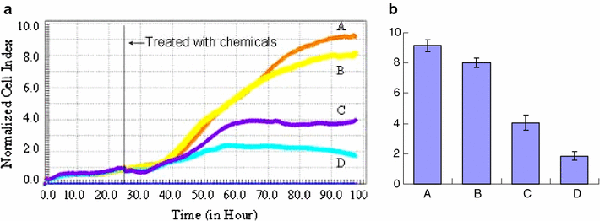
aDynamic response of SMMC-7721 cells exposure to: (A) chemical-free (control); (B) 2.0 μg/mL of Ni nanoparticles; (C) 50 μmol/L of quercetin and (D) 50 μmol/L of quercetin together with 2.0 μg/mL of Ni nanoparticles.bThe relevant cell index after treatment with chemicals for 72 h
Conclusion
In summary, in this study the cellular effect and in vitro cytotoxicity of SMMC-7721 cancer cells treated with anticancer agents accompanying with functionalized Ni nanoparticles capped with positively charged tetraheptylammonium have been investigated. The morphologies of SMMC-7721 cancer cells in response to various treatments have been explored by microscopy techniques. The enhancement effect of these Ni nanoparticles on the drug uptake of quercetin on SMMC-7721 cancer cells has been observed and their relevant effects on cell proliferation have been evaluated by MTT and RT-CES assay. The results suggest that Ni nanoparticles and quercetin have synergic effect on SMMC-7721 cells, and Ni nanoparticles can efficiently enhance the permeation and uptake of quercetin into cancer cells, implying the great potential of Ni nanoparticles in cancer biomedical and chemotherapeutic applications.
Acknowledgments
We gratefully acknowledge the support from the National Natural Science Foundation of China (90713023, 20675014, 20535010), National Basic Research Program of China (No. 2010CB732404), Ministry of Science & Technology of China (2007AA022007), and the Natural Science Foundation of Jiangsu Province (BK2008149).
References
- Kamat PV. Chem. 1993. p. 267. COI number [1:CAS:528:DyaK3sXmvFOnsA%3D%3D] [DOI]
- Lewis LN. Chem. 1993. p. 2693. COI number [1:CAS:528:DyaK3sXmsFOhs7Y%3D] [DOI]
- Lin XM, Sorensen CM, Klabunde KJ, Hadjipanayis GC. Langmuir. 1998. p. 7140. COI number [1:CAS:528:DyaK1cXntlansr8%3D] [DOI]
- Wang G, Huang T, Murray RW, Menard L, Nuzzo RG. J. 2005. p. 812. COI number [1:CAS:528:DC%2BD2MXkvVOmsQ%3D%3D] [DOI] [PubMed]
- Allen TM, Cullis PR. Science. 2004. p. 1818. COI number [1:CAS:528:DC%2BD2cXitFehu70%3D]; Bibcode number [2004Sci...303.1818A] [DOI] [PubMed]
- Everts M, Saini V, Leddon JL, Kok RJ, Stoff-Khalili M, Preuss MA, Millican CL, Perkins G, Brown JM, Bagaria H, Nikles DE, Johnson DT, Zharov VP, Curiel DT. Nano. 2006. p. 587. COI number [1:CAS:528:DC%2BD28Xit1OrsLs%3D]; Bibcode number [2006NanoL...6..587E] [DOI] [PubMed]
- Jain TK, Morales MA, Sahoo SK, Leslie-Pelecky DL, Labhasetwar V. Mol. 2005. p. 194. COI number [1:CAS:528:DC%2BD2MXjtl2jt7Y%3D] [DOI] [PubMed]
- Sonvico F, Mornet S, Vasseur S, Dubernet C, Jaillard D, Degrouard J, Hoebeke J, Duguet E, Colombo P, Couvreur P. Bioconjugate Chem. 2005. p. 1181. COI number [1:CAS:528:DC%2BD2MXpsleqsrs%3D] [DOI] [PubMed]
- Tiefenauer LX, Kuhne G, Andres RY. Bioconjugate Chem. 1993. p. 347. COI number [1:CAS:528:DyaK3sXlslKktrY%3D] [DOI] [PubMed]
- Duarte J, Perez-Palencia R, Vargas F, Ocete MA, Perez-Vizcaino F, Zarzuelo A. Br. 2001. p. 117. COI number [1:CAS:528:DC%2BD3MXjs1ais70%3D] [DOI] [PMC free article] [PubMed]
- Formica JV, Regelson W. Food Chem. 1995. p. 1061. COI number [1:CAS:528:DyaK28Xht1Cgsw%3D%3D] [DOI] [PubMed]
- Hertog MGL Holland PCH Eur 19965063.COI number [1:STN:280:DyaK283hvFeitQ%3D%3D]8641249
- Yeon JK, Yong CB, Kuen TS, Jin SJ. Biochem. 2006. p. 1268. [DOI] [PubMed]
- Huang SL, Hsu CL, Yen GC. Life Sci. 2006. p. 203. COI number [1:CAS:528:DC%2BD28XkvVGis7k%3D] [DOI] [PubMed]
- Kang TB, Liang NC. Biochem. 1997. p. 1013. COI number [1:CAS:528:DyaK2sXntFelt7Y%3D] [DOI] [PubMed]
- Tan WF, Lin LP, Li MH, Zhang YX, Tong TG, Xiao D. Eur. 2003. p. 255. COI number [1:CAS:528:DC%2BD3sXotVGh] [DOI] [PubMed]
- Ana MO, Victor CD. Bioelectrochemistry. 2004. p. 133. [DOI] [PubMed]
- Ana MO, Victor CD. Bioelectrochemistry. 2004. p. 143. [DOI] [PubMed]
- Howard PH, Arlen DK, Craig EL. J. 1994. p. 325. [DOI]
- Kilmartin PA, Zou H, Waterhouse AL. J. 2001. p. 1957. COI number [1:CAS:528:DC%2BD3MXhs1ymsrk%3D] [DOI] [PubMed]
- Xing JZ, Zhu LJ, Gabos S, Xie L. Toxicol. In Vitro. 2006. p. 995. COI number [1:CAS:528:DC%2BD28Xmt1Oru70%3D] [DOI] [PubMed]
- Xing JZ, Zhu L, Jackson JA, Gabos S, Sun XJ, Wang XB, Xu X. Chem. 2005. p. 154. COI number [1:CAS:528:DC%2BD2MXps1Ogtw%3D%3D] [DOI] [PubMed]
- Zhu J, Wang XB, Xu X, Abassi YA. J. Immunol. Methods. 2006. p. 25. COI number [1:CAS:528:DC%2BD28XhtlGmsLY%3D] [DOI] [PubMed]
- Mossman TJ. Immunol. Methods. 1983. p. 55. [DOI]
- Guo D, Wu C, Li X, Jiang H, Wang X, Chen B. J. 2008. p. 2301. COI number [1:CAS:528:DC%2BD1cXmslWru78%3D] [DOI] [PubMed]
- Nifli AP, Theodoropoulos PA, Munier S, Castagnino C, Roussakis E, Katerinopoulos HE, Vercauteren J, Castanas E. J. 2007. p. 2873. COI number [1:CAS:528:DC%2BD2sXjsVOrsLo%3D] [DOI] [PubMed]
- Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, Balogh L, Orr BG, Baker JR, Holl MB. Bioconjugate Chem. 2004. p. 774. COI number [1:CAS:528:DC%2BD2cXlsF2isbg%3D] [DOI] [PubMed]
- Hong S, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MB. Bioconjugate Chem. 2006. p. 728. COI number [1:CAS:528:DC%2BD28XktVSksr0%3D] [DOI] [PubMed]
- Mecke A, Lee DK, Ramamoorthy A, Orr BG, Holl MB. Biophys. 2005. p. 4043. COI number [1:CAS:528:DC%2BD2MXhtlWms77K] [DOI] [PMC free article] [PubMed]


