Abstract
Aging is believed to be among the most important contributors to atherosclerosis, through mechanisms that remain largely obscure. Serum levels of tumor necrosis factor (TNF) rise with aging and have been correlated with the incidence of myocardial infarction. We therefore sought to determine whether genetic variation in the TNF receptor-1 gene (TNFR1) contributes to aging-related atherosclerosis in humans and whether Tnfr1 expression aggravates aging-related atherosclerosis in mice. With 1330 subjects from a coronary angiography database, we performed a case–control association study of coronary artery disease (CAD) with 16 TNFR1 single-nucleotide polymorphisms (SNPs). Two TNFR1 SNPs significantly associated with CAD in subjects >55 years old, and this association was supported by analysis of a set of 759 independent CAD cases. In multiple linear regression analysis, accounting for TNFR1 SNP rs4149573 significantly altered the relationship between aging and CAD index among 1811 subjects from the coronary angiography database. To confirm that TNFR1 contributes to aging-dependent atherosclerosis, we grafted carotid arteries from 18- and 2-month-old wild-type (WT) and Tnfr1−/− mice into congenic apolipoprotein E-deficient (Apoe−/−) mice and harvested grafts from 1 to 7 weeks post-operatively. Aged WT arteries developed accelerated atherosclerosis associated with enhanced TNFR1 expression, enhanced macrophage recruitment, reduced smooth muscle cell proliferation and collagen content, augmented apoptosis and plaque hemorrhage. In contrast, aged Tnfr1−/− arteries developed atherosclerosis that was indistinguishable from that in young Tnfr1−/− arteries and significantly less than that observed in aged WT arteries. We conclude that TNFR1 polymorphisms associate with aging-related CAD in humans, and TNFR1 contributes to aging-dependent atherosclerosis in mice.
INTRODUCTION
Aging has been proposed to be among the most important risk factors for atherosclerosis (1), at least in part because of myriad aging-associated changes in gene expression in the arterial wall (2,3). Aging-associated arterial changes common to humans and rodents may predispose aged arteries to accelerated atherosclerosis and include neointimal hyperplasia, medial thickening, endothelial dysfunction that augments monocyte/endothelial adherence, enhanced endothelial cell apoptosis and diminished vascular cell replicative capacity (2). Diet-induced atherosclerosis is, indeed, accelerated in aged animals (3), but the role of arterial aging in this phenomenon remains unclear.
Among multiple pro-atherogenic arterial wall gene products, tumor necrosis factor receptor-1 (TNFR1) (4) may attain particular significance in the context of aging. Even in the absence of atherosclerosis, arterial TNF expression increases with aging (5) and may engender the increased expression of intercellular adhesion molecule-1 (ICAM-1), matrix metalloproteinase-2 (MMP-2) and monocyte chemoattractant protein-1 (MCP-1) seen in aged rat arteries (6). However, despite substantial aging-related increases in arterial wall expression of its two atherogenic agonists (TNF and lymphotoxin-α) (7), arterial wall TNFR1 is not downregulated with aging (5). Serum TNF concentrations also rise with aging in humans (8) and have been shown to predict recurrent myocardial infarction (MI) (9) and mortality in aged men and women (10). In aged apolipoprotein E-deficient (Apoe−/−) mice, the absence of TNFR1 reduces atherosclerosis of the aorta, but not the brachiocephalic artery (4); however, these studies could not evaluate the atherogenic role of aging, either systemically or in the arterial wall. Poirier et al. (11) found that, in humans, TNFR1 polymorphisms were associated with MI; however, this study could not evaluate the role of TNFR1 variants in aging-related MI or atherosclerosis.
To determine whether TNFR1 contributes to aging-dependent atherosclerosis, we used both human coronary artery disease (CAD) genetic association studies and a mouse model for assessing atherogenicity of arterial wall gene products (4). Human studies were conducted to determine genetic effects on the relationship between aging and atherosclerosis in two independent cohorts comprising subjects spanning the age range of 18–91 years (12). Our mouse atherosclerosis studies were conducted by transplanting into young Apoe−/− mice carotid arteries from aged or young mice that were either wild-type (WT) or Tnfr1−/− (4).
RESULTS
TNFR1 single-nucleotide polymorphisms associate with atherosclerosis in aged subjects
To test the hypothesis that TNFR1 (TNFRSF1A) single-nucleotide polymorphisms (SNPs) associate with aging-related atherosclerosis in humans, we used two sample sets we have described previously: CATHGEN (CATHeterization GENetics), a cohort of subjects aged 18–91 years who underwent coronary angiography at Duke University Medical Center, and GENECARD (GENetics of Early-onset Coronary Artery Disease), a cohort comprising subjects with familial, early-onset CAD, as well as their unaffected family members (12). A comparison of the clinical characteristics of CATHGEN cases and controls and GENECARD probands is presented in Table 1. All three case groups (CATHGEN ‘young affected’, ‘older affected’ and GENECARD probands) manifest a risk factor profile consistent with CAD compared with controls: male gender, smoking, clinical history of increased prevalence of diabetes and hypertension and lower HDL cholesterol levels. The clinical characteristics also demonstrate risk factor relationships consistent with the age differences among the case groups and controls, with increased systolic blood pressure in the older groups compared with the younger. The GENECARD probands exhibit more pro-atherogenic lipoprotein profiles, highlighting the strong genetic component to variability in lipoprotein cholesterol levels.
Table 1.
Clinical characteristics of CATHGEN subjects and GENECARD probands (mean ± SD)
| CATHGEN |
GENECARD (US probands) (n = 759) |
||||
|---|---|---|---|---|---|
| Young affected (n = 656) |
Older affected (n = 264) |
Older normal (n = 410) |
Random sample (n = 481) |
||
| Age, years | 46 ± 6 | 66 ± 7 | 70 ± 6 | 65 ± 16 | 48 ± 10 |
| CADia | 58 ± 22 | 82 ± 9 | 6 ± 9 | 30 ± 23 | NA |
| %Caucasian | 71 | 87 | 75 | 80 | 86 |
| %Male | 79 | 72 | 43 | 59 | 69 |
| Family history of CADb, % | 55 | 43 | 26 | 38 | 100 |
| BMI, kg/m2 | 31 ± 6 | 28 ± 6 | 29 ± 7 | 29 ± 7 | 30 ± 7 |
| Ever-smoked, % | 69 | 52 | 40 | 47 | 78 |
| History of diabetesc, % | 33 | 31 | 21 | 28 | 25 |
| History of hypertensionc, % | 70 | 75 | 67 | 69 | 63 |
| History of MIc, % | 52 | 38 | 0 | 36 | 63 |
| Mean systolic blood pressure, mmHg | 141 ± 24 | 151 ± 26 | 150 ± 24 | 145 ± 26 | 142 ± 25 |
| Mean diastolic blood pressure, mmHg | 78 ± 14 | 78 ± 13 | 77 ± 14 | 76 ± 15 | 78 ± 14 |
| Total cholesterol, mmol/l | 4.99 ± 1.61 | 4.47 ± 1.06 | 4.95 ± 1.26 | 4.65 ± 1.21 | 5.17 ± 1.50 |
| LDL, mmol/l | 2.84 ± 1.12 | 2.54 ± 0.88 | 2.77 ± 0.93 | 2.64 ± 0.97 | 3.16 ± 1.53 |
| HDL, mmol/l | 1.05 ± 0.32 | 1.16 ± 0.36 | 1.35 ± 0.48 | 1.20 ± 0.42 | 1.14 ± 0.83 |
aCADi is a numerical summary of coronary angiographic evidence of atherosclerosis, with 0 representing no atherosclerosis and 100 representing hemodynamically severe left main disease (12).
bFamily history by self-report for CATHGEN, and by extensive pedigree investigation for GENECARD.
cHealth history data were collected at the time of angiography (CATHGEN) or by medical record review (GENECARD).
To associate TNFR1 SNPs with CAD, we initially genotyped 1330 CATHGEN subjects for a total of 16 TNFR1 SNPs, selected from the TNFR1 gene as well as untranslated regions 3000 bp upstream and downstream of the gene, as described in Materials and Methods. The 1330 subjects, described in Table 1, were stratified as described in Materials and Methods by CAD age of onset: young affected ≤55 years; older affected >55 years; ‘older normal’ ≥60 years without clinically or angiographically evident CAD. This age stratification at 55 years was based on the incidence curves of CAD and estimates of genetic effect at various ages (13–15). In analyses adjusted for gender and CAD risk factors, as described in Materials and Methods, we found that in subjects ≥56 years old, two of the TNFR1 SNPs associated significantly with CAD index (CADi) (12) (Supplementary Material, Table S1 and Fig. S1, underlined): rs4149578 and rs4149573, for which P-values remained significant after Bonferroni correction for multiple comparisons (P < 0.003). For these TNFR1 SNPs, the frequency of minor alleles was higher in CAD cases. Consequently, the minor alleles constitute ‘risk’ alleles, for which the odds ratios (95% confidence intervals) were 2.3 (1.4–4.0) and 2.4 (1.4–4.3), respectively, for rs4149578 and rs4149573.
To corroborate the associations we observed between TNFR1 SNPs and CADi in the CATHGEN data set, we analyzed TNFR1 genotypes in the GENECARD data set, using two approaches. First, to evaluate the consistency of the effect of TNFR1 SNP minor alleles in terms of the odds ratio magnitude and P-value, we compared GENECARD probands and CATHGEN older normal controls. In this analysis, the comparison of GENECARD cases with CATHGEN controls produced findings congruent with those we obtained using CATHGEN cases: the odds ratios (with 95% confidence intervals) for rs4149578 and rs4149573 were similar at 1.6 (1.03–2.6) and 1.7 (1.1–2.7), respectively; however, these results were not significant after the adjustment for multiple comparisons (Supplementary Material, Table S1). In our second approach, we sought to support inferences from CATHGEN with data completely independent from CATHGEN. To do so, we used the GENECARD data set to perform family-based association tests. Although these tests suggested only modest evidence for the association of TNFR1 with CAD, it is notable, nonetheless, that they suggest evidence for the association between CAD and the TNFR1 R92Q polymorphism—the SNP originally associated with MI, as found by Poirier et al. (11) (P < 0.05; Supplementary Material, Table S2). Together, data from GENECARD and Poirier et al. (11) support the association between CAD and TNFR1 discerned in CATHGEN. Moreover, in conjunction with CATHGEN older normal controls, our GENECARD data also support the association found in CATHGEN between two TNFR1 SNPs and aging-associated atherosclerosis (Supplementary Material, Table S1).
TNFR1 genotype modifies the effect of age on atherosclerosis
In addition to the association between TNFR1 SNPs and CADi in aged adults, we found the expected (16–18), strong positive correlation of age with CADi (Table 2). Together, these findings suggest the intriguing possibility that the well-known relationship between atherosclerosis risk and age could be modified, in part, by TNFR1 polymorphisms engendering changes in TNFR1 expression levels and/or function.
Table 2.
Effects of age and TNFR1 rs4149573 genotype on CADi in 1811 CATHGEN subjects
| Linear regression model component | Linear regression model [regression coefficient (SE), and P-value for the Wald test] |
||
|---|---|---|---|
| Clinical variables only | Clinical variables + SNP genotype | Clinical variables + SNP genotype + age × genotype | |
| Age | 0.65 (0.06), <0.0001 | 0.65 (0.06), <0.0001 | 0.58 (0.06), <0.0001 |
| rs4149573 CG/GG genotypes | Not in model | −1.33 (2.34), 0.57 | −25.32 (9.53), 0.0079 |
| Age × genotype interaction | Not in model | Not in model | 0.44 (0.14), 0.0032 |
| Intercept | 16.27 (6.22), 0.0090 | 17.54 (6.72), 0.0091 | 20.55 (6.24), 0.0010 |
| Gender (F) | −11.42 (6.26), <0.0001 | −11.45 (1.54), <0.0001 | −11.55 (1.54), <0.0001 |
| Race (AA) | 2.78 (1.94), 0.15 | 2.79 (1.95), 0.15 | 2.51 (1.95), 0.20 |
| History of diabetesa | 6.51 (1.83), 0.0004 | 6.59 (1.82), 0.0003 | 6.62 (1.83), 0.0003 |
| History of smoking | 5.88 (1.53), 0.0001 | 5.87 (1.53), 0.0001 | 5.90 (1.52), 0.0001 |
| BMI (kg/m2) | −0.19 (0.11), 0.09 | −0.19 (0.11), 0.09 | −0.18 (0.11), 0.11 |
| History of hypertension | 2.53 (1.79), 0.16 | 2.55 (1.79), 0.15 | 2.45 (1.77), 0.17 |
| History of hyperlipidemia | 12.09 (1.68), <0.0001 | 12.08 (1.69), <0.0001 | 12.15 (1.67), <0.0001 |
SNP genotype, TNFR1 rs4149573 genotype; age × genotype, age-by-genotype interaction term; F, female; AA, African American.
aDetermined by clinical history.
To test this possibility, we fitted a weighted regression model adjusted for CAD risk factors and sampling weights for all 1811 CATHGEN subjects on the outcome of CADi, and the following dependent variables: age, TNFR1 genotype for each of the two SNPs significantly associated with atherosclerosis (coded as a dominant model) and an age-by-genotype interaction term (Table 2). Although age was significantly associated with CADi in these linear regression analyses, the coefficient for the age-by-genotype interaction term was not significantly different from zero in models that included rs4149578 alone. However, for the model that included rs4149573, the linear regression coefficient for the age-by-genotype interaction was, indeed, significantly different from zero (P = 0.0032). It is important to note that if the interaction between age and rs4149573 genotype is not taken into account [i.e. if we consider only the average age in the CATHGEN cohort (∼65 years)], the association between rs4149573 and CADi is not detectable (Table 2). Therefore, these findings suggest that the relationship between CADi and aging is modified by the genotype of the TNFR1 SNP rs4149573.
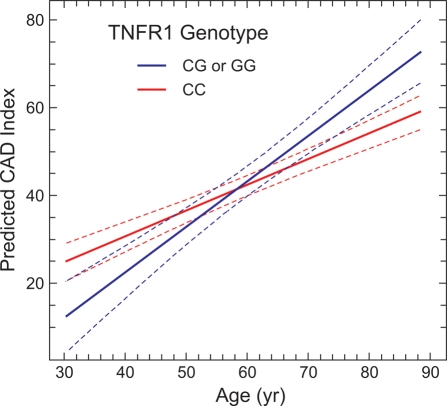
To demonstrate graphically the effect of TNFR1 SNP rs4149573 on aging-related atherosclerosis in the CATHGEN cohort, we fit CADi regression lines for the major and minor alleles of rs4149573 across the age range of subjects from 30 to 90 years of age (Fig. 1). The presence of the G (‘risk’) allele alters the predicted relationship between CADi and age, with the highest level of CADi observed in G allele carriers at the oldest age. Thus, these modeling data support the association we observed between TNFR1 SNP rs4149573 and CADi in our comparison of older affected versus older normal subjects (Fig. 1), and they illustrate that genetic variation in TNFR1 may contribute to aging-related atherosclerosis in humans.
Figure 1.
TNFR1 genotype affects the relationship between aging and atherosclerosis. Fitted regression lines (±95% confidence intervals) are plotted for CADi (adjusted for gender and known CAD risk factors) as a function of age, for CATHGEN cohort groups defined by genotype at the TNFR1 SNP rs4149573: (i) those with the ‘risk’ alleles GG or CG (blue line) and (ii) those with the more prevalent allele CC (red line). CADi was determined angiographically, by observers unaware of genotypes, for 1811 CATHGEN subjects. The model was fit using SAS version 9.1 software.
Arterial aging promotes atherogenesis in mice
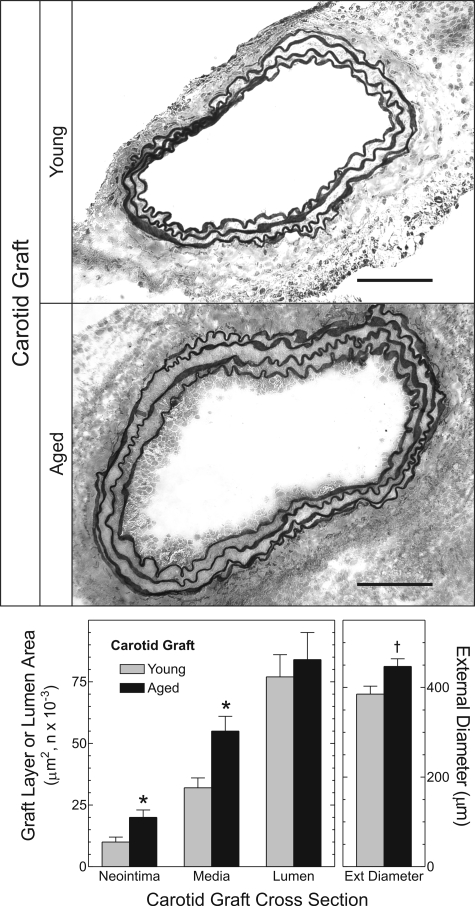
To determine whether TNFR1 contributes to aging-dependent atherosclerosis, as suggested by our human studies, we developed a mouse model that focuses on the influence of arterial wall aging on atherogenesis. This model employs orthotopic transplantation into Apoe−/− mice of the right common carotid artery from congenic WT C57BL/6 mice that were either young (10 ± 2 weeks old) or aged (79 ± 2 weeks old). Consequently, this model allows us to dissect effects of arterial aging from effects of prolonged exposure to elevated plasma lipoprotein levels. Prior to and within 1 week of implantation, the young and aged carotid arteries were morphologically indistinguishable: luminal areas of young and aged specimens, respectively, were 0.134 ± 0.007 and 0.140 ± 0.004 mm2; medial areas were 0.021 ± 0.001 and 0.021 ± 0.003 mm2 (n = 5 per group). Prior to grafting, aged and young carotid arteries also demonstrated equivalent expression of smooth muscle cell (SMC) actin and apoE (Supplementary Material, Fig. S2). However, within 2 weeks of grafting, atherogenesis proceeded more rapidly in aged than in young arteries. Young carotid arteries developed only scattered intimal macrophages and mild medial expansion. In contrast, aged arteries developed asymmetrical intimal lesions and considerable medial thickening consequent to macrophage infiltration (Figs 2 and 3). Neointimal and medial areas of aged arteries were 1.9- and 1.7-fold greater, respectively, than those of young arteries (P < 0.02; Fig. 2). In addition, outward remodeling was 20% greater in aged than in young arteries (P < 0.03; Fig. 2).
Figure 2.
Arterial wall aging accelerates atherogenesis. Ten-week-old Apoe−/− mice underwent common carotid interposition grafting with common carotid arteries from congenic WT mice that were aged either 10 weeks or 18 months. Grafts were harvested 2 weeks post-operatively, embedded and frozen in OCT compound, then cross-sectioned and stained with a modified connective tissue stain. Computerized morphometry yielded the indicated dimensions (n = 5 per group), as described in Materials and Methods. Compared with WT: *P < 0.02 or †P < 0.03. Scale bar: 100 μm (original magnification ×220).
Figure 3.
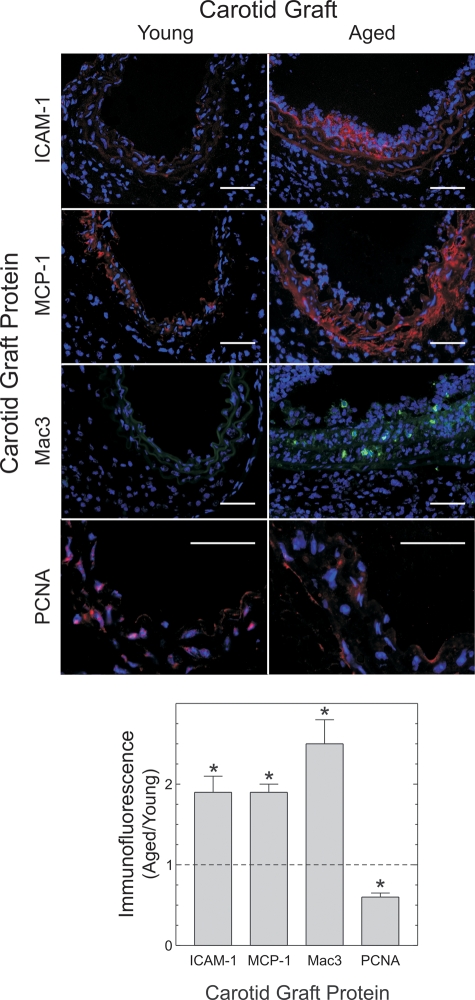
Aged arteries demonstrate greater macrophage recruitment and less SMC proliferation than young arteries during early atherogenesis. Two-week-old grafts from Figure 2 were immunostained for the indicated proteins and counterstained for nuclear DNA (blue). Protein immunofluorescence was then normalized to nuclear fluorescence within each microscopic field, as described in Materials and Methods. The means ± SE from four or more arteries are plotted. Compared with young specimens: *P < 0.05. Scale bars: 50 μm (original magnification ×440).
Mechanisms by which arterial aging may accelerate atherosclerosis include the activity of gene products upregulated by nuclear factor-κB (NFκB), a pro-inflammatory transcription factor whose p65 subunit is itself upregulated with aging in human endothelial cells (19). Two such NFκB-regulated gene products include the chemokine MCP-1 and the adhesion molecule ICAM-1, both of which serve vital roles in the recruitment of monocytes to, and the retention of monocytes in, the arterial intima (20). Indeed, expression of ICAM-1 and MCP-1 protein in aged artery grafts was 1.9 ± 0.4- and 1.9 ± 0.2-fold greater, respectively, than that observed in young artery grafts (P < 0.02; Fig. 3). This higher level of chemokine and adhesion molecule expression in aged artery grafts would be expected to enhance macrophage infiltration of aged, compared with young artery grafts—and such was the case: the prevalence of macrophages was 2.5 ± 0.5-fold higher in aged than in young artery grafts (P < 0.04; Fig. 3).
Although inflammatory changes and macrophage recruitment were greater in aged than in young arteries, proliferation of medial SMCs was greater in young than in aged arteries (Fig. 3). Indeed, compared with aged arteries, young arteries showed a 65% higher prevalence of medial cells that stained for proliferating cell nuclear antigen (PCNA). This finding accords with previous in vitro studies demonstrating that SMCs from aged arteries proliferate more slowly than SMCs from young arteries (2).
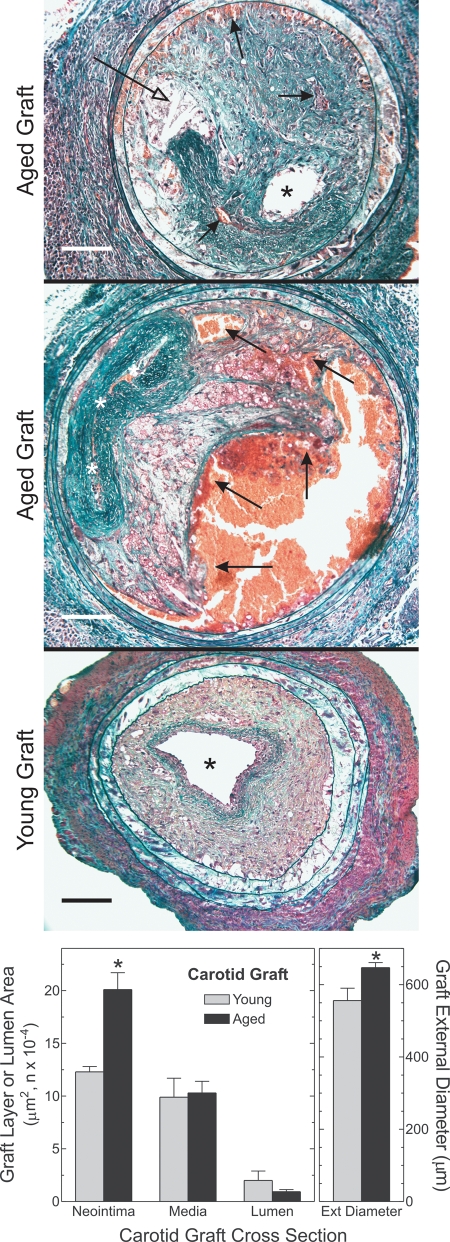
The enhancement of atherosclerosis in aged arteries remained evident even 7 weeks post-operatively, when atherosclerosis was far advanced in both aged and young carotid arteries. Neointimal area was 63% greater in aged than in young carotid grafts (P < 0.03), luminal area was 2-fold smaller and arterial external diameter was 20% greater (reflecting greater outward remodeling) (Fig. 4).
Figure 4.
Arterial wall aging exacerbates plaque hemorrhage and arterial outward remodeling. Carotid grafts were harvested 7 weeks post-operatively, after perfusion–fixation. Paraffin sections were stained with a modified connective tissue stain (see Materials and Methods). Computerized morphometry yielded the indicated dimensions from six or more arteries of each type. Compared with young grafts: *P < 0.01. Open arrows, closed arrows and asterisks indicate cholesterol clefts, plaque hemorrhage and lumen, respectively. Scale bars: 100 μm (original magnification ×200). The middle image was concatenated with Image-Pro Plus® (Media Cybernetics).
Plaque hemorrhage also distinguished advanced plaques of aged arteries from those of young arteries (Fig. 4). We found intra-plaque (yet extravascular) erythrocytes in six out of eight aged specimens, but in none of nine young specimens (P < 0.05). Whether plaque rupture occurs in mouse arteries remains a controversial issue (21). However, the possibility of plaque rupture as a cause of plaque hemorrhages in the aged carotid grafts is suggested not only by proximity of the plaque hemorrhages to the lumen, but also by the presence of ‘buried fibrous caps’ within atheromata of the aged specimens (Fig. 4) (21).
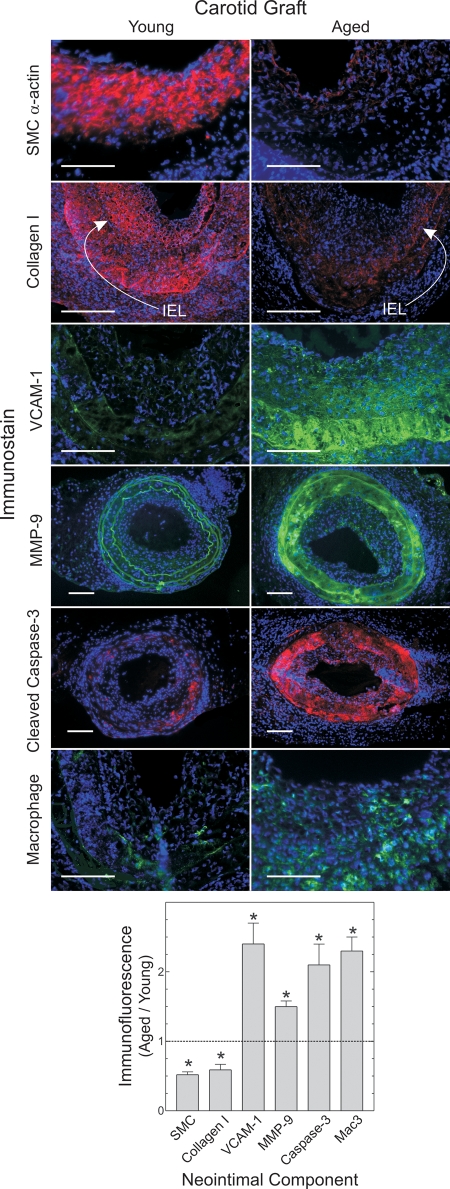
Atheromata in aged arteries, to a greater degree than atheromata in young arteries, demonstrated several features predisposing to plaque instability (21,22). In aged artery plaques, the prevalence of SMCs was 50 ± 30% less than in young arteries—a difference particularly noticeable in the fibrous cap region of the plaque (P < 0.03; Fig. 5) and congruent with the reduced medial PCNA staining we observed in 2-week-old aged grafts (Fig. 3). Conversely, the prevalence of macrophages in aged artery plaques was 130 ± 30% greater than in young artery plaques (P < 0.03; Fig. 5). Perhaps as a consequence of increased macrophage prevalence, or perhaps as a consequence of diminished anti-oxidant gene expression in aged arteries (3), aged arteries also demonstrated 2.5 ± 0.5-fold greater neointimal and medial nitrotyrosine than young arteries (Supplementary Material, Fig. S3). Consistent with diminished SMC prevalence in the plaque, aged artery plaques also demonstrated 40 ± 10% lower collagen density than plaques in young arteries (P < 0.04; Fig. 5). Aged artery plaques also expressed 2.4 ± 0.5-fold more vascular cell adhesion molecule-1 (VCAM-1) than young artery plaques and contained 1.5 ± 0.2-fold more matrix metalloproteinase-9 (MMP-9) (P < 0.04; Fig. 5)—both findings consonant with the increased macrophage prevalence in plaques from aged arteries. Lastly, the prevalence of apoptotic cells was higher in plaques of aged, compared with young arteries, particularly in the fibrous cap region, as reflected by 2.1 ± 0.7-fold greater intensity of cleaved caspase-3 immunofluorescence per cell (Fig. 5).
Figure 5.
Aged artery grafts demonstrate diminished SMC and collagen content, but greater macrophage prevalence and apoptosis than young artery grafts. Carotid grafts harvested 7 weeks post-operatively were immunostained for the indicated proteins and counterstained for DNA (blue). Data were quantitated from four or more arteries, as in Figure 3. Compared with young grafts: *P < 0.01. Arrows indicate locations of the internal elastic laminae (IEL). Scale bars: 100 μm (original magnification ×220 or ×440).
Tnfr1 is upregulated in aged arteries
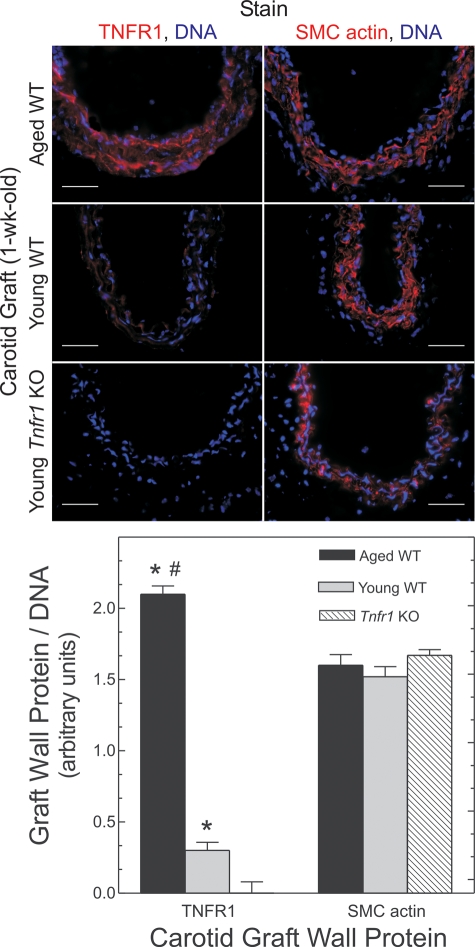
Because aged carotids demonstrate greater expression of MCP-1 and ICAM-1 during early stages of atherogenesis, and because arterial wall expression of these genes depends largely upon TNFR1 signaling in arterial wall cells (4), we reasoned that TNFR1 could contribute to the accelerated atherosclerosis observed in aged arteries—particularly if aged arteries express higher levels of TNFR1 than young arteries. To test this possibility, we examined carotid grafts harvested 1 week post-operatively, before intimal macrophages could be detected (data not shown). In these pre-atherosclerotic arteries, TNFR1 expression was 7 ± 3-fold greater in aged than in young specimens (P = 0.01; Fig. 6). In contrast, SMC actin expression was indistinguishable between pre-atherosclerotic arteries from aged and young mice (Fig. 6).
Figure 6.
Aging upregulates arterial TNFR1 expression. Carotid grafts from mice of the indicated age and genotype were harvested 1 week post-operatively, immunostained for the indicated proteins and counterstained for DNA (blue). Data were quantitated as in Figure 3; means ± SE from four or more arteries are presented. Relative to Tnfr1−/− (‘KO’) grafts: *P < 10−4; relative to young WT grafts: #P < 10−3. Lumen is upward. Scale bars: 50 μm (original magnification ×440).
TNFR1 contributes to aging-dependent atherosclerosis
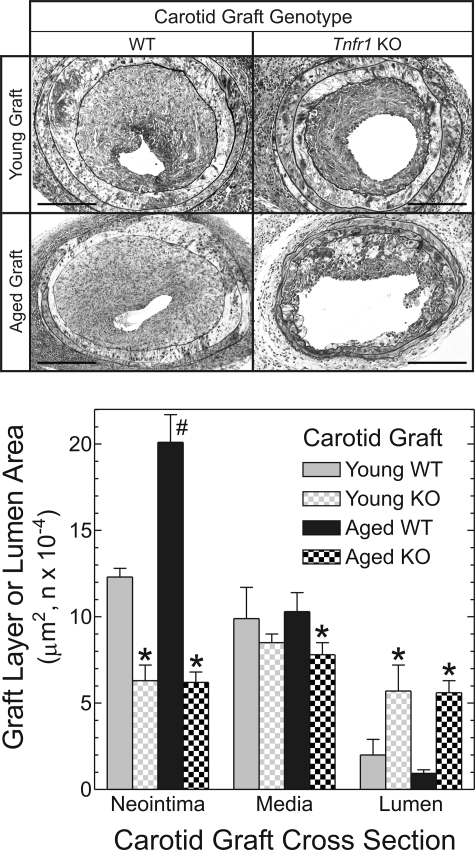
If age-related enhancement of arterial wall TNFR1 expression is causally related to aging-promoted acceleration of atherosclerosis, then elimination of TNFR1 from the arterial wall should substantially reduce aging-dependent atherogenesis. To test this expectation, we repeated our carotid grafting experiments with aged and young carotid artery donors that were deficient in TNFR1 (from Tnfr1−/− mice). Young Tnfr1−/− carotid grafts developed only 50% as much neointimal area as young WT carotid grafts, and only 60% as much luminal stenosis (Fig. 7), as we found previously (4). However, in striking contrast to WT arteries, there was no aging-dependent atherosclerosis in Tnfr1−/− arteries: neointimal, medial and lumen areas were equivalent in aged and young Tnfr1−/− grafts (Fig. 7). Unlike WT arteries, aged and young atherosclerotic Tnfr1−/− arteries also demonstrated equivalent prevalence of SMC-like cells, macrophages and apoptotic cells, as well as equivalent levels of VCAM-1 and collagen I protein expression (data not shown). It is also noteworthy that no plaque hemorrhage could be detected in aged Tnfr1−/− carotid grafts. Furthermore, Tnfr1-dependent differences between aged carotid arteries were greater than Tnfr1-dependent differences between young arteries, and substantially greater than aging-dependent differences observed in WT arteries (Fig. 7). Thus, arterial wall expression of TNFR1 indeed appears to contribute to aging-dependent atherosclerosis.
Figure 7.
Tnfr1 deficiency abrogates aging-dependent atherosclerosis. Carotid grafts from mice of the indicated age and genotype were harvested 7 weeks post-operatively, and processed as in Figure 4. Computerized morphometry data are plotted from five or more arteries of each type (Tnfr1 KO, ‘KO’). Compared with cognate WT grafts: *P < 0.02. Compared with young WT grafts: #P < 0.02. Scale bar: 100 μm (original magnification ×220).
DISCUSSION
Through complementary approaches in humans and mice, our studies implicate TNFR1 in the pathogenesis of aging-related atherosclerosis. Two independent human cohorts support the association of TNFR1 SNPs with CAD, and at least one TNFR1 SNP appears to modify the effect of age on CAD. These association data in humans resonate with our mouse data, which demonstrate not only that arterial wall aging accelerates atherosclerosis, but also that this aging-dependent acceleration is abrogated when arterial wall cells lack TNFR1. These novel findings comport cogently with our data regarding arterial wall TNFR1 expression, which increases both with aging and with the earliest stages of atherogenesis.
We built our human study on the findings of Poirier et al. (11), by genotyping the same set of TNFR1 SNPs identified among premature MI patients by that group, and then extending that set of SNPs with tagSNPs so as to cover common genetic variation throughout the entire human TNFR1. Poirier et al. used single-strand conformational polymorphism and sequencing analysis on only ∼26% of the total TNFR1 gene, predominantly on coding sequences, to identify CAD-associated TNFR1 SNPs, and subsequently found that the 92Q allele of TNFR1 (rs4149584) was associated with increased risk of MI among UK and French residents of European descent. Although we observed evidence of 92Q allele transmission to affected siblings in GENECARD, our CAD cases from CATHGEN and GENECARD did not demonstrate greater frequency of the 92Q allele than our controls, even though the absolute 92Q frequency in our cohorts was similar to that observed by Poirier et al. in their control groups (1.6–2%; Supplementary Material, Table S3) (11).
Because Poirier et al. did not fully sequence the introns of TNFR1 (11), they did not assess the two TNFR1 tagSNPs most closely associated with CAD in our case–control studies. Moreover, these two TNFR1 tagSNPs were not in linkage disequilibrium (LD) with any of the SNPs associated with MI by Poirier et al. (11) (Supplementary Material, Fig. S4). Consequently, their data can be used neither to validate nor to refute our findings. However, the inability of Poirier et al. to find association between MI and TNFR1 SNP rs12426675 may, at first, appear to conflict with our analyses (Supplementary Material, Table S2) (11). Nonetheless, it is important to note that this inter-analysis discrepancy may be attributable to differences in the age range of subjects studied: our strongest CAD associations for TNFR1 SNPs occurred in subjects aged ≥56 years (range 56–91), whereas the mean age studied by Poirier et al. was 56 years (range 28–72) (11). As Figure 1 illustrates, the interaction among CADi, age and TNFR1 genotype could obscure an SNP/CAD association if only a limited range of ages was studied. The importance for genetic association studies of accurately taking age into account has only recently been illustrated, in a study of age-varying association between a ROBO1 SNP and obesity (23). Therefore, together with results from Poirier et al., our results lend support to the notion that genetically complex diseases such as CAD may have a spectrum of genetic susceptibility variants: from rare coding polymorphisms more easily identified in strongly genetic cases such as early-onset familial cases, to common polymorphisms identified only in large case–control samples containing subjects of the requisite age ranges.
The importance of age in atherosclerosis risk often prompts study designs to incorporate age, as we did by selecting cases and controls on the basis of both age and CADi. However, if one considers the age variable only categorically, by strata, and not continuously, then one may obscure or completely miss genetic associations characterized by age-dependent associations (23). For this reason, we have attempted to account for the entire range of age in our CAD cases by implementing a novel study design in which the case–control sample set is augmented by a random sample of subjects from the rest of the CATHGEN cohort. This approach is similar to the cohort sampling approach that has been shown to be an efficient method for detecting gene–environment interactions (24).
In our human genetic studies, we initially associated TNFR1 SNPs with CAD in older subjects (≥56 years old) and subsequently confirmed these associations in the GENECARD study of early-onset CAD. Because GENECARD CAD probands were younger than older affected CATHGEN CAD cases, the association of TNFR1 SNPs with CADi in both GENECARD and CATHGEN older affected cohorts might, at first, seem to negate the inference that TNFR1 SNPs associate with aging-dependent atherosclerosis. However, although they are chronologically young (by definition), GENECARD probands—by virtue of early-onset or aggressive atherosclerosis—may be considered biologically ‘older’, a general concept that continues to gain credibility (25,26). Alternatively, the association between CADi and TNFR1 genotype in GENECARD probands could result from issues pertinent to familial transmission of CAD in this young cohort of subjects. To test whether this possibility might obtain in our CATHGEN “young affected” cohort, we performed analyses stratified by family history. The results of our study were not changed in the stratified analysis, i.e. in young affected subjects with or without a family history of CAD, there was no significant association between CADi and TNFR1 genotype (data not shown), despite a 55% prevalence of CAD family history in this sample. Thus, with regard to increasing the risk of atherosclerosis, the relationship between age, TNFR1 genotype and CAD family history remains to be elucidated.
The association we found between three TNFR1 SNPs and CAD in aged humans serves to highlight the potential importance of TNFR1 in aging-dependent atherosclerosis. Although these intronic TNFR1 SNPs are not yet known to affect TNFR1 expression or TNFR1 function, at least one SNP could conceivably augment TNFR1 expression. Compared with the major allele of TNFR1 SNP rs4149573, the risk (G) allele would be predicted to lose the ability to bind myoneurin, a transcriptional regulator believed to reduce gene expression (27). Additionally, it is possible that the TNFR1 alleles that actually affect TNFR1 expression or TNFR1 function were not discerned by our analysis of TNFR1 tagSNPs, but exist nonetheless in LD with the SNPs that we did associate with aging-related atherosclerosis. Review of HapMap LD patterns in the region surrounding TNFR1 does not suggest that there are SNPs in high LD with the identified TNFR1 SNPs (Supplementary Material, Fig. S4). Furthermore, the association in GENECARD families between CADi and coding polymorphisms identified by Poirier et al. suggests that both rare and common SNPs contribute to the association of TNFR1 with CAD. Thus, our association analysis serves less to implicate specific TNFR1 alleles in aging-related atherogenesis than it does to implicate TNFR1 generally in aging-related atherogenesis. Having associated TNFR1 with aging-related atherosclerosis, we tested the causality of this association in mice, using the limit case of Tnfr1 expression differences: Tnfr1−/− and WT arteries.
Our mouse atherosclerosis model allows us to distinguish aging of the arterial wall from aging of other systems believed to be pertinent to atherogenesis (3,25). Importantly, genotype-specific results obtained with this model accord with results obtained in standard aortic atherosclerosis experiments (4). Assessed as expression of chemokine and adhesion molecules or macrophage prevalence, aged arteries demonstrated enhanced macrophage recruitment reminiscent of that observed with senescent endothelial cells (2) and consistent with the upregulation of MCP-1 observed in aged, non-atherosclerotic rat aortas (6,28). Macrophage recruitment in young arteries appears to depend significantly upon arterial wall TNFR1, which promotes the preponderance of (NFκB-dependent) MCP-1, ICAM-1 and VCAM-1 expression in the artery (4). Enhancement of macrophage recruitment in aged arteries may, therefore, plausibly be attributed to aging- and atherosclerosis-dependent upregulation of arterial TNFR1, as we observed, and/or arterial TNF (28), the pro-atherogenic actions of which are mediated principally by TNFR1 (4,29). Indeed, the elimination of enhanced macrophage recruitment by the elimination of TNFR1 in aged arteries (Fig. 7) strongly supports this inference.
Augmentation of aging-dependent atherosclerosis by TNFR1 accords well mechanistically with what is currently known about both TNFR1 signaling and effects of aging on vascular cells. TNFR1 signals through NAD(P)H oxidase in SMCs (30) as well as in other cells; oxidative signaling is augmented in aging arteries (28); and TNFR1 oligomerization and signaling are augmented by oxidative stress (31). Aging has been associated with diminished arterial expression of anti-oxidant genes (3), as well as enhanced endothelial expression of NAD(P)H oxidase and NFκB (19), the (pro-inflammatory) activity of which is potentiated by reactive oxygen species (28). That oxidative signaling contributes to aging-dependent atherosclerosis is highlighted by the ability of the NAD(P)H oxidase inhibitor apocynin to mitigate aging-dependent atherosclerosis (3).
If TNFR1 indeed contributes to aging-dependent atherosclerosis in humans, we would predict that anti-TNF therapies should mitigate atherosclerosis in aging animals. Using vascular inflammatory markers in aged rats, Csiszar et al. (32) have provided data congruent with this prediction. However, limited human data available thus far do not support this prediction. Anti-TNF therapy given as etanercept provided no benefit in terms of death or heart failure hospitalizations to 683 subjects with NYHA Class II–IV chronic heart failure in the Randomized Etanercept Worldwide Evaluation (RENEWAL) trial, which studied subjects with predominantly (∼60%) ischemic cardiomyopathy (33). MI incidence was not reported in this trial, however; median follow-up was only 5.7–12.9 months at the time RENEWAL's component trials were prematurely terminated, and the trial's results may have been confounded by excess prevalence of diabetes in the etanercept group (33). Moreover, it is not yet known whether RENEWAL used optimal dosing or agent for anti-TNF therapy, whether CAD was so far advanced in RENEWAL subjects that anti-TNF therapy could not reduce MI, or whether a pharmacogenomic interaction might exist between TNFR1 genotype and etanercept efficacy. It is also worth noting that although antagonizing TNFR1 activity may prove anti-atherogenic, neutralizing TNF—as with etanercept—may not. TNF activates both TNFR1, which is pro-atherogenic (4), and TNFR2, which, by promoting endothelial cell survival, may be anti-atherogenic (29). Our findings suggest that expression of TNFR1, at least in the arterial wall, contributes to aging-dependent atherosclerosis. Whether specific pharmacological antagonism of TNFR1 will prove anti-atherogenic remains to be determined.
MATERIALS AND METHODS
CAD case–control studies
CATHGEN participants were recruited sequentially through the cardiac catheterization laboratories at Duke University Hospital (Durham, NC, USA) with approval from the Duke Institutional Review Board. All participants undergoing catheterization were offered participation in the study and they provided informed consent. Medical history and clinical data were collected and stored in the Duke Information System for Cardiovascular Care database maintained at the Duke Clinical Research Institute (34). The full CATHGEN cohort comprised 8449 subjects. For this study, controls and cases were chosen from the cohort on the basis of their CADi, a numerical summary of coronary angiographic data that incorporates the extent and anatomical distribution of CAD (35), as we have reported previously (12,36–39). CADi predicts clinical outcome more accurately than just the extent of CAD (40). We designated those subjects with a CADi ≥32 as ‘affected with CAD’ (41). However, for patients >55 years old, we designated subjects with a CADi ≥74 as ‘affected with CAD’ to adjust for the higher baseline extent of CAD in this older group. Medical records were reviewed to determine the age of onset of CAD (the age at first documented coronary revascularization, MI or cardiac catheterization showing the above-defined CAD indices). CATHGEN cases were stratified into the following groups: (a) young affected (age of onset ≤55 years, n = 656), (b) older affected (age of onset >55, n = 264) and (c) older normal controls (subjects ≥60 years old, with no CAD on angiography and no history of cerebro- or peripheral vascular disease, MI or coronary revascularization, n = 410). These age limits were based on the incidence curves of CAD and the estimates of genetic effect at various ages (13–15). We performed analyses for all subjects and for whites only.
The case–control selection scheme described above identified CATHGEN subjects at the extremes of the distributions for CADi and age. In order to test for an age-by-genotype interaction with CAD severity as the outcome, we selected an additional random sample of 481 CATHGEN subjects from across all strata of age and CADi. We assigned sampling probabilities as a function of CADi, age (in 5 year increments) and the total number of CATHGEN subjects with that CADi—age combination. Sampling probabilities were ∼1.0 in the CADi/age categories defining case–control subsets. A total of 1330 individuals were selected for the case–control subsets defined above as young affected, older affected and older normal. Including these subjects and the 481 subjects from the CATHGEN ‘random sample’, we constituted a sample (n = 1811) that spans the entire range of age (18–91 years) and CADi (Table 1). Consequently, applying these sampling probabilities as observation weights allows an unbiased analysis of age and genotype effects on the CADi in the CATHGEN cohort.
GENECARD is a multicenter, international study of families with at least two siblings with early-onset CAD as documented in the medical record and as we have reported previously (42,43). Enrollment spanned March 1998 through 31 March 2002. All subjects signed consent forms approved by local Institutional Review Boards. The characteristics of the probands in this study group are summarized in Table 1 and in our previous work (42,43). Our collection includes, from all families, a large number of subjects designated as ‘unaffected by CAD’—signifying probands' siblings and relatives who have not been diagnosed with CAD, and are >55 years old (men) or >60 years old (women), for a total of 2954 subjects in 1101 families. For the purposes of providing an additional CAD case comparison group, we selected the 759 probands from the US-based GENECARD collection sites. These individuals were compared with CATHGEN controls.
Selection of polymorphisms to genotype in TNFR1 (TNFRSF1A)
The HapMap (http://www.hapmap.org) and SeattleSNPs (http://pga.gs.washington.edu/) databases were used to identify SNPs which represent all of the variation (tagSNPs) within the TNFRSF1A gene (TNFR1) as well as within 3000 bp both upstream and downstream of the gene (to account for putative promoter and downstream regulatory elements). A total of 11 validated tagSNPs were identified with a minor allele frequency of ≥5% among Caucasian Americans of European descent. LDSelect (44) and Tagger (45) were used to determine that these SNPs, using an r2 cutoff of 0.7, represent 11 LD bins. Each of these tagSNPs was genotyped in our CATHGEN sample. In addition, 10 novel TNFRSF1A polymorphisms were previously identified within subjects with premature MI (Supplementary Material, Table S3) (11), and we incorporated these into our study: 3 were already among the tagSNPs we selected; 2 were found to be monomorphic (and were, therefore, not included in the analysis); 5 SNPs were added to our 11 validated tagSNPs, so that we genotyped a total of 16 TNFR1 SNPs.
Genotyping methods
Genomic DNA for CATHGEN and GENECARD was extracted from whole blood using the Puregene system (Gentra Systems, Minneapolis, MN, USA) in the Duke Center for Human Genetics DNABank. All SNPs were genotyped at the Duke Center for Human Genetics genotyping laboratory using TaqMan allelic discrimination assays. A total of 15 quality control samples—composed of six reference genotype controls in duplicate, two CEPH (Centre d'Étude du Polymorphisme Humain) pedigree individuals and one no-template sample—were included in each quadrant of the 384-well plate. SNPs that showed mismatches on quality control samples were reviewed by an independent genotyping supervisor for potential genotyping errors. All SNPs examined were successfully genotyped for ≥95% of the individuals in the study. Among SNPs that passed genotyping quality control, error rates (determined by blinded duplicate samples) were <0.2%.
Statistical analysis of genetic epidemiological data
All SNPs were tested for deviations (P ≤ 0.0001) from Hardy–Weinberg equilibrium (HWE) in the affected and unaffected groups. None of the SNPs assayed showed deviation from HWE. LD between pairs of SNPs was assessed using the Graphical Overview of Linkage Disequilibrium package (46). SAS 9.1 (SAS Institute, Cary, NC, USA) was used for statistical analysis. Association between CAD and TNFR1 SNPs was evaluated using multivariable logistic regression modeling in CATHGEN cases, GENECARD probands and CATHGEN controls adjusted for covariates of race, sex and known CAD risk factors (history of hypertension, history of diabetes mellitus, body mass index (BMI), history of dyslipidemia and smoking history). These adjustments could hypothetically allow us to control for competing genetic pathways that are independent risk factors for CAD, thereby allowing us to detect a separate CAD genetic effect. We fitted the regression models for all subjects with an indicator for race in the model, and also for white subjects only. The results were similar for both analyses (data not shown).
To determine whether the relationship between CADi and genotype varies across the age range in our study, we performed analyses to evaluate age-by-genotype interactions. We used regression analysis as implemented in PROC SURVEYREG in SAS version 9.1 (SAS Institute), with CADi as the dependent variable. Each observation was weighted by the sampling probabilities defined above. The regression models included CAD risk factors, race, age, genotype and an age-by-genotype interaction term. A significant regression coefficient for the interaction term suggests that the relationship between CADi and aging is modified by genotype. Using the sample weights also allows unbiased estimates of genetic effects for the entire Duke Catheterization cohort.
We also performed family-based association analysis for CAD and our 16 TNFR1 SNPs in GENECARD families using the association in the presence of linkage (APL) test (47) and the pedigree disequilibrium test (PDT) (48), both of which test the hypothesis that there is distortion of transmission to affected children. The APL test uses nuclear families (including affected sibling pair families) with appropriate inference in regions of linkage when parents are missing. The APL test was performed on the full GENECARD data set of 1101 families. The PDT test uses any type of pedigree, but requires genotypes for parents or unaffected siblings to evaluate allelic transmission; 234 GENECARD families were appropriate for PDT analysis. Thus, the APL and PDT may detect slightly different effects in different subsets of the data, such as individuals with one or more parents or unaffected siblings available to provide a DNA sample.
Mouse atherosclerosis studies
All mouse experiments complied with Duke University Institutional Animal Care and Use Committee guidelines. WT, Tnfr1−/− and Apoe−/− mouse lines were congenic in the C57BL/6 background, obtained from Jackson Laboratories, and interbred with each other every fifth generation to minimize genetic drift. PCR genotyping was performed by protocols specified by Jackson Laboratories. All mice were fed Purina Breeder Chow.
Carotid interposition grafting was performed as we described previously (4). The right common carotid artery from WT or Tnfr1−/− ‘donor’ mice (∼8 mm long) was transplanted orthotopically into Apoe−/− ‘recipient’ mice. Anesthesia for both donor and recipient mice was achieved with pentobarbital (50 mg/kg, i.p.). Because the recipient common carotid was ligated and cut after end-to-side anastomoses were completed, all right carotid blood flow was conducted through the graft. Donor mice were gender-matched across surgical groups, stratified by age: ‘young’ (10 ± 2 weeks old) or ‘aged’ (79 ± 2 weeks old). Whereas young donor mice weighed less than aged donors (21 ± 3 versus 33 ± 8 gm), dimensions of the right common carotid artery (cross-sectional areas of the lumen and media) were indistinguishable between young and aged cohorts (data not shown). The Apoe−/− recipient mice were only young (10 ± 2 weeks old); they were gender-matched across surgical groups and had equivalent weights (22 ± 4 g). After carotid interposition grafting, Apoe−/− recipient mice were housed singly, fed normal chow and sacrificed at 1, 2 or 7 weeks post-operatively. Carotid grafts were harvested as described (4), either after perfusion fixation at 80 mmHg (for paraffin-embedded specimens used for morphometry) or after PBS perfusion, embedding in OCT compound and freezing (for immunostaining).
Analyses of carotid artery grafts
For morphometry, we stained 5 μm sections of paraffin-embedded grafts with a modified Masson's trichrome and Verhoeff's elastic tissue stain, as described (49), to render collagen green, elastin and nuclei black, and cytoplasm red. With Scion Image™, neointimal area was calculated from ×220 photomicrographs after tracing perimeters of the endothelium and the internal elastic lamina; medial area was measured similarly between the internal and external elastic laminae; arterial external diameter was calculated from the perimeter of the external elastic lamina, dividing by π. Identification of intra-plaque erythrocytes and specimen morphometry were performed by observers blinded to specimen identity.
Immunofluorescence was performed as described (50), with IgGs from the following sources: goat or rabbit IgG targeting ICAM-1 (sc-7897), VCAM-1 (sc-8304), MCP-1 (sc-1784), TNFR1 (sc-7895), MMP-9 (sc-6841), apoE (sc-6385) or MMP-9 (sc-6841) from Santa Cruz Biotechnology, Inc; mouse IgG2a and IgG1, respectively, targeting PCNA and collagen I, and rabbit IgG targeting nitrotyrosine from Sigma-Aldrich, Inc; rabbit IgG targeting cleaved (activated) caspase-3 from Cell Signaling Technology, Inc.; FITC-conjugated rat M3/84 IgG1(κ) targeting Mac3 (macrophage antigen) from BD Pharmingen, Inc. Non-immune goat or rabbit IgG, isotype control mouse IgG or FITC-conjugated rat IgG1κ was used on cognate sections to determine non-specific fluorescence. Cy3-conjugated 1A4 IgG (Sigma) was used to detect SMC α-actin; the DNA-binding fluorophore Hoechst 33342 (10 μg/ml) was added to secondary antibody incubations. To minimize elastic laminae fluorescence, we used 0.2% gelatin in both blocking and IgG diluent buffer: 20 mmol/l Tris–Cl (pH 7.5)/125 mmol/l NaCl/0.1% (vol./vol.) Tween-20. Single microscopic fields were imaged for multiple fluorophores, and protein immunofluorescence was quantitated by normalizing to nuclear DNA fluorescence (as a measure of cellularity), exactly as described (50). All specimens stained within a single batch were imaged with identical CCD camera settings. In all cases, non-specific fluorescence (with non-immune primary IgG) was subtracted from total fluorescence (with specific primary IgG) to obtain antigen-specific fluorescence. Immunofluorescence was quantitated by observers blinded to specimen identity. Before surgery, there was equivalent apoE expression in carotid arteries from mice of both ages and genotypes used (Supplementary Material, Fig. S2; data not shown).
Statistical analyses of carotid artery grafts
Data are presented as mean ± SD in the text, and ±SE in the figures. Morphometry and protein expression data were analyzed by two-way ANOVA with Tukey's post hoc test for multiple comparisons. Proportions (for plaque hemorrhage) were analyzed by Fisher's exact test.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institutes of Health [grant numbers HL73005, HL77185 and AG25462 (N.J.F.); HL073389, HL73042 and AG028716 (E.R.H. and W.E.K.); HL089412 (J.J.C.); and HL72842 (K.P.)].
Supplementary Material
REFERENCES
- 1.Lakatta E.G., Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises. Part I: aging arteries: a ‘set up’ for vascular disease. Circulation. 2003;107:139–146. doi: 10.1161/01.cir.0000048892.83521.58. doi:10.1161/01.CIR.0000048892.83521.58. [DOI] [PubMed] [Google Scholar]
- 2.Minamino T., Komuro I. Vascular cell senescence: contribution to atherosclerosis. Circ. Res. 2007;100:15–26. doi: 10.1161/01.RES.0000256837.40544.4a. doi:10.1161/01.RES.0000256837.40544.4a. [DOI] [PubMed] [Google Scholar]
- 3.Collins A.R., Lyon C.J., Xia X., Liu J.Z., Tangirala R.K., Yin F., Boyadjian R., Bikineyeva A., Pratico D., Harrison D.G., et al. Age-accelerated atherosclerosis correlates with failure to upregulate antioxidant genes. Circ. Res. 2009;104:e42–e54. doi: 10.1161/CIRCRESAHA.108.188771. doi:10.1161/CIRCRESAHA.108.188771. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Peppel K., Sivashanmugam P., Orman E.S., Brian L., Exum S.T., Freedman N.J. Expression of tumor necrosis factor receptor-1 in arterial wall cells promotes atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007;27:1087–1094. doi: 10.1161/ATVBAHA.0000261548.49790.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csiszar A., Ungvari Z., Koller A., Edwards J.G., Kaley G. Proinflammatory phenotype of coronary arteries promotes endothelial apoptosis in aging. Physiol. Genomics. 2004;17:21–30. doi: 10.1152/physiolgenomics.00136.2003. [DOI] [PubMed] [Google Scholar]
- 6.Spinetti G., Wang M., Monticone R., Zhang J., Zhao D., Lakatta E.G. Rat aortic MCP-1 and its receptor CCR2 increase with age and alter vascular smooth muscle cell function. Arterioscler. Thromb. Vasc. Biol. 2004;24:1397–1402. doi: 10.1161/01.ATV.0000134529.65173.08. doi:10.1161/01.ATV.0000134529.65173.08. [DOI] [PubMed] [Google Scholar]
- 7.Schreyer S.A., Vick C.M., LeBoeuf R.C. Loss of lymphotoxin-alpha but not tumor necrosis factor-alpha reduces atherosclerosis in mice. J. Biol. Chem. 2002;277:12364–12368. doi: 10.1074/jbc.M111727200. doi:10.1074/jbc.M111727200. [DOI] [PubMed] [Google Scholar]
- 8.Bruunsgaard H., Pedersen B.K. Age-related inflammatory cytokines and disease. Immunol. Allergy Clin. North Am. 2003;23:15–39. doi: 10.1016/s0889-8561(02)00056-5. doi:10.1016/S0889-8561(02)00056-5. [DOI] [PubMed] [Google Scholar]
- 9.Ridker P.M., Rifai N., Pfeffer M., Sacks F., Lepage S., Braunwald E. Elevation of tumor necrosis factor-alpha and increased risk of recurrent coronary events after myocardial infarction. Circulation. 2000;101:2149–2153. doi: 10.1161/01.cir.101.18.2149. [DOI] [PubMed] [Google Scholar]
- 10.Bruunsgaard H., Andersen-Ranberg K., Hjelmborg J.B., Pedersen B.K., Jeune B. Elevated levels of tumor necrosis factor alpha and mortality in centenarians. Am. J. Med. 2003;115:278–283. doi: 10.1016/s0002-9343(03)00329-2. doi:10.1016/S0002-9343(03)00329-2. [DOI] [PubMed] [Google Scholar]
- 11.Poirier O., Nicaud V., Gariepy J., Courbon D., Elbaz A., Morrison C., Kee F., Evans A., Arveiler D., Ducimetiere P., et al. Polymorphism R92Q of the tumour necrosis factor receptor 1 gene is associated with myocardial infarction and carotid intima-media thickness—the ECTIM, AXA, EVA and GENIC studies. Eur. J. Hum. Genet. 2004;12:213–219. doi: 10.1038/sj.ejhg.5201143. doi:10.1038/sj.ejhg.5201143. [DOI] [PubMed] [Google Scholar]
- 12.Shah S.H., Freedman N.J., Zhang L., Crosslin D.R., Stone D.H., Haynes C., Johnson J., Nelson S., Wang L., Connelly J.J., et al. Neuropeptide Y gene polymorphisms confer risk of early-onset atherosclerosis. PLoS Genet. 2009;5:e1000318. doi: 10.1371/journal.pgen.1000318. doi:10.1371/journal.pgen.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shea S., Ottman R., Gabrieli C., Stein Z., Nichols A. Family history as an independent risk factor for coronary artery disease. J. Am. Coll. Cardiol. 1984;4:793–801. doi: 10.1016/s0735-1097(84)80408-8. [DOI] [PubMed] [Google Scholar]
- 14.Rissanen A.M. Familial occurrence of coronary heart disease: effect of age at diagnosis. Am. J. Cardiol. 1979;44:60–66. doi: 10.1016/0002-9149(79)90251-0. doi:10.1016/0002-9149(79)90251-0. [DOI] [PubMed] [Google Scholar]
- 15.Marenberg M.E., Risch N., Berkman L.F., Floderus B., de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. doi:10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 16.Moss A.J. Profile of high risk in people known to have coronary heart disease: a review. Circulation. 1975;52:III147–III154. [PubMed] [Google Scholar]
- 17.Lerner D.J., Kannel W.B. Patterns of coronary heart disease morbidity and mortality in the sexes: a 26-year follow-up of the Framingham population. Am. Heart J. 1986;111:383–390. doi: 10.1016/0002-8703(86)90155-9. doi:10.1016/0002-8703(86)90155-9. [DOI] [PubMed] [Google Scholar]
- 18.Wilson P.W., Castelli W.P., Kannel W.B. Coronary risk prediction in adults (the Framingham Heart Study) Am. J. Cardiol. 1987;59:91G–94G. doi: 10.1016/0002-9149(87)90165-2. doi:10.1016/0002-9149(87)90165-2. [DOI] [PubMed] [Google Scholar]
- 19.Donato A.J., Eskurza I., Silver A.E., Levy A.S., Pierce G.L., Gates P.E., Seals D.R. Direct evidence of endothelial oxidative stress with aging in humans: relation to impaired endothelium-dependent dilation and upregulation of nuclear factor-kappaB. Circ. Res. 2007;100:1659–1666. doi: 10.1161/01.RES.0000269183.13937.e8. doi:10.1161/01.RES.0000269183.13937.e8. [DOI] [PubMed] [Google Scholar]
- 20.Hansson G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. doi:10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 21.Jackson C.L., Bennett M.R., Biessen E.A., Johnson J.L., Krams R. Assessment of unstable atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 2007;27:714–720. doi: 10.1161/01.ATV.0000261873.86623.e1. doi:10.1161/01.ATV.0000261873.86623.e1. [DOI] [PubMed] [Google Scholar]
- 22.Rekhter M.D., Hicks G.W., Brammer D.W., Hallak H., Kindt E., Chen J., Rosebury W.S., Anderson M.K., Kuipers P.J., Ryan M.J. Hypercholesterolemia causes mechanical weakening of rabbit atheroma: local collagen loss as a prerequisite of plaque rupture. Circ. Res. 2000;86:101–108. doi: 10.1161/01.res.86.1.101. [DOI] [PubMed] [Google Scholar]
- 23.Lasky-Su J., Lyon H.N., Emilsson V., Heid I.M., Molony C., Raby B.A., Lazarus R., Klanderman B., Soto-Quiros M.E., Avila L., et al. On the replication of genetic associations: timing can be everything! Am. J. Hum. Genet. 2008;82:849–858. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bureau A., Diallo M.S., Ordovas J.M., Cupples L.A. Estimating interaction between genetic and environmental risk factors: efficiency of sampling designs within a cohort. Epidemiology. 2008;19:83–93. doi: 10.1097/EDE.0b013e31815c4d0e. doi:10.1097/EDE.0b013e31815c4d0e. [DOI] [PubMed] [Google Scholar]
- 25.Rauscher F.M., Goldschmidt-Clermont P.J., Davis B.H., Wang T., Gregg D., Ramaswami P., Pippen A.M., Annex B.H., Dong C., Taylor D.A. Aging, progenitor cell exhaustion, and atherosclerosis. Circulation. 2003;108:457–463. doi: 10.1161/01.CIR.0000082924.75945.48. doi:10.1161/01.CIR.0000082924.75945.48. [DOI] [PubMed] [Google Scholar]
- 26.Hlatky M.A., Greenland P., Arnett D.K., Ballantyne C.M., Criqui M.H., Elkind M.S., Go A.S., Harrell F.E., Jr, Howard B.V., Howard V.J., et al. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–2416. doi: 10.1161/CIRCULATIONAHA.109.192278. doi:10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cifuentes-Diaz C., Bitoun M., Goudou D., Seddiqi N., Romero N., Rieger F., Perin J.P., Alliel P.M. Neuromuscular expression of the BTB/POZ and zinc finger protein myoneurin. Muscle Nerve. 2004;29:59–65. doi: 10.1002/mus.10526. doi:10.1002/mus.10526. [DOI] [PubMed] [Google Scholar]
- 28.Csiszar A., Wang M., Lakatta E.G., Ungvari Z.I. Inflammation and endothelial dysfunction during aging: role of NF-κB. J. Appl. Physiol. 2008;105:1333–1341. doi: 10.1152/japplphysiol.90470.2008. doi:10.1152/japplphysiol.90470.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., Sivashanmugam P., Wu J.H., Brian L., Exum S.T., Freedman N.J., Peppel K. Tumor necrosis factor receptor-2 signaling attenuates vein graft neointima formation by promoting endothelial recovery. Arterioscler. Thromb. Vasc. Biol. 2008;28:284–289. doi: 10.1161/ATVBAHA.107.151613. doi:10.1161/ATVBAHA.107.151613. [DOI] [PubMed] [Google Scholar]
- 30.De Keulenaer G.W., Alexander R.W., Ushio-Fukai M., Ishizaka N., Griendling K.K. Tumour necrosis factor alpha activates a p22phox-based NADH oxidase in vascular smooth muscle. Biochem. J. 1998;329:653–657. doi: 10.1042/bj3290653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozsoy H.Z., Sivasubramanian N., Wieder E.D., Pedersen S., Mann D.L. Oxidative stress promotes ligand-independent and enhanced ligand-dependent tumor necrosis factor receptor signaling. J. Biol. Chem. 2008;283:23419–23428. doi: 10.1074/jbc.M802967200. doi:10.1074/jbc.M802967200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csiszar A., Labinskyy N., Smith K., Rivera A., Orosz Z., Ungvari Z. Vasculoprotective effects of anti-tumor necrosis factor-alpha treatment in aging. Am. J. Pathol. 2007;170:388–398. doi: 10.2353/ajpath.2007.060708. doi:10.2353/ajpath.2007.060708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann D.L., McMurray J.J., Packer M., Swedberg K., Borer J.S., Colucci W.S., Djian J., Drexler H., Feldman A., Kober L., et al. Targeted anticytokine therapy in patients with chronic heart failure: results of the Randomized Etanercept Worldwide Evaluation (RENEWAL) Circulation. 2004;109:1594–1602. doi: 10.1161/01.CIR.0000124490.27666.B2. doi:10.1161/01.CIR.0000124490.27666.B2. [DOI] [PubMed] [Google Scholar]
- 34.Fortin D.F., Califf R.M., Pryor D.B., Mark D.B. The way of the future redux. Am. J. Cardiol. 1995;76:1177. doi: 10.1016/s0002-9149(99)80331-2. doi:10.1016/S0002-9149(99)80331-2. [DOI] [PubMed] [Google Scholar]
- 35.Smith L.R., Harrell F.E., Jr, Rankin J.S., Califf R.M., Pryor D.B., Muhlbaier L.H., Lee K.L., Mark D.B., Jones R.H., Oldham H.N., et al. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84:III245–III253.. [PubMed] [Google Scholar]
- 36.Connelly J.J., Shah S.H., Doss J.F., Gadson S., Nelson S., Crosslin D.R., Hale A.B., Lou X., Wang T., Haynes C., et al. Genetic and functional association of FAM5C with myocardial infarction. BMC Med. Genet. 2008;9:33. doi: 10.1186/1471-2350-9-33. doi:10.1186/1471-2350-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connelly J.J., Wang T., Cox J.E., Haynes C., Wang L., Shah S.H., Crosslin D.R., Hale A.B., Nelson S., Crossman D.C., et al. GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet. 2006;2:1265–1273. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosslin D.R., Shah S.H., Nelson S.C., Haynes C.S., Connelly J.J., Gadson S., Goldschmidt-Clermont P.J., Vance J.M., Rose J., Granger C.B., et al. Genetic effects in the leukotriene biosynthesis pathway and association with atherosclerosis. Hum. Genet. 2009;125:217–229. doi: 10.1007/s00439-008-0619-0. doi:10.1007/s00439-008-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Hauser E.R., Shah S.H., Pericak-Vance M.A., Haynes C., Crosslin D., Harris M., Nelson S., Hale A.B., Granger C.B., et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am. J. Hum. Genet. 2007;80:650–663. doi: 10.1086/512981. doi:10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong D.F., Shaw L.K., Harrell F.E., Muhlbaier L.H., Lee K.L., Califf R.M., Jones R.H. Predicting survival from the coronary arteriogram: an experience-based statistical index of coronary artery disease severity. J. Am. Coll. Cardiol. 2002;39(Suppl. A):327A. (Abstract) [Google Scholar]
- 41.Felker G.M., Shaw L.K., O'Connor C.M. A standardized definition of ischemic cardiomyopathy for use in clinical research. J. Am. Coll. Cardiol. 2002;39:210–218. doi: 10.1016/s0735-1097(01)01738-7. doi:10.1016/S0735-1097(01)01738-7. [DOI] [PubMed] [Google Scholar]
- 42.Hauser E.R., Crossman D.C., Granger C.B., Haines J.L., Jones C.J., Mooser V., McAdam B., Winkelmann B.R., Wiseman A.H., Muhlestein J.B., et al. A genome-wide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am. J. Hum. Genet. 2004;75:436–447. doi: 10.1086/423900. doi:10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauser E.R., Mooser V., Crossman D.C., Haines J.L., Jones C.H., Winkelmann B.R., Schmidt S., Scott W.K., Roses A.D., Pericak-Vance M.A., et al. Design of the Genetics of Early Onset Cardiovascular Disease (GENECARD) study. Am. Heart J. 2003;145:602–613. doi: 10.1067/mhj.2003.13. doi:10.1067/mhj.2003.13. [DOI] [PubMed] [Google Scholar]
- 44.Carlson C.S., Eberle M.A., Rieder M.J., Yi Q., Kruglyak L., Nickerson D.A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. doi:10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. doi:10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 46.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97. doi: 10.1038/ng786. doi:10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 47.Martin E.R., Bass M.P., Hauser E.R., Kaplan N.L. Accounting for linkage in family-based tests of association with missing parental genotypes. Am. J. Hum. Genet. 2003;73:1016. doi: 10.1086/378779. doi:10.1086/378779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin E.R., Monks S.A., Warren L.L., Kaplan N.L. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am. J. Hum. Genet. 2000;67:146. doi: 10.1086/302957. doi:10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L., Hagen P.O., Kisslo J., Peppel K., Freedman N.J. Neointimal hyperplasia rapidly reaches steady state in a novel murine vein graft model. J. Vasc. Surg. 2002;36:824–832. [PubMed] [Google Scholar]
- 50.Zhang L., Peppel K., Brian L., Chien L., Freedman N.J. Vein graft neointimal hyperplasia is exacerbated by tumor necrosis factor receptor-1 signaling in graft-intrinsic cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:2277–2283. doi: 10.1161/01.ATV.0000147766.68987.0d. doi:10.1161/01.ATV.0000147766.68987.0d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.