Abstract
Constituents of tobacco smoke can cause DNA double-strand breaks (DSBs), leading to tumorigenesis. The NBS1 gene product is a vital component in DSB detection and repair, thus genetic variations may influence cancer development. We examined the associations between NBS1 polymorphisms and haplotypes and newly incident smoking-related cancers in three case–control studies (Los Angeles: 611 lung and 601 upper aero-digestive tract (UADT) cancer cases and 1040 controls; Memorial Sloan-Kettering Cancer Center: 227 bladder cancer cases and 211 controls and Taixing, China: 218 esophagus, 206 stomach, 204 liver cancer cases and 415 controls). rs1061302 was associated with cancers of the lung [adjusted odds ratio (ORadj) = 1.6, 95% confidence interval (CI): 1.2, 2.4], larynx (ORadj = 0.56, 95% CI: 0.32, 0.97) and liver (ORadj = 1.7, 95% CI: 1.0, 2.9). Additionally, positive associations were found for rs709816 with bladder cancer (ORadj = 4.2, 95% CI: 1.4, 12) and rs1063054 with lung cancer (ORadj = 1.6, 95% CI: 1.0, 2.3). Some associations in lung and stomach cancers varied with smoking status. CAC haplotype was positively associated with smoking-related cancers: lung (ORadj = 1.7, 95% CI: 1.1, 2.9) and UADT (ORadj = 2.0, 95% CI: 1.1, 3.7), specifically, oropharynx (ORadj = 2.1, 95% CI: 1.0, 4.2) and larynx (ORadj = 4.8, 95% CI: 1.7, 14). Bayesian false-discovery probabilities were calculated to assess Type I error. It appears that NBS1 polymorphisms and haplotypes may be associated with smoking-related cancers and that these associations may differ by smoking status. Our findings also suggest that single-nucleotide polymorphisms located in the binding region of the MRE-RAD50-NBS1 complex or microRNA targeted pathways may influence tumor development. These hypotheses should be further examined in functional studies.
Introduction
Tobacco smoke is associated with many cancers (1) and is known to contain >80 definite, probable or possible carcinogenic substances (1,2), which can induce DNA single-strand breaks and double-strand breaks (DSBs) (3). DSBs pose a major threat to a cell’s genomic integrity (4). Unrepaired or defectively repaired chromosomal irregularities may lead to cell apoptosis or tumorigenesis (5).
The NBS1 (HUGO name: NBN) gene, a component of the MRE–RAD50–NBS1 (MRN) complex, plays a critical role in the DSB repair pathway, functioning both in the non-homologous end joining pathway as a sensor for DNA damage and in the homologous recombination pathway by participating in DNA repair and the intra-S phase cell cycle checkpoint (6,7). Increased tumor expression of NBS1 has been found in smoking-related cancer sites including lung, liver, esophagus and head and neck (8,9). Studies in mice have shown that mice heterozygous for NBS1-null mutations develop tumors affecting the lung, liver, mammary gland and prostate (10,11). In humans, rare mutations in NBS1 cause Nijmegen Breakage Syndrome, a disorder resulting in microcephaly, immunodeficiency, chromosome instability and increased risk of cancer (12,13). Thus, it could be hypothesized that more common genetic variations in NBS1 could influence cancer development; to date, at least 15 studies have investigated this potential association (14–28).
A meta-analysis of the commonly investigated rs1805794 polymorphism, a nonsynonymous coding single-nucleotide polymorphism (SNP), showed a slight positive association between all cancers and the variant genotype [odds ratio (OR) = 1.06, 95% confidence interval (CI): 1.00, 1.12] (29). Given that the NBS1 gene spans ∼50 kb and codes for a 754 amino acid (a.a.) protein (6,30,31), it is possible that additional SNPs within the region may influence smoking-related cancer development. These associations may also be modified by smoking status, although this possibility has rarely been investigated in published literature. We used data from three case–control studies to investigate potential associations between NBS1 gene SNPs (rs709816, rs1061302 and rs1063054), its haplotypes and nine smoking-related cancer sites: lung, upper aero-digestive tract (UADT) (oropharynx, larynx, nasopharynx and esophagus), stomach, liver, bladder and kidney.
Materials and methods
Details regarding the Los Angeles (LA) (32,33); Taixing, China (34–36) and Memorial Sloan-Kettering Cancer Center (MSKCC) (37,38) studies were previously published. All study participants signed informed consents. All studies were approved by the Institutional Review Boards of the respective institutions.
The LA study was a population-based case–control study conducted from 1999 to 2004. Eligible participants were (i) residents of LA County at the time of recruitment (for controls) or diagnosis (for cases); (ii) 18–65 years of age at recruitment and (iii) fluent in English or Spanish. Newly diagnosed cases were identified using the rapid ascertainment system of the Cancer Surveillance Program for LA County (32). Controls were identified through a formal algorithm from a list of households within the neighborhood of each individual case and individually matched to cases on age (10 years intervals) and gender. Among eligible cases, recruitment rates were 39% (lung cancer) and 46% (UADT cancer). Seventy-nine percent of contacted eligible controls participated. Ultimately, the study enrolled 611 lung and 601 UADT cancer cases and 1040 population-based controls who did not have lung or UADT cancer.
The Taixing study was a population-based case–control study conducted in Taixing City, Jiangsu Province, China, from 1 June 2000 to 30 December 2000. Eligibility criteria included residency in Taixing City for ≥10 years and ≥20 years of age at recruitment. All newly diagnosed and pathologically or clinically confirmed esophageal, stomach and liver cancer cases were identified from the Taixing Tumor Registry at the Taixing Center for Disease Control and Prevention. Of those identified, 65% (stomach), 57% (liver) and 67% (esophagus) consented to participate in the study. Population-based healthy controls were randomly selected from a list of residents frequency matched to cases on gender, age group (5 years intervals) and residential village (or residential block in the city). The participation rate for controls was 89%. This study included 218 esophageal, 206 stomach and 204 liver cancer cases and 415 controls.
The MSKCC study was a hospital-based case–control study conducted from 1 August 1993 to 30 June 1997. Eligible bladder and kidney cancer cases were newly diagnosed or undergoing a surgical procedure for their relevant cancer at MSKCC. Cases had a pathologically confirmed diagnosis were in stable medical condition and lived in the USA for at least 1 year. Ninety-five percent of cases agreed to participate. Hospital-based controls were recruited from the MSKCC blood bank or were patients with a negative cancer diagnosis at MSKCC who had resided in the USA for at least 1 year and were in stable medical condition. The participation rate for controls was 92%. During the 4 years study period, a total of 227 cases with bladder cancer, 30 cases with kidney cancer and 211 controls were interviewed.
Data and biological specimen collection
All data were collected in-person by trained interviewers. Study-specific standardized questionnaires ascertained information regarding demographic factors, cigarette smoking, passive smoking, alcohol consumption, history of occupational and environmental exposures, family history of cancer, dietary factors (food frequency questionnaire), medical history and questions regarding cancer site-specific environmental exposures.
Biological specimens were collected from interviewed participants in the forms of buccal cells (39) (LA study) and peripheral blood samples (Taixing and MSKCC studies). The percentages of interviewed participants providing buccal cells were 89% for controls and 89, 68, 88 and 90% for lung, oropharyngeal/nasopharyngeal, laryngeal and esophageal cancer cases, respectively. Blood samples in the Taixing study were obtained from 97.5% of controls, 95% of stomach and liver cancer cases and 94% of esophageal cancer cases. Eighty-one percent of bladder cancer cases, 67% of kidney cancer cases and 85% of MSKCC study controls provided blood samples. All biological specimens were transported and stored in freezers at −70°C in the Molecular Epidemiology Laboratory at the UCLA School of Public Health.
SNP selection and genotyping by TaqMan and SNPlex assays
Candidate SNPs in NBS1 were selected according to the following a priori criteria: (i) genomic context suggesting the possibility of functionality (i.e. located in an exon or 3′ untranslated region); (ii) previously reported minor allele frequency >5% in National Center for Biotechnology Information's database of single nucleotide polymorphism (40) and (iii) previously associated with a disease outcome including smoking-related cancer. Based on these criteria, five SNPs were selected and genotyped for this study.
All DNA samples were isolated from biological specimens using a modified phenol–chloroform method and assayed for purity and concentration by spectrometry (39). SNP genotyping was performed using TaqMan and SNPlex platforms from Applied Biosystems (ABI) (Foster City, CA). For both platforms, aliquots of cases’ and controls’ DNA were randomized onto polymerase chain reaction plates. All five SNPs were genotyped on the SNPlex platform (41,42). Detection was performed on the ABI Analyzer 3730 and data interpretation was performed with ABI Genemapper software v4.0. The UCLA Genotyping Core, when using the SNPlex genotyping assay, on average achieves call, reproducibility and concordance rates of 96, 99.7 and 99.8%, respectively.
SNPs rs1061302, rs1063053, rs1063054 and rs2735383 were also genotyped using TaqMan; details regarding the protocol were previously reported (43). End-point fluorescence was read using the ABI Primer 7900HT instrument and genotypes were scored using SDS 2.3 Allelic Discrimination Software. For quality control, 5% of randomly selected samples were re-genotyped to evaluate reproducibility; concordance was >99%. All SNPs had automatic call rates ≥96%. Laboratory researchers were blinded to the disease status and identity of quality control samples. For SNPs assayed on both platforms in our present study, we used the TaqMan genotyping results because of its higher call rates, which maximized the precision of our estimates. Concordance for the SNPs genotyped on both platforms had average Cohen's kappa coefficient of 0.919.
Preliminary analysis of data indicated that the genotypes of rs1063054, rs1063053 and rs2735383 were in high linkage disequilibrium (LD) (r2 > 0.99). We therefore only present the results for one of these SNPs (rs1063054) to avoid redundant reporting.
Statistical analysis
Unconditional logistic regression models were used to obtain ORs and 95% CIs with SAS v9.1.3 software (SAS Institute, Cary, NC). Tests for Hardy–Weinberg equilibrium and differences in variant allele frequencies were evaluated for SNPs using Pearson’s chi-squared test. For lung and UADT cancers (LA study), we adjusted for age (<34, 35–36, 37–38, 39–40, 41–42, 43–44, 45–46, 47–48, 49–50, 51–52, 53–54, 55–56, 57–58 and 59–62), gender, race/ethnicity (White, Asian American, African American, Mexican–American and other), educational level (years, continuous) and tobacco smoking (pack years, continuous). Controls who were >3 years younger than the youngest case or 3 years older than the oldest case were excluded from the analysis: 11 lung cancer controls and 1 UADT cancer control were excluded. For UADT cancers, alcohol drinking (drinks per day, continuous) was included in the statistical models. Analyses for the Taixing study were adjusted for age (continuous), gender, education (four level, ordinal), smoking (pack-years, continuous) and alcohol drinking (never, occasionally, often or every day). The regression model additionally included helicobacter pylori infection status or HBsAg status for stomach and liver cancer analyses, respectively. For bladder and kidney cancers, we adjusted for gender, age (<55, 55 to <60, 60 to <65 and ≥65), race (White/non-White), smoking (never/ever) and years of education (continuous). If the direction of associations was consistent across all cancer sites, we pooled data from all studies to examine the association between NBS1 SNPs and any smoking-related cancer, adjusting for study location/ethnicity, age (continuous), gender and smoking status. We used varying cut-points for adjustment of potential confounding variables determined from our previous publications (32–38) and the different distributions of the covariates in the control populations for each specific cancer sites. However, when using the same cut-points for potential confounding factors, the observed associations were consistent.
We evaluated the associations of SNPs and cancers according to unrestricted genotypes and genotypes following three genetic models (log-additive, recessive and dominant). For all SNPs, the ancestral allele as indicated by database of single nucleotide polymorphism (40,44) was considered wild-type and the homozygous wild-type genotype was considered the referent genotype. ORs for these SNPs across levels of tobacco smoking were evaluated using unconditional logistic regression, adjusting for the previously mentioned covariates. Tests for interaction between each SNP and smoking status on the multiplicative scale were conducted using the likelihood ratio test to compare the fit of the full regression model to the full regression model with the product of SNP genotypes and smoking status (ever versus never). To measure departures from multiplicativity, the ratio of adjusted odds ratios (RORadj) was calculated by dividing the joint OR [genotype and smoking adjusted odds ratio (ORadj)] by the product of the ORadj for separate ORs (genotype ORadj × smoking ORadj).
Haplotype analyses were conducted using HPlus v2.5 (45,46). Haplotypes are presented in the order of 5′–3′ (rs709816, rs1061302 and rs1063054) and variant alleles within the haplotypes are specified by an underline. The most common haplotype served as the referent, which happened to be the wild-type allele for all three SNPs. On account of limited DNA, rs709816 was not genotyped in the Taixing study. Therefore, the haplotypes for the esophagus, stomach and liver cancer sites are presented with two alleles.
To account for false-positive findings due to multiple hypothesis testing, we calculated the Bayesian false-discovery probability (BFDP) (47). Due to the likelihood that the DSB repair pathway plays a role in smoking-related carcinogenesis, we considered a prior probability range from 0.01 to 0.10, an ORadj of 1.5 between cancer and genotype, and an ORadj of 2.5 for associations stratified by smoking status. To determine noteworthy findings, the BFDP threshold was set to 0.8, where false non-discovery rate is four times as costly as a false discovery.
Results
Baseline characteristics of the study participants are presented in Table I. The LA study was the most ethnically heterogeneous. The percentage of self-reported smoking was greatest among lung and bladder cancer cases. The allelic distributions for all investigated NBS1 SNPs among the control populations did not vary much between racial/ethnic groups, with the exception of rs709816, where among non-White populations, the variant allele was more frequently distributed (supplementary Table I is available at Carcinogenesis Online). The distribution of rs709816 among the White MSKCC control population deviated from Hardy–Weinberg equilibrium (P = 0.023) but the remaining SNPs were in Hardy–Weinberg equilibrium among all study controls. SNPs rs709816 and rs1061302 are synonymous and rs1063054, as well as the two SNPs in LD with rs1063054 (rs1063053 and rs2735383), are located in the 3′ untranslated region.
Table I.
Baseline characteristics of cases and controls, stratified by study
| LA study |
Taixing study |
MSKCC study |
||||||||
| Lung | UADT | Controls | Esophagus | Stomach | Liver | Controls | Bladder | Kidney | Controls | |
| Total, N | 611 | 601 | 1040 | 218 | 206 | 204 | 415 | 227 | 30 | 211 |
| Mean age | 52.2 | 50.4 | 49.9 | 60.6 | 61.5 | 53.9 | 57.7 | 64.3 | 59.5 | 41.8 |
| Gender (%) | ||||||||||
| Male | 303 (49.6) | 454 (75.5) | 623 (59.9) | 141 (64.7) | 138 (67.0) | 159 (77.9) | 287 (69.2) | 189 (83.3) | 21 (70.0) | 152 (72.0) |
| Female | 308 (50.4) | 147 (24.5) | 417 (40.1) | 77 (35.3) | 68 (33.0) | 45 (22.1) | 128 (30.8) | 38 (16.7) | 9 (30.0) | 46 (21.8) |
| Missing | — | — | — | — | — | — | — | — | — | 13 (6.2) |
| Ethnicity (%) | ||||||||||
| White | 359 (58.8) | 341 (56.7) | 634 (61.0) | 206 (90.8) | 24 (80.0) | 191 (90.5) | ||||
| African American | 53 (8.7) | 70 (11.7) | 150 (14.4) | |||||||
| Mexican American | 96 (15.7) | 69 (11.5) | 102 (9.8) | |||||||
| Asian American | 70 (11.5) | 64 (10.7) | 62 (6.0) | |||||||
| Chinese | 218 (100.0) | 206 (100.0) | 204 (100.0) | 415 (100.0) | ||||||
| Other | 32 (5.2) | 55 (9.2) | 91 (8.8) | 16 (7.1) | 6 (20.0) | 6 (2.8) | ||||
| Missing | 1 (0.2) | 2 (0.3) | 1 (0.1) | — | — | — | — | 5 (2.2) | — | 14 (6.6) |
| Education (%) | ||||||||||
| ≤12 years | 265 (43.4) | 273 (45.4) | 300 (28.9) | 215 (98.6) | 203 (98.5) | 204 (100.0) | 405 (97.6) | 87 (38.3) | 12 (40.0) | 34 (16.1) |
| >12 years | 346 (56.6) | 328 (54.6) | 739 (71.1) | 0 (0.0) | 3 (1.5) | 0 (0.0) | 10 (2.4) | 139 (61.2) | 18 (60.0) | 177 (83.9) |
| Missing | — | — | 1 (0.1) | 3 (1.4) | — | — | — | 1 (0.4) | — | — |
| Smoking (%) | ||||||||||
| Never | 110 (18.0) | 182 (30.3) | 492 (47.3) | 94 (43.1) | 92 (44.7) | 85 (41.7) | 217 (52.3) | 39 (17.2) | 10 (33.3) | 107 (50.7) |
| Ever | 501 (82.0) | 419 (69.7) | 548 (52.7) | 117 (53.7) | 109 (52.9) | 107 (52.5) | 197 (47.5) | 184 (81.1) | 20 (66.7) | 89 (42.2) |
| Missing | — | — | — | 7 (3.2) | 5 (2.4) | 12 (5.9) | 1 (0.2) | 4 (1.8) | — | 15 (7.1) |
| Drinking (%) | ||||||||||
| Never | 170 (27.8) | 117 (19.5) | 264 (25.4) | 116 (53.2) | 111 (53.9) | 87 (42.7) | 207 (49.9) | 31 (13.7) | 9 (30.0) | 36 (17.1) |
| Ever | 440 (72.0) | 482 (80.2) | 772 (74.2) | 95 (43.6) | 90 (43.7) | 105 (51.5) | 205 (49.4) | 187 (82.4) | 21 (70.0) | 161 (76.3) |
| Missing | 1 (0.2) | 2 (0.3) | 4 (0.4) | 7 (3.2) | 5 (2.4) | 12 (5.9) | 3 (0.7) | 9 (4.0) | — | 14 (6.6) |
Table II presents the associations between NBS1 polymorphisms and cancer sites. We found that the rs1061302 heterozygous genotype was associated with lung cancer (ORadj = 0.74; 95% CI: 0.56, 0.98); however, a linear trend was not detected and we found a positive association between the homozygous variant and lung cancer (ORadj = 1.6; 95% CI: 1.0, 2.4). The rs1061302 variant allele was inversely associated with laryngeal cancer (ORadj = 0.56; 95% CI: 0.32, 0.97). The rs1063054 homozygous variant was associated with lung and liver cancers and rs709816 was positively associated with bladder cancer, suggesting recessive models of inheritance for these two SNPs. No robust associations were detected between NBS1 SNPs and cancers of the nasopharynx and kidney (data not shown).
Table II.
Associations between NBS1 SNPs and smoking-related cancers, stratified by cancer site
| SNP | rs709816 (D399D) |
rs1061302 (P672P) |
rs1063054 (3' UTR) |
|||||
| Case/control | ORadj (95% CI)a | Genotype | Case/control | ORadj (95% CI)a | Genotype | Case/control | ORadj (95% CI)a | |
| LA study | ||||||||
| Lung | ||||||||
| TT | 156/278 | 1.0 | AA | 265/407 | 1.0 | AA | 241/411 | 1.0 |
| TC | 230/421 | 0.88 (0.65, 1.2) | AG | 196/414 | 0.74 (0.56, 0.98) | AC | 226/427 | 0.88 (0.67, 1.2) |
| CC | 127/194 | 1.0 (0.70, 1.5) | GG | 72/95 | 1.6 (1.0, 2.4) | CC | 72/80 | 1.5 (0.96, 2.3) |
| Ptrend | 0.97 | Ptrend | 0.45 | Ptrend | 0.39 | |||
| Dominant | 357/615 | 0.92 (0.68, 1.2) | Dominant | 268/509 | 0.88 (0.68, 1.2) | Dominant | 298/507 | 0.96 (0.74, 1.2) |
| Recessive | 127/194 | 1.1 (0.81, 1.6) | Recessive | 72/95 | 1.8 (1.2, 2.7) | Recessive | 72/80 | 1.6 (1.0, 2.3) |
| UADT (squamous) | ||||||||
| TT | 107/283 | 1.0 | AA | 174/413 | 1.0 | AA | 170/415 | 1.0 |
| TC | 194/423 | 1.1 (0.81, 1.5) | AG | 184/415 | 1.0 (0.79, 1.3) | AC | 200/430 | 1.1 (0.86, 1.5) |
| CC | 80/196 | 0.95 (0.64, 1.4) | GG | 38/97 | 0.82 (0.52, 1.3) | CC | 33/82 | 0.95 (0.59, 1.5) |
| Ptrend | 0.90 | Ptrend | 0.60 | Ptrend | 0.60 | |||
| Dominant | 274/619 | 1.1 (0.79, 1.4) | Dominant | 222/512 | 0.99 (0.76, 1.3) | Dominant | 233/512 | 1.0 (0.84, 1.4) |
| Recessive | 80/196 | 0.89 (0.64, 1.3) | Recessive | 38/97 | 0.81 (0.52, 1.2) | Recessive | 33/82 | 0.89 (0.57, 1.4) |
| UADT stratified | ||||||||
| Oropharynx | ||||||||
| TT | 63/283 | 1.0 | AA | 97/413 | 1.0 | AA | 94/415 | 1.0 |
| TC | 117/423 | 1.2 (0.83, 1.7) | AG | 114/415 | 1.2 (0.84, 1.6) | AC | 119/430 | 1.2 (0.86, 1.6) |
| CC | 45/196 | 1.0 (0.65, 1.7) | GG | 24/97 | 0.96 (0.56, 1.7) | CC | 27/82 | 1.4 (0.81, 2.3) |
| Ptrend | 0.73 | Ptrend | 0.74 | Ptrend | 0.19 | |||
| Dominant | 162/619 | 1.2 (0.82, 1.7) | Dominant | 138/512 | 1.1 (0.82, 1.5) | Dominant | 146/512 | 1.2 (0.88, 1.6) |
| Recessive | 45/196 | 0.92 (0.62, 1.4) | Recessive | 24/97 | 0.89 (0.54, 1.5) | Recessive | 27/82 | 1.2 (0.75, 2.0) |
| Larynx | ||||||||
| TT | 24/283 | 1.0 | AA | 41/413 | 1.0 | AA | 36/415 | 1.0 |
| TC | 35/423 | 0.65 (0.34, 1.2) | AG | 28/415 | 0.54 (0.30, 0.98) | AC | 39/430 | 0.91 (0.53, 1.6) |
| CC | 17/196 | 0.49 (0.21, 1.1) | GG | 7/97 | 0.62 (0.24, 1.6) | CC | 2/82 | 0.25 (0.05, 1.2) |
| Ptrend | 0.080 | Ptrend | 0.079 | Ptrend | 0.16 | |||
| Dominant | 52/619 | 0.60 (0.33, 1.1) | Dominant | 35/512 | 0.56 (0.32, 0.97) | Dominant | 41/512 | 0.80 (0.47, 1.4) |
| Recessive | 17/196 | 0.66 (0.33, 1.3) | Recessive | 7/97 | 0.82 (0.32, 2.1) | Recessive | 2/82 | 0.28 (0.06, 1.2) |
| MSKCC | ||||||||
| Bladder | ||||||||
| TT | 40/63 | 1.0 | AA | 75/85 | 1.0 | AA | 90/88 | 1.0 |
| TC | 79/58 | 1.0 (0.43, 2.4) | AG | 83/58 | 0.97 (0.48, 2.0) | AC | 76/60 | 1.3 (0.64, 2.6) |
| CC | 29/29 | 4.2 (1.3, 14) | GG | 19/24 | 1.7 (0.56, 5.4) | CC | 10/20 | 2.3 (0.57, 9.2) |
| Ptrend | 0.037 | Ptrend | 0.50 | Ptrend | 0.24 | |||
| Dominant | 108/87 | 1.5 (0.68, 3.2) | Dominant | 102/82 | 1.1 (0.56, 2.1) | Dominant | 86/80 | 1.4 (0.71, 2.7) |
| Recessive | 29/29 | 4.2 (1.4, 12) | Recessive | 19/24 | 1.8 (0.59, 5.3) | Recessive | 10/20 | 2.1 (0.54, 8.1) |
| Taixing studyb | ||||||||
| Esophagus | ||||||||
| AA | 72/140 | 1.0 | AA | 74/152 | 1.0 | |||
| AG | 93/169 | 0.97 (0.63, 1.5) | AC | 95/178 | 1.0 (0.78, 1.6) | |||
| GG | 36/69 | 1.0 (0.59, 1.8) | CC | 32/52 | 1.4 (0.78, 2.5) | |||
| Ptrend | 0.89 | Ptrend | 0.33 | |||||
| Dominant | 129/238 | 0.98 (0.66, 1.5) | Dominant | 127/230 | 1.1 (0.75, 1.7) | |||
| Recessive | 36/69 | 1.0 (0.63, 1.7) | Recessive | 32/52 | 1.4 (0.80, 2.3) | |||
| Stomach | ||||||||
| AA | 54/140 | 1.0 | AA | 66/152 | 1.0 | |||
| AG | 96/169 | 1.3 (0.82, 2.0) | AC | 97/178 | 1.2 (0.77, 1.8) | |||
| GG | 38/69 | 1.2 (0.70, 2.2) | CC | 26/52 | 1.2 (0.63, 2.2) | |||
| Ptrend | 0.37 | Ptrend | 0.61 | |||||
| Dominant | 134/238 | 1.3 (0.82, 2.0) | Dominant | 123/230 | 1.2 (0.79, 1.8) | |||
| Recessive | 38/69 | 1.1 (0.65, 1.7) | Recessive | 26/52 | 1.1 (0.60, 1.9) | |||
| Liver | ||||||||
| AA | 52/140 | 1.0 | AA | 67/152 | 1.0 | |||
| AG | 94/169 | 1.2 (0.75, 2.0) | AC | 90/178 | 1.1 (0.72, 1.8) | |||
| GG | 39/69 | 1.9 (1.1, 3.5) | CC | 32/52 | 1.8 (0.98, 3.4) | |||
| Ptrend | 0.034 | Ptrend | 0.092 | |||||
| Dominant | 133/238 | 1.4 (0.89, 2.2) | Dominant | 122/230 | 1.3 (0.83, 1.9) | |||
| Recessive | 39/69 | 1.7 (1.0, 2.9) | Recessive | 32/52 | 1.7 (0.96, 3.0) | |||
UTR, untranslated region.
LA study: lung cancer adjusted for gender, smoking, education, race and age; UADT cancers additionally adjusted for alcohol drinking; MSKCC Study: bladder cancers adjusted for gender, smoking, race, education and age. Taixing study: esophageal cancer adjusted for gender, smoking, age, alcohol drinking and education; stomach and liver cancers additionally adjusted for helicobacter pylori infection and HBsAg status, respectively.
rs709816 (D399) not genotyped in Taixing study.
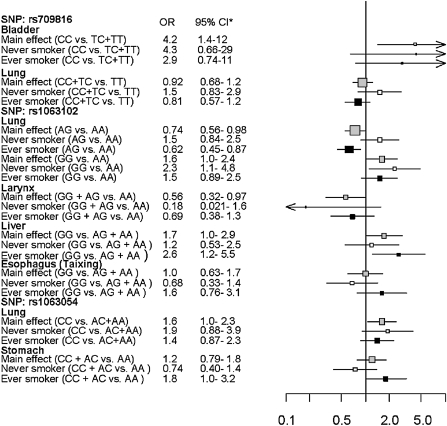
Associations stratified by smoking status are presented in Figure 1; supplementary Table II (available at Carcinogenesis Online). Across all cancer sites, a pooled estimate among never-smokers indicated that rs709816 was positively associated with smoking-related cancers under the dominant inheritance model (ORadj = 1.4; 95% CI: 0.98, 2.0) (data not shown). The inverse association between the heterozygous rs1061302 genotype and lung cancer persisted in ever-smokers (ORadj = 0.62; 95% CI: 0.45, 0.87) (Figure 1), and the estimated OR for this genotype was lower for smokers than for never smokers (RORadj = 0.41; 95% CI: 0.22, 0.77) (Table III); test for heterogeneity of ORadj: P = 0.021. The change in the lung cancer OR with smoking status among homozygous variants was similar but imprecise (RORadj = 0.56; 95% CI: 0.24, 1.4). Noteworthy changes in ORs were also found when using the dominant model for both rs709816 and smoking with lung cancer and for rs1063054 and smoking with stomach cancer (Pinteraction = 0.024 and 0.046, respectively).
Fig. 1.
Selected associations between NBS1 SNPs and smoking-related cancer sites, stratified by smoking status. Grey boxes represent association between SNP and cancer site, white boxes represent findings for never-smokers and black boxes represent findings for ever-smokers. *LA study: lung cancer adjusted for gender, smoking, education, race and age; UADT cancers additionally adjusted for alcohol drinking; MSKCC study: bladder cancer adjusted for gender, smoking, race, education and age. Taixing study: esophageal cancer adjusted for gender, smoking, age, alcohol drinking and education; stomach and liver cancers additionally adjusted for helicobacter pylori infection and HBsAg status, respectively.
Table III.
Modifications of SNPs and smoking status on the association of selected SNPs and smoking-related cancer sites
| Cases/controls |
OR (95% CI)a |
||||||
| Lung | |||||||
| rs1061302 | Genotype | A/A | A/G | G/G | A/A | A/G | G/G |
| Smoking status | Never | 34/194 | 39/189 | 19/47 | 1.0 | 1.4 (0.82, 2.4) | 2.4 (1.2, 4.9) |
| Ever | 231/213 | 157/225 | 53/48 | 1.7 (1.0, 2.8) | 1.0 (0.60, 1.7) | 2.3 (1.2, 4.3) | |
| Genotype-specific RORadj (95% CI) | 0.41 (0.22, 0.77)b | 0.56 (0.24, 1.4)c | |||||
| Genotype-combined RORadj (95% CI) | 1.0 (0.92, 1.2)d | ||||||
| rs709816 | T/T | T/C and C/C | T/T | T/C and C/C | |||
| Smoking status | Never | 17/137 | 72/285 | 1.0 | 1.6 (0.90, 3.0) | ||
| Ever | 139/141 | 285/330 | 2.0 (1.1, 3.8) | 1.5 (0.82, 2.8) | |||
| RORadj (95% CI) | 0.46 (0.23, 0.90) | ||||||
| Stomach | |||||||
| rs1063054 | A/A | A/C & C/C | A/A | A/C & C/C | |||
| Smoking status | Never | 33/71 | 51/125 | 1.0 | 0.77 (0.43, 1.4) | ||
| Ever | 30/80 | 70/105 | 0.64 (0.27, 1.5) | 1.1 (0.50, 2.5) | |||
| RORadj (95% CI) | 2.3 (1.0, 5.3) | ||||||
Lung cancer adjusted for gender, smoking, education, race and age; stomach cancer adjusted for gender, smoking, age, alcohol drinking, helicobacter pylori infection and education.
RORadj (95% CI) for rs1061302 A/G genotype and smoking, with A/A plus never-smoker as referent.
RORadj (95% CI) for rs1061302 G/G genotype and smoking, with A/A plus never-smoker as referent.
RORadj (95% CI) for rs1061302 genotypes-combined and smoking, with A/A plus never-smoker as referent.
The LD estimates for the SNPs in our study are as follows: r2 = 0.55 for rs709816 and rs1061302, r2 = 0.25 for rs709816 and rs1063054 and r2 = 0.43 for rs1061302 and rs1063054. We found that the CGC haplotype was inversely associated with laryngeal cancer ORadj = 0.45 (95% CI: 0.23, 0.86) (Table IV). The CAC haplotype was positively associated with cancers of the lung, UADT, oropharynx and larynx. In never-smokers, the CGC haplotype was positively associated with lung cancer, whereas among ever-smokers, the GA and GC haplotypes were positively associated with liver and stomach cancers, respectively. Additional NBS1 haplotype and smoking-related cancer findings are presented in supplementary Table III (available at Carcinogenesis Online). Because of the multiple hypotheses that were tested, we considered the BFDP. We found that when using a prior probability of 10%, 11 of the observed associations remained noteworthy (supplementary Table IV is available at Carcinogenesis Online). When we reduced the prior probability to 5%, the associations between lung cancer and the rs1061302 heterozygous variant (BFDP = 0.65) and the CAC haplotype (BFDP = 0.71) among ever-smokers remained noteworthy.
Table IV.
Selective associations between NBS1 haplotypesa and smoking-related cancers
| Haplotypea | All |
Never-smoker |
Ever-smoker |
|||||||||
| Frequency |
Frequency |
Frequency |
||||||||||
| Cases | Controls | ORadj (95% CI)b | Pvalue | Cases | Controls | ORadj (95% CI)b | Pvalue | Cases | Controls | ORadj (95% CI)b | Pvalue | |
| LA study | ||||||||||||
| Lung | ||||||||||||
| TAA | 0.45 | 0.49 | 1.0 | 0.39 | 0.50 | 1.0 | 0.47 | 0.48 | 1.0 | |||
| CGC | 0.23 | 0.25 | 1.0 (0.81, 1.3) | 0.91 | 0.30 | 0.24 | 1.6 (1.0, 2.4) | 0.031 | 0.22 | 0.25 | 0.83 (0.53, 1.3) | 0.40 |
| CAA | 0.14 | 0.11 | 1.3 (0.96, 1.7) | 0.098 | 0.16 | 0.10 | 1.4 (0.84, 2.3) | 0.203 | 0.13 | 0.12 | 1.26 (0.90, 1.8) | 0.19 |
| CAC | 0.045 | 0.016 | 1.7 (1.1, 2.9) | 0.033 | 0.015 | 0.014 | 0.67 (0.12, 3.6) | 0.647 | 0.052 | 0.019 | 2.2 (1.3, 3.8) | <0.01 |
| UADT | ||||||||||||
| TAA | 0.47 | 0.50 | 1.0 | 0.46 | 0.50 | 1.0 | 0.48 | 0.48 | 1.0 | |||
| CAC | 0.036 | 0.017 | 2.0 (1.1, 3.7) | 0.021 | 0.038 | 0.014 | 2.4 (0.93, 6.2) | 0.069 | 0.036 | 0.019 | 1.9 (0.87, 3.9) | 0.11 |
| Oropharyx | ||||||||||||
| TAA | 0.47 | 0.50 | 1.0 | 0.48 | 0.50 | 1.0 | 0.47 | 0.49 | 1.0 | |||
| CAC | 0.030 | 0.017 | 2.1 (1.0, 4.2) | 0.039 | 0.014 | 0.014 | 1.2 (0.26, 5.6) | 0.82 | 0.040 | 0.019 | 2.7 (1.2, 6.1) | 0.021 |
| Larynx | ||||||||||||
| TAA | 0.51 | 0.50 | 1.0 | 0.33 | 0.50 | 1.0 | 0.52 | 0.49 | 1.0 | |||
| CGC | 0.17 | 0.25 | 0.45 (0.23, 0.86) | 0.016 | 0.056 | 0.24 | 0.36 (0.05, 2.37) | 0.29 | 0.19 | 0.25 | 0.52 (0.28, 0.97) | 0.041 |
| CAC | 0.064 | 0.017 | 4.8 (1.7, 14) | <0.01 | <0.01 | 0.014 | — | 0.12 | 0.051 | 0.019 | 3.7 (1.2, 11) | 0.020 |
| MSKCC | ||||||||||||
| Bladder | ||||||||||||
| TAA | 0.48 | 0.55 | 1.0 | 0.26 | 0.56 | 1.0 | 0.50 | 0.54 | 1.0 | |||
| CAA | 0.13 | 0.062 | 3.6 (1.0, 13) | 0.045 | 0.24 | 0.066 | 2.0 (0.55, 7.5) | 0.29 | <0.01 | <0.01 | — | |
| Taixing studyc | ||||||||||||
| Stomach | ||||||||||||
| AA | 0.51 | 0.56 | 1.0 | 0.49 | 0.51 | 1.0 | 0.52 | 0.60 | 1.0 | |||
| GC | 0.38 | 0.34 | 1.1 (0.85, 1.5) | 0.40 | 0.34 | 0.37 | 0.86 (0.56, 1.3) | 0.50 | 0.41 | 0.31 | 1.5 (0.99, 2.3) | 0.055 |
| GA | 0.086 | 0.070 | 1.3 (0.74, 2.2) | 0.39 | 0.11 | 0.078 | 1.3 (0.64, 2.8) | 0.43 | 0.068 | 0.062 | 1.2 (0.51, 2.6) | 0.72 |
| Liver | ||||||||||||
| AA | 0.52 | 0.56 | 1.0 | 0.47 | 0.53 | 1.0 | 0.55 | 0.60 | 1.0 | |||
| GC | 0.37 | 0.33 | 1.4 (0.98, 1.9) | 0.066 | 0.40 | 0.35 | 1.4 (0.89, 2.2) | 0.14 | 0.34 | 0.31 | 1.3 (0.81, 2.1) | 0.29 |
| GA | 0.087 | 0.067 | 1.6 (0.91, 2.8) | 0.11 | 0.088 | 0.076 | 1.1 (0.44, 2.6) | 0.88 | 0.086 | 0.058 | 2.3 (1.1, 4.8) | 0.025 |
LA study: Lung cancer adjusted for gender, smoking, education, race and age; UADT cancers additionally adjusted for alcohol drinking; MSKCC study: bladder cancer adjusted for gender, smoking, race, education and age. Taixing study: esophageal cancer adjusted for gender, smoking, age, alcohol drinking and education; stomach and liver cancers additionally adjusted for helicobacter pylori infection and HBsAg status, respectively.
Haplotypes in order of 5′–3′ (rs709816, rs1061302 and rs1063054).
Taixing study includes only two SNPs for haplotype analysis because rs709816 was not genotyped.
Discussion
We found that NBS1 polymorphisms and haplotypes are associated with incidence of smoking-related cancers and that these associations may be modified by smoking status, supporting the notion that genetic variation within NBS1 plays a role in tobacco-related carcinogenesis. To our knowledge, many of these relationships have not been previously reported for cancers of the stomach, liver and UADT subsites (esophagus, oropharynx, larynx and nasopharynx).
We observed positive associations with the CAC haplotype and cancers of the lung, oropharynx and larynx. In our study, we did not find consistent associations with rs709816 and rs1063054 in smoking-related cancer sites. However, haplotype analyses showed consistent positive associations across three of the four evaluated cancer sites, suggesting that either the variant C alleles in rs709816 and rs1063054 have a combined effect in tobacco-related tumorigenesis or that there is some other causal allele in LD with the CAC haplotype. In addition, when stratified by smoking, we found that this positive association persisted among ever-smokers. Given that the associations were observed in cancer sites with direct contact to smoke inhalation (such as the oral cavity and lung), the CAC haplotype may be involved in field cancerization (48).
To our knowledge, these two SNPs have rarely been investigated in smoking-related cancers (14). We are aware of one study investigating NBS1 haplotypes (rs1063045 and rs1805794, which are in LD with rs1061302) in lung cancer, in which the investigators found a positive association between lung cancer and the haplotype with recessive allele variants A and G (16). While it is still possible these findings are in error given the rarity of the haplotype, the CAC haplotype should be evaluated in studies with larger sample sizes and additional cancer sites, such as the esophagus, stomach and liver. The potential effect modification from smoking should be examined as well.
The potential functionality of rs709816 and rs1063054 remains unclear. However, the microRNA Target Site (PolymiRTS) database designates rs2735383, which is in LD (r2 > 0.99) with rs1063054, as one of the most probable targets for microRNA, miR-499 (49,50), offering the possibility that alleles at this locus may influence NBS1 gene regulation.
SNP rs1061302 was associated with cancers of the lung, larynx and liver in our study. Two NBS1 SNPs not included in our study, rs1805794 and rs1063045, were previously examined in relation to smoking-related cancers (14–20,22–24,26–28). LD estimates from the Haploview program (51) using HapMap data of the Centre d'Etude du Polymorphisme Humain (CEPH) population (52) showed that both of these SNPs are in high LD (r2 > 0.99) with each other and with rs1061302. A recent meta-analysis of four bladder cancer studies reported that the variant allele of rs1805794 was positively associated with bladder cancer (OR = 1.15; 95% CI: 1.01, 1.31) (53). A meta-analysis of three lung cancer studies detected no association with this cancer site (OR = 0.98; 95% CI: 0.51, 1.89) (53). Since rs1805794 is in high LD with rs1061302, we expected to find similar results in our study. For bladder cancer, our point estimates for heterozygote and homozygote variant allele carriers of rs1061302 were in the positive direction but the CIs included the null. Similar positive associations were seen for smokers and non-smokers. The large CIs may be due to small sample size.
Interestingly, we found that the association between rs1061302 and lung cancer differed by smoking status, which may explain why we observed associations where the previous meta-analysis did not. For never-smokers, there was a clear dose–response relationship for the variant allele of rs1061302 and lung cancer. For smokers, rs1061302 heterozygotes showed an inverse relationship with lung cancer. Thus, it is possible that these smoking-dependent associations were masked in the previous meta-analysis by combining smokers and non-smokers. Additionally, our BFDP correction suggests that the observed rs1061302 association in smokers and non-smokers were probably not due to false discoveries (supplementary Table II is available at Carcinogenesis Online).
There are no published reports between rs1061302 and laryngeal or liver cancer. We observed that rs1061302 was inversely associated with laryngeal cancer and positively associated with liver cancer. The different direction of associations may reflect modification by major risk factors. Hepatitis infections are the primary risk factors for liver cancer (54), whereas alcohol and smoking are considered risk factors for laryngeal and liver cancers (55). To date, however, alcohol has not been established as a known risk factor for lung cancer (56).
The associations between NBS1 polymorphisms and smoking-related cancers may be modified by smoking status, suggesting different tumorigenic pathways among smokers and never-smokers (57). Our interaction analyses (Table III) support the idea that smoking status modifies the rate ratio for the associations for rs1061302 and rs709816 with lung cancer and for rs1063054 with stomach cancer. A study among a cohort of smokers found that rs6998169 was inversely associated with gene methylation (58). Although our SNPs do not appear to be in LD with rs6998169, these findings suggest that genetic variation in NBS1 among smokers may alter the development of lung cancer by decreasing DNA methylation.
The NBS1 protein has three known functional regions: N-terminus (a.a. position 1–196), central region (a.a. position 278–343) and C-terminus (a.a. position 665–693) (59). The C-terminus is the binding site for the MRN complex (59) and DSBs cannot be detected in the non-homologous end joining pathway or repaired by the homologous recombination pathway without this complex (6,7). Given that rs1061302 codes for a.a. 672 in the C-terminus and that we found associations between rs1061302 and lung, larynx and liver cancers, SNPs in LD with rs1061302 or within this region may inadvertently affect proper binding of the MRN complex and alter its ability to accurately repair or detect DNA DSBs. Although rs1061302 is a synonymous polymorphism, the altered nucleotide may affect messenger RNA stability (60,61), splicing (62) or the translation rate (63). Confirmation studies are needed to determine the true ‘functional’ SNP.
Our candidate SNP selection approach, which relied heavily on previously reported SNPs, is a limitation of this study. We may have missed important genetic variations in NBS1. However, an analysis of SNP genotype data from the International HapMap project (52) suggests that the SNPs included in our study may be adequate proxies for the majority of genetic variation in NBS1. We used LDSelect (64,65) to bin SNPs in the NBS1 gene region (all exons, introns and 2 kb flanking sequence) that were in high LD (r2 ≥ 0.80) in the CEPH population. The rs1061302 SNP captured information on 31 other SNPs, rs1063054 captured information on 13 additional SNPs and rs709816 captured information on another 9 SNPs. Thus, of the 63 NBS1 SNPs with a minor allele frequency >5% genotyped in the HapMap project, our three selected SNPs captured information on ∼89% of the SNPs.
The small sample sizes or low participation rates observed for some of the cancer sites may be a consequence of low survival for many of these cancers (liver, stomach and esophagus) (66). If NBS1 polymorphisms are associated with survival, our study findings may be affected by survival bias. To our knowledge, only a breast cancer study investigated such an hypothesis and found no association among rs709816, rs1805794, rs1063045 and rs1061302 in breast cancer survival (P = 0.65, 0.24, 0.53 and 0.40, respectively) (67). Additionally, the small sample sizes of the Taixing City and MSKCC studies limited the precision of our estimates and assessment of effect modification by smoking status. Subsequent studies evaluating SNP function or potential SNP–SNP interactions between NBS1 and other genes in the non-homologous end joining or homologous recombination pathway may elucidate potential biological mechanisms.
Ours is the first study that we are aware of to investigate the association between NBS1 SNPs and haplotypes with nine smoking-related cancers. We had a range of racial/ethnic groups, enabling us to examine NBS1 SNPs in different populations, and our sample size was fairly large for lung and UADT cancer sites. We took several precautions to minimize genotype misclassification and our quality control assessment indicated a high degree of accuracy. We also used the BFDP correction to account for multiple comparisons. Our findings suggest that NBS1 is probably influential in the development of lung, UADT, stomach, liver and bladder cancers and that smoking status may alter the direction of the association. The association of the CAC haplotype with cancers of the lung, UADT, oropharynx and larynx indicates that this haplotype or SNPs in LD with this haplotype may alter tobacco smoking-related DSB repair ability. The role of SNPs that influence microRNA function or those that code for the C-terminus portion of NBS1 should be considered and evaluated in future studies.
Supplementary material
Supplementary Tables I–IV can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (ES06718, ES01167, CA90833, CA077954, CA09142, CA96134, DA11386); Seymour Family Gift for Innovative Investigator-Initiated Research in Bladder Cancer; Alper Research Center for Environmental Genomics of the University of California, Los Angeles Jonsson Comprehensive Cancer Center.
Supplementary Material
Acknowledgments
We are indebted to all studies’ participants for their time and dedication.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- a.a.
amino acid
- ORadj
adjusted odds ratio
- BFDP
Bayesian false-discovery probability
- CI
confidence interval
- DSB
double-strand break
- LA
Los Angeles
- LD
linkage disequilibrium
- MSKCC
Memorial Sloan-Kettering Cancer Center
- RORadj
ratio of adjusted odds ratio
- SNP
single-nucleotide polymorphism
- UADT
upper aero-digestive tract
References
- 1.IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco Smoke and Involuntary Smoking. Lyon: IARC; 2004. [PMC free article] [PubMed] [Google Scholar]
- 2.Smith CJ, et al. IARC carcinogens reported in cigarette mainstream smoke and their calculated log P values. Food. Chem. Toxicol. 2003;41:807–817. doi: 10.1016/s0278-6915(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 3.Pouget J-P, et al. General aspects of the cellular response to low- and high-LET radiation. Eur. J. Nucl. Med. 2001;28:541–561. doi: 10.1007/s002590100484. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer P, et al. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis. 2000;15:289–302. doi: 10.1093/mutage/15.4.289. [DOI] [PubMed] [Google Scholar]
- 5.Khanna KK, et al. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 6.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 7.Koberle B, et al. DNA repair and cancer. In: Knowles M, Selby P, editors. Introduction to the Cellular and Molecular Biology of Cancer. New York, NY: Oxford Press; 2005. pp. 61–77. [Google Scholar]
- 8.Chen Y-C, et al. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 2005;280:32505–32511. doi: 10.1074/jbc.M501449200. [DOI] [PubMed] [Google Scholar]
- 9.Yang M-H, et al. Increased NBS1 expression is a marker of aggressive head and neck cancer and overexpression of NBS1 contributes to transformation. Clin. Cancer Res. 2006;12:507–515. doi: 10.1158/1078-0432.CCR-05-1231. [DOI] [PubMed] [Google Scholar]
- 10.Dumon-Jones V, et al. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- 11.Zhang Y, et al. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006;16:45–54. doi: 10.1038/sj.cr.7310007. [DOI] [PubMed] [Google Scholar]
- 12.van der Burgt I, et al. Nijmegen breakage syndrome. J. Med. Genet. 1996;33:153–156. doi: 10.1136/jmg.33.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Online Mendelian Inheritance in Man (OMIM) Johns Hopkins University. < http://www.ncbi.nlm.nih.gov/omim/251260> (15 January 2010, date last accessed) [Google Scholar]
- 14.Choudhury A, et al. Analysis of variants in DNA damage signalling genes in bladder cancer. BMC Med. Genet. 2008;9:69. doi: 10.1186/1471-2350-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa JD, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28:1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 16.Lan Q, et al. Smoky coal exposure, NBS1 polymorphisms, p53 protein accumulation, and lung cancer risk in Xuan Wei, China. Lung Cancer. 2005;49:317–323. doi: 10.1016/j.lungcan.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Landi S, et al. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006;66:11062–11069. doi: 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 18.Margulis V, et al. Genetic susceptibility to renal cell carcinoma: the role of DNA double-strand break repair pathway. Cancer Epidemiol. Biomarkers Prev. 2008;17:2366–2373. doi: 10.1158/1055-9965.EPI-08-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matullo G, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 20.Medina PP, et al. Screening of homologous recombination gene polymorphisms in lung cancer patients reveals an association of the NBS1-185Gln variant and p53 gene mutations. Cancer Epidemiol. Biomarkers Prev. 2003;12:699–704. [PubMed] [Google Scholar]
- 21.Mosor M, et al. Polymorphisms and haplotypes of the NBS1 gene in childhood acute leukaemia. Eur. J. Cancer. 2008;44:2226–2232. doi: 10.1016/j.ejca.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Ryk C, et al. Influence of polymorphism in DNA repair and defence genes on p53 mutations in bladder tumours. Cancer Lett. 2006;241:142–149. doi: 10.1016/j.canlet.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Ryk C, et al. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54:285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Sanyal S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 25.Sobti RC, et al. Effect of NBS1 gene polymorphism on the risk of cervix carcinoma in a northern Indian population. Int. J. Biol. Markers. 2008;23:133–139. doi: 10.1177/172460080802300301. [DOI] [PubMed] [Google Scholar]
- 26.Stern MC, et al. Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res. 2009;69:6857–6864. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zienolddiny S, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am. J. Hum. Genet. 2006;78:464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, et al. Association between the NBS1 E185Q polymorphism and cancer risk: a meta-analysis. BMC Cancer. 2009;9:124. doi: 10.1186/1471-2407-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuura S, et al. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- 31.Varon R, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 32.Cui Y, et al. Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int. J. Cancer. 2006;118:714–720. doi: 10.1002/ijc.21413. [DOI] [PubMed] [Google Scholar]
- 33.Hashibe M, et al. Marijuana use and the risk of lung and upper aerodigestive tract cancers: results of a population-based case-control study. Cancer Epidemiol. Biomarkers Prev. 2006;15:1829–1834. doi: 10.1158/1055-9965.EPI-06-0330. [DOI] [PubMed] [Google Scholar]
- 34.Mu LN, et al. Green tea drinking and multigenetic index on the risk of stomach cancer in a Chinese population. Int. J. Cancer. 2005;116:972–983. doi: 10.1002/ijc.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu LN, et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms and the risk of primary hepatocellular carcinoma (HCC) in a Chinese population. Cancer Causes Control. 2007;18:665–675. doi: 10.1007/s10552-007-9012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H, et al. Dietary mineral and trace element intake and squamous cell carcinoma of the esophagus in a Chinese population. Nutr. Cancer. 2006;55:63–70. doi: 10.1207/s15327914nc5501_8. [DOI] [PubMed] [Google Scholar]
- 37.Cao W, et al. Tobacco smoking, GSTP1 polymorphism, and bladder carcinoma. Cancer. 2005;104:2400–2408. doi: 10.1002/cncr.21446. [DOI] [PubMed] [Google Scholar]
- 38.Hung RJ, et al. Protective effects of plasma carotenoids on the risk of bladder cancer. J. Urol. 2006;176:1192–1197. doi: 10.1016/j.juro.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Cao W, et al. Comparison of methods for DNA extraction from paraffin-embedded tissues and buccal cells. Cancer Detect. Prev. 2003;27:397–404. doi: 10.1016/s0361-090x(03)00103-x. [DOI] [PubMed] [Google Scholar]
- 40.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobler AR, et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J. Biomol. Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YC, et al. A case-control study of the association of the polymorphisms and haplotypes of DNA ligase I with lung and upper-aerodigestive-tract cancers. Int. J. Cancer. 2008;122:1630–1638. doi: 10.1002/ijc.23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SL, et al. Associations between variants of the 8q24 chromosome and nine smoking-related cancer sites. Cancer Epidemiol. Biomarkers Prev. 2008;17:3193–3202. doi: 10.1158/1055-9965.EPI-08-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Database of Single Nucleotide Polymorphism (dbSNP). dbSNP accession: {ss3291922, ss44843758, ss44891596, ss44893481, ss44911227}, (dbSNP Build ID: 130) Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine. http://www.ncbi.nlm.nih.gov/snp/ [Google Scholar]
- 45.Li SS, et al. Estimating haplotype frequencies and standard errors for multiple single nucleotide polymorphisms. Biostatistics. 2003;4:513–522. doi: 10.1093/biostatistics/4.4.513. [DOI] [PubMed] [Google Scholar]
- 46.Zhao LP, et al. A method for the assessment of disease associations with single-nucleotide polymorphism haplotypes and environmental variables in case-control studies. Am. J. Hum. Genet. 2003;72:1231–1250. doi: 10.1086/375140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slaughter DP, et al. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 49.Cui Y. PolymiRTS Database. http://compbio.uthsc.edu/miRSNP/ (3 February 2010, date last accessed) [Google Scholar]
- 50.Bao L, et al. PolymiRTS database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 52.The International HapMap Consortium. The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 53.Vineis P, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J. Natl Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 54.London WT, et al. Liver cancer. In: Schottenfeld D, Fraumeni J, editors. Cancer Epidemiology and Prevention. New York, NY: Oxford University Press; 2006. pp. 763–786. [Google Scholar]
- 55.Poschl G, et al. Alcohol and cancer. Alcohol Alcohol. 2004;39:155–165. doi: 10.1093/alcalc/agh057. [DOI] [PubMed] [Google Scholar]
- 56.Baan R, et al. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 57.Sun S, et al. Lung cancer in never smokers—a different disease. Nat. Rev. Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 58.Leng S, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer Res. 2008;68:3049–3056. doi: 10.1158/0008-5472.CAN-07-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi J, et al. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst.) 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 60.Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 61.Wang D, et al. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet. Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 62.Nielsen KB, et al. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am. J. Hum. Genet. 2007;80:416–432. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 64.Genome Variation Server. Seattle, WA: SeattleSNPs Program for Genomic Applications. < http://gvs.gs.washington.edu/GVS/index.jsp> (October 12 2009, date last accessed) [Google Scholar]
- 65.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 67.Goode EL, et al. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res. 2002;62:3052–3057. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.