Abstract
Recent studies have identified single nucleotide polymorphisms (SNPs) that significantly modify breast cancer risk in BRCA1 and BRCA2 mutation carriers. Since these risk modifiers were originally identified as genetic risk factors for breast cancer in genome-wide association studies (GWASs), additional risk modifiers for BRCA1 and BRCA2 may be identified from promising signals discovered in breast cancer GWAS. A total of 350 SNPs identified as candidate breast cancer risk factors (P < 1 × 10−3) in two breast cancer GWAS studies were genotyped in 3451 BRCA1 and 2006 BRCA2 mutation carriers from nine centers. Associations with breast cancer risk were assessed using Cox models weighted for penetrance. Eight SNPs in BRCA1 carriers and 12 SNPs in BRCA2 carriers, representing an enrichment over the number expected, were significantly associated with breast cancer risk (Ptrend < 0.01). The minor alleles of rs6138178 in SNRPB and rs6602595 in CAMK1D displayed the strongest associations in BRCA1 carriers (HR = 0.78, 95% CI: 0.69–0.90, Ptrend = 3.6 × 10−4 and HR = 1.25, 95% CI: 1.10–1.41, Ptrend = 4.2 × 10−4), whereas rs9393597 in LOC134997 and rs12652447 in FBXL7 showed the strongest associations in BRCA2 carriers (HR = 1.55, 95% CI: 1.25–1.92, Ptrend = 6 × 10−5 and HR = 1.37, 95% CI: 1.16–1.62, Ptrend = 1.7 × 10−4). The magnitude and direction of the associations were consistent with the original GWAS. In subsequent risk assessment studies, the loci appeared to interact multiplicatively for breast cancer risk in BRCA1 and BRCA2 carriers. Promising candidate SNPs from GWAS were identified as modifiers of breast cancer risk in BRCA1 and BRCA2 carriers. Upon further validation, these SNPs together with other genetic and environmental factors may improve breast cancer risk assessment in these populations.
INTRODUCTION
Deleterious germline mutations in the BRCA1 and BRCA2 genes confer high risks of breast and ovarian cancer. However, there is a substantial variation in breast cancer risks among carriers of BRCA1 and BRCA2 mutations, due to a combination of genetic and lifestyle factors, with penetrance estimated at between 40 and 87% by age 70 (1–7). In addition, there is significant variability in the age of onset of cancer and the site of cancer in these populations (1–3). Studies of high-risk families quantifying the extent of risk variation have suggested that other genetic factors may modify the risk of breast cancer associated with BRCA1 and BRCA2 mutations (4–6).
Initial efforts to identify genetic modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers through candidate gene studies established that homozygosity for the RAD51 135G>C allele is associated with increased breast cancer risk in BRCA2 mutation carriers, but not in BRCA1 carriers (7–10). Subsequent efforts have focused on evaluation of common breast cancer susceptibility loci that harbor single nucleotide polymorphisms (SNPs) exhibiting genome-wide significant (P < 1 × 10−7) associations with breast cancer (11–17). Studies of six of these SNPs by the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) recently showed that SNPs in the FGFR2, MAP3K1, TNRC9, LSP1 and 2q35 loci but not the 8q24 locus were significantly associated with breast cancer risk in BRCA2 mutation carriers (18,19). In contrast, only SNPs in the TNRC9 and 2q35 loci exhibited significant associations with risk in BRCA1 carriers (18,19). Since ∼70% of breast tumors in BRCA1 carriers are estrogen receptor (ER) negative, this finding is consistent with the observation that these SNPs are more strongly associated with ER positive than ER negative breast cancer in the general population (20). Together, these variants account for 0.7 and 3.7% of the genetic modifying variance of breast cancer risk in BRCA1 and BRCA2 carriers, respectively (21). Thus, a large proportion of the genetic variability underlying breast cancer in BRCA1 and BRCA2 carriers remains to be elucidated.
These findings suggest that selection of SNPs displaying highly significant associations with breast cancer in genome-wide association study (GWAS) of unselected breast cancers is an efficient method for identification of modifiers of breast cancer risk in BRCA1 and BRCA2 carriers. Under this model, which relies on selection of small numbers of high-quality candidate SNPs displaying highly significant associations with the phenotype of interest, it may be possible to avoid the need for additional GWAS and to reduce the number of samples required to identify variants significantly associated with the related disease phenotype. In order to test this model and to identify additional common genetic variants associated with breast cancer risk in BRCA1 and BRCA2 carriers, we selected 295 SNPs highly significantly associated with breast cancer risk (P < 10−3) in breast cancer GWAS conducted in the UK (UK1) (11,15) and by the Cancer Genetic Markers of Susceptibility (CGEMS) consortium (12,17), along with 55 SNPs selected from candidate SNPs displaying association with breast cancer risk in a Mayo Clinic breast cancer case–control study. These variants were genotyped in BRCA1 and BRCA2 mutation carriers from nine studies involved in CIMBA. Here we demonstrate that a number of SNPs associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers can be identified using this model and we evaluate the influence of the SNPs on age-related risk of breast cancer to show the potential utility of risk modifiers for cancer risk assessment in the BRCA1 and BRCA2 carrier populations.
RESULTS
Genotyping data for 333 SNPs on 3451 BRCA1 and 2006 BRCA2 carriers from nine centers were available for analysis after exclusion of 225 samples and 17 SNPs during quality assessment. A total of 3030 carriers from the nine study groups were diagnosed with breast cancer, while 2427 additional carriers were unaffected at the date of last observation (n = 1807), were diagnosed with ovarian cancer (n = 412) or had undergone a bilateral prophylactic mastectomy (n = 208). From the overall analysis of individual SNPs, a total of 8 displayed significant associations (P < 0.01) with breast cancer risk in BRCA1 mutation carriers and 12 exhibited associations (P < 0.01) with breast cancer in BRCA2 mutation carriers. In contrast, only three were expected to be associated with breast cancer in BRCA1 and BRCA2 carriers by chance. The estimated HRs and P-values of the SNPs exhibiting significant associations (P < 0.01) are shown in Table 1.
Table 1.
Associations between GWAS SNPs and risk of breast cancer (P trend ≤ 0.01) in BRCA1 and BRCA2 mutation carriers
| rsID | Gene name | HR (95% CI) (per allele) | Ptrend | FDR qvalue | HR (95% CI) |
P2df | |
|---|---|---|---|---|---|---|---|
| One copy | Two copies | ||||||
| BRCA1 carriers | |||||||
| rs6138178 | SNRPB | 0.78 (0.69–0.90) | 3.6 × 10−4 | 0.07 | 0.75 (0.63–0.90) | 0.66 (0.49–0.90) | 0.0015 |
| rs6602595 | CAMK1D | 1.25 (1.10–1.41) | 4.2 × 10−4 | 0.07 | 1.23 (1.02–1.48) | 1.56 (1.21–2.01) | 0.0019 |
| rs6590224 | KIRREL3 | 1.26 (1.10–1.45) | 0.0013 | 0.13 | 1.25 (1.05–1.50) | 1.61 (1.10–2.37) | 0.0056 |
| rs1048635 | TRIM45 | 1.50 (1.17–1.94) | 0.0015 | 0.13 | 1.59 (1.22–2.09) | 1.15 (0.24–5.44) | 0.0029 |
| rs7629693 | HMGN2P7 | 0.83 (0.73–0.94) | 0.0041 | 0.25 | 0.88 (0.74–1.05) | 0.65 (0.49–0.86) | 0.0104 |
| rs6005863 | HSCB | 0.85 (0.76–0.95) | 0.0049 | 0.25 | 1.01 (0.84–1.22) | 0.66 (0.52–0.85) | 0.0007 |
| rs4415084 | MRPS30 | 0.84 (0.75–0.95) | 0.0052 | 0.25 | 0.80 (0.67–0.96) | 0.73 (0.57–0.93) | 0.015 |
| rs2292326 | NECAB2 | 1.27 (1.06–1.52) | 0.0087 | 0.36 | 1.31 (1.07–1.61) | 1.33 (0.68–2.57) | 0.026 |
| BRCA2 carriers | |||||||
| rs9393597 | LOC134997 | 1.55 (1.25–1.92) | 6.0 × 10−5 | 0.018 | 1.56 (1.18–2.08) | 2.33 (1.23–4.39) | 0.00030 |
| rs12652447 | FBXL7 | 1.37 (1.16–1.62) | 1.7 × 10−4 | 0.028 | 1.25 (0.96–1.62) | 1.92 (1.39–2.67) | 0.00041 |
| rs6837016 | LOC729902 | 0.76 (0.63–0.90) | 0.0020 | 0.20 | 0.68 (0.53–0.88) | 0.60 (0.42–0.85) | 0.0046 |
| rs909116 | TNNT3 | 0.79 (0.68–0.92) | 0.0025 | 0.20 | 0.88 (0.68–1.14) | 0.61 (0.45–0.84) | 0.0078 |
| rs2067980 | MRPS30 | 1.42 (1.12–1.80) | 0.0036 | 0.22 | 1.31 (1.02–1.69) | 3.86 (1.40–10.6) | 0.0053 |
| rs4660891 | MAST2 | 1.32 (1.09–1.60) | 0.0040 | 0.22 | 1.50 (1.18–1.92) | 1.44 (0.88–2.35) | 0.0040 |
| rs10941679 | MRPS30 | 1.31 (1.08–1.58) | 0.0054 | 0.26 | 1.33 (1.06–1.68) | 1.65 (1.00–2.72) | 0.019 |
| rs1533802 | LOC339778 | 0.79 (0.67–0.94) | 0.0072 | 0.28 | 0.89 (0.70–1.13) | 0.50 (0.32–0.78) | 0.0090 |
| rs17827708 | DSEL | 0.74 (0.59–0.93) | 0.0093 | 0.28 | 0.70 (0.54–0.90) | 0.77 (0.37–1.62) | 0.018 |
| rs7185203 | CMIP | 0.79 (0.67–0.95) | 0.010 | 0.28 | 0.84 (0.66–1.07) | 0.59 (0.40–0.88) | 0.032 |
| rs6504950 | STXBP4 | 1.27 (1.06–1.53) | 0.010 | 0.028 | 1.31 (1.04–1.65) | 1.53 (0.94–2.49) | 0.034 |
| rs7090828 | PCBD1 | 0.81 (0.69–0.95) | 0.010 | 0.28 | 0.93 (0.72–1.21) | 0.62 (0.45–0.87) | 0.012 |
FDR, false discovery rate; 2df, two degree of freedom model.
Several SNPs recently identified and validated as genetic risk factors for breast cancer in the general population through breast cancer GWAS were included in this study and were evaluated as modifiers of breast cancer risk in BRCA1 and BRCA2 carriers. rs4415084 from the MRPS30 locus on chromosome 5p12 (14) (P = 0.005) and rs2107425 from H19 (11) (P = 0.015) were significantly associated with breast cancer in BRCA1 carriers, whereas the MRPS30 SNPs, rs10941679 (P = 0.0054) and rs2067980 (P = 0.0036), but not rs4415084, were associated with breast cancer in BRCA2 carriers (Table 1; Supplementary Material, Table S1). In addition, rs6504950 from the STXBP4/COX11 locus at 17q23.2 (15), rs16886165 from the MAP3K1 locus (11), rs6735174 from the TNP1 locus at 2q35 in high LD (r2 = 0.80) with rs13387042 (17) and rs2107425 from the H19 locus (11) displayed marginally significant associations (P < 0.05) in BRCA2 carriers, but not in BRCA1 carriers (Table 1; Supplementary Material, Table S5). In contrast, rs1357245 from NEK10 at 3p24 (15), rs4973768 from SLC4A7 at 3p24 (15) and rs999737 from RAD51L1 at 1p11.2 (17) did not display significant associations with breast cancer risk in either BRCA1 or BRCA2 carriers (Table 1; Supplementary Material, Table S1 and S5).
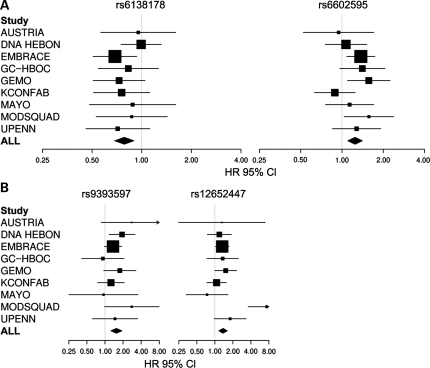
The SNPs showing the strongest associations with breast cancer in BRCA1 carriers in this study were rs6138178 in SNRPB and rs6602595 in CAMK1D (HR = 0.78; 95% CI: 0.69–0.90, Ptrend = 0.00036, and HR = 1.25; 95% CI: 1.10–1.41, Ptrend = 0.00042, respectively). When applying false discovery rate (FDR) to account for multiple testing, these SNPs showed probabilities of false positive associations of less than 7% (q = 0.07) in BRCA1 carriers (Table 1). For both SNPs, the estimated risk for heterozygotes and homozygotes were consistent with a multiplicative model (rs6138178 HR(het) = 0.75; 95% CI: 0.63–0.90 and HR(hom) = 0.66; 95% CI: 0.49–0.90) (rs6602595 HR(het) = 1.23; 95% CI: 1.02–1.48 and HR(hom) = 1.56; 95% CI: 1.21–2.01). No apparent heterogeneity in the study-specific HR was observed for rs6138178 (Phet = 0.82) or rs6602595 (Phet = 0.27) for BRCA1 mutation carriers (Fig. 1).
Figure 1.
Study-specific per allele hazards ratio estimates for selected SNPs in BRCA1 and BRCA2 mutation carriers. (A) HR estimates for rs6138178 from the SNRPB locus and rs6602595 from the CAMK1D locus. (B) HR estimates for rs12652447 from the FBXL7 locus and rs9393597 from the LOC134997 locus. The area of each square is proportional to the sample size of each study. Horizontal lines represent the 95% CI for each study.
To investigate whether the inclusion of ovarian cancer cases influenced associations with breast cancer risk in BRCA1 mutation carriers, analyses were performed after excluding all 315 individuals diagnosed with ovarian cancer only or ovarian cancer prior to breast cancer. A total of 32 SNPs displayed significant associations with breast cancer including rs6138178 in SNRPB and rs6602595 in CAMK1D (Ptrend = 0.03 and 0.004, respectively). Interestingly, rs6590224 from the KIRREL3 locus was also highly significantly associated with risk (HR = 1.45; 95% CI: 1.23–1.71, Ptrend = 1.0 × 10−5) in the absence of the ovarian cases (Supplementary Material, Table S2). Next, the influence of inclusion of prevalent cases was assessed by exclusion of all cases diagnosed with breast cancer more than 5 years before recruitment. A total of 21 SNPs including rs6138178 in SNRPB (Ptrend = 0.006) and rs6602595 in CAMK1D (Ptrend = 0.017) displayed significance in the absence of the prevalent cases (Supplementary Material, Table S3). In addition, because risk-reducing oophorectomy (RRO) reduces the risk of breast cancer in BRCA1 carriers by up to 50% (21), we included RRO as a time-dependent covariate in the analysis. Little influence on the associations with breast cancer risk was observed (Supplementary Material, Table S4).
In BRCA2 carriers, rs9393597 in LOC134997 (HR = 1.57; 95% CI: 1.25–1.92, Ptrend = 6 × 10−5) and rs12652447 in FBXL7 (HR = 1.37; 95% CI: 1.16–1.62, Ptrend = 1.7 × 10−4) displayed the strongest associations with breast cancer risk (Table 1). When applying FDR, rs9393597 in LOC134997 and rs12652447 in FBXL7 showed probabilities of false positive associations of less than 5% (q < 0.05) (Table 1). For both SNPs, the estimated risk in homozygotes was higher than in heterozygotes. While there was no evidence of heterogeneity in the per-allele HRs for rs9393597 (Phet = 0.48) among BRCA2 mutation carriers from the nine centers, some evidence of heterogeneity for rs12652447 (Phet = 0.002) was observed (Fig. 1). When repeating the analysis after exclusion of the Austria, Mayo and ModSquad studies that contributed less than 100 BRCA2 carriers (Table 2), there was no evidence of heterogeneity (Phet = 0.66) and little change in the per-allele HR (1.37–1.33).
Table 2.
Age distribution and breast cancer (affected and unaffected) status of 5457 eligible BRCA1 and BRCA2 mutation carriers from CIMBA nine study centers
| BRCA1 |
BRCA2 |
|||
|---|---|---|---|---|
| n | Agea (SD) | n | Agea (SD) | |
| Austria | ||||
| Affected | 92 | 39.6 (8.4) | 30 | 40.7 (7.3) |
| Unaffected | 93 | 38.9 (10.3) | 15 | 43.0 (9.6) |
| DNA Hebon | ||||
| Affected | 285 | 39.6 (9.0) | 103 | 43.5 (9.4) |
| Unaffected | 290 | 43.7 (14.0) | 168 | 43.8 (13.5) |
| EMBRACE | ||||
| Affected | 421 | 40.5 (8.5) | 318 | 44.2 (8.6) |
| Unaffected | 394 | 41.4 (11.8) | 326 | 44.2 (11.9) |
| GC-HBOC | ||||
| Affected | 261 | 39.4 (9.9) | 121 | 46.7 (10.0) |
| Unaffected | 137 | 41.2 (13.0) | 42 | 45.7 (12.7) |
| GEMO | ||||
| Affected | 230 | 40.9 (9.5) | 151 | 43.5 (10.2) |
| Unaffected | 178 | 42.9 (12.2) | 75 | 44.3 (11.7) |
| kConFab | ||||
| Affected | 250 | 41.3 (9.3) | 203 | 44.2 (9.9) |
| Unaffected | 199 | 42.9 (11.9) | 150 | 44.9 (13.0) |
| Mayo | ||||
| Affected | 108 | 44.0 (10.0) | 58 | 45.2 (9.9) |
| Unaffected | 103 | 46.7 (12.0) | 38 | 47.8 (12.6) |
| MODSQUAD | ||||
| Affected | 110 | 40.8 (9.6) | 60 | 44.9 (10.3) |
| Unaffected | 48 | 42.4 (12.0) | 31 | 42.0 (13.1) |
| UPENN | ||||
| Affected | 154 | 39.4 (8.9) | 75 | 42.3 (10.1) |
| Unaffected | 98 | 43.5 (11.9) | 42 | 43.1 (10.8) |
| All Subjects | ||||
| Affectedb | 1911 | 40.5 (9.2) | 1119 | 44.2 (9.6) |
| Unaffectedc | 1540 | 42.6 (12.4) | 887 | 44.3 (12.5) |
| Total | 3451 | 41.4 (10.6) | 2006 | 44.2 (10.9) |
aAges are summarized as mean age; SD: standard deviation.
bAffected refers to carriers diagnosed with breast cancer.
cUnaffected (earliest of age at last follow up, age at ovarian cancer diagnosis, age at prophylactic mastectomy; age 80).
The sensitivity of the SNP associations with breast cancer to the presence of ovarian cancer cases and prevalent breast cancer cases in BRCA2 carriers was also assessed. After exclusion of all ovarian cancer cases, 30 SNPs displayed significance (P < 0.05) including rs9393597 in LOC134997 (Ptrend = 0.008) and rs12652447 in FBXL7 (Ptrend = 0.0006) (Supplementary Material, Table S6). Similarly, exclusion of prevalent cases yielded 29 SNPs displaying significant associations (P < 0.05) with breast cancer, including rs9393597 in LOC134997 (Ptrend = 0.0002) and rs12652447 in FBXL7 (Ptrend = 0.005) (Supplementary Material, Table S7). Likewise, little effect on the associations with breast cancer was observed after multivariate analysis including RRO as a time-dependent covariate (Supplementary Material, Table S8).
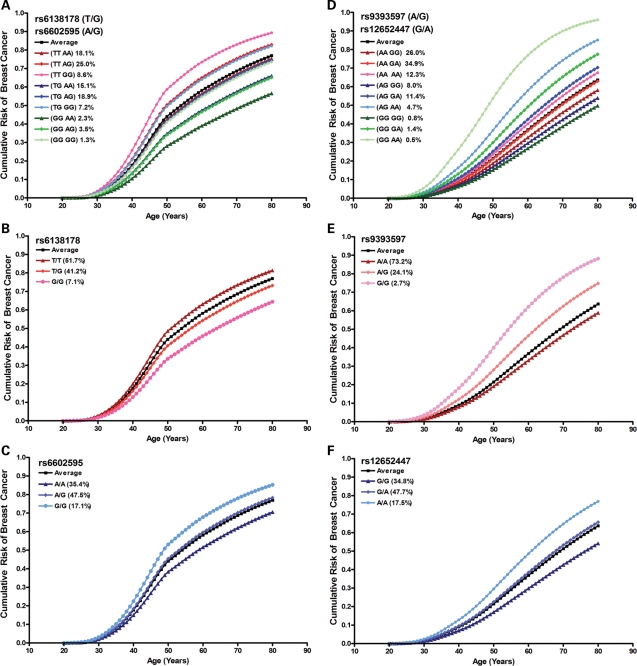
It has previously been postulated that genetic modifiers of breast cancer risk may prove useful for refined cancer risk assessment for BRCA1 and BRCA2 carriers (18). Thus, in subsequent risk assessment studies, we investigated the possible application of SNPs with strongest associations to cancer risk assessment in BRCA1 and BRCA2 carriers. Initially, the proportion of risk-modifying variance in BRCA1 mutation carriers explained by SNRPB rs6138178 and CAMK1D rs6602595 was estimated at 1.6 and 2.0%, respectively. Subsequently, assessment of associations between combinations of these SNPs and breast cancer risk showed that a model containing only the two ordinal SNP effects performed as well as more complex models (Table 3). Under this simple model, the (TT,GG) genotype had the highest HR of 1.81, whereas the (TG,AG) and (GG,GG) genotypes had the same average risk as observed in all BRCA1 carriers (Table 3). Figure 2 shows the estimated magnitude of the effects of the individual SNPs and the combined SNPs under the simple ordinal model on the cumulative incidence of breast cancer.
Table 3.
Association of combinations of rs6138178 and rs6602595 among BRCA1 carriers and combinations of rs9393597 and rs12652447 among BRCA2 carriers and breast cancer risk
| SNP1 | SNP2 | Frequency (%) | HR | 95% CI | P-value |
|---|---|---|---|---|---|
| BRCA1 | |||||
| rs6138178 | rs6602595 | ||||
| TT | AA | 18.1 | 1.00 | (Reference) | |
| TT | AG | 25.0 | 1.34 | 1.04–1.73 | 0.0251 |
| TT | GG | 8.6 | 1.81 | 1.29–2.54 | 0.0006 |
| TG | AA | 15.1 | 0.84 | 0.63–1.12 | 0.2376 |
| TG | AG | 18.9 | 0.98 | 0.74–1.29 | 0.8650 |
| TG | GG | 7.2 | 1.19 | 0.81–1.76 | 0.3767 |
| GG | AA | 2.3 | 0.79 | 0.44–1.44 | 0.4461 |
| GG | AG | 3.5 | 0.80 | 0.50–1.27 | 0.3371 |
| GG | GG | 1.3 | 0.96 | 0.48–1.92 | 0.9069 |
| BRCA2 | |||||
| rs9393597 | rs12652447 | ||||
| AA | GG | 26.0 | 1.00 | (Reference) | |
| AA | GA | 34.9 | 1.13 | 0.83–1.53 | 0.4437 |
| AA | AA | 12.3 | 1.52 | 1.03–2.22 | 0.0331 |
| AG | GG | 8.0 | 1.06 | 0.63–1.78 | 0.8235 |
| AG | GA | 11.4 | 1.82 | 1.22–2.73 | 0.0036 |
| AG | AA | 4.7 | 4.23 | 2.25–7.94 | <0.0001 |
| GG | GG | 0.8 | 2.65 | 0.91–7.69 | 0.0738 |
| GG | GA | 1.4 | 2.14 | 0.85–5.40 | 0.1065 |
Figure 2.
Age-related modifying effects of breast cancer risk by selected SNPs in BRCA1 and BRCA2 mutation carriers. Cumulative risk modifying effects of rs6138178 and rs6602595 combined (A), rs6138178 (B) and rs6602595 (C) genotypes in BRCA1 mutation carriers. Cumulative risk modifying effects of rs9393597 and rs12652447 combined (D), rs9393597 (E) and rs12652447 (F) genotypes in BRCA2 mutation carriers. All possible genotypes of the loci and their frequencies in BRCA1 or BRCA2 mutation carriers are shown. ‘Average’ represents the cumulative breast cancer risk over all possible modifying effects among BRCA1 or BRCA2 mutation carriers.
Similarly, the proportion of risk-modifying variance in BRCA2 mutation carriers predicted to be explained by LOC134997 rs9393597 and FXBL7 rs12652447 was 3.9 and 2.9%, respectively. Here a model including the two ordinal SNP effects together with a multiplicative interaction fit the observed data better than the simple model that included only the two ordinal SNP effects (P = 0.032). Figure 2 shows the estimated magnitude of the effects of the individual and the combined SNPs under this model on the cumulative incidence of breast cancer. BRCA2 mutation carriers homozygous for the minor risk alleles at both loci (GG,AA) and with (AG,AA), (GG,GA), (AG,GA) and (AA,AA) genotypes had higher age-related cumulative breast cancer risk than the average incidence in the BRCA2 carriers, consistent with multiplicative interactions between disease alleles (Table 3 and Fig. 2). Approximately 31% of BRCA2 mutation carriers were predicted to have HRs in excess of 1.5 (Table 3 and Fig. 2).
DISCUSSION
By evaluating candidate genetic risk factors for breast cancer risk from two recent breast cancer GWAS, we have identified several potential modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. In particular, rs6138178 in SNRPB and rs6602595 in CAMK1D were significantly associated with breast cancer in BRCA1 carriers, while rs9393597 in LOC134997 and rs12652447 in FBXL7 were significantly associated with breast cancer in BRCA2 carriers, each reaching P < 0.001. Of these, the rare allele of rs6138178 in SNRPB was associated with a reduced risk of breast cancer, whereas the rare alleles of rs6602595 in CAMK1D, rs9393597 in LOC134997 and rs12652447 in FBXL7 were all associated with increased risk. Importantly, the HR estimates for these four SNPs were quite consistent across the nine study groups (Fig. 1). These observations and the finding that the SNPs remained significantly associated with breast cancer risk after exclusion of ovarian cases, after exclusion of prevalent cases, and after evaluation of the influence of RRO, support a role for these variants as risk modifiers.
Although none of the associations for SNPs evaluated in this study reached ‘genome-wide’ levels of significance, there was an excess of association significant at the P < 0.01 level (8 in BRCA1 carriers, 12 in BRCA2 carriers, compared with 3 expected by chance). Also only 1 of the 8 SNPs in BRCA1 carriers and 2 of the 12 SNPs in BRCA2 carriers were previously established in breast cancer loci (all in 5p12). Since previous CIMBA studies have shown that the majority of established breast cancer SNPs are associated with breast cancer risk in BRCA2 carriers and, in some cases, BRCA1 carriers, the suggestion is that some of the other significant SNPs reported here may reflect true associations with breast cancer in BRCA1 and BRCA2 mutation carriers. The results also suggest that SNPs in GWAS that do not reach genome levels of significance (P < 1 × 10−7) for association with breast cancer may still account for effects in a BRCA1 or BRCA2 mutant background. Thus, evaluation of larger numbers of SNPs displaying moderate associations in GWAS, such as P < 1 × 10−3, may be an efficient strategy for identifying further genetic modifiers in BRCA1 and BRCA2 carriers. This approach may also be a useful strategy for identifying genetic modifiers for familial forms of other diseases, such as hereditary nonpolyposis colorectal cancer. Further studies with larger numbers of BRCA1 and BRCA2 carriers will be needed to establish which particular SNP associations can be replicated, and to estimate precisely the risks associated with each locus. In addition, the results raise the possibility that certain SNPs that show modest associations in case–control studies may show stronger effects in BRCA1 or BRCA2 carriers because breast cancer in BRCA1 or BRCA2 carriers may have less heterogeneity. Thus, additional studies using unselected breast cancer cases, perhaps in subgroups of cases defined by pathology, should be undertaken to determine if these SNPs are genetic risk factors for breast cancer in non-carriers.
None of the four most strongly associated SNPs are putative functional variants. The rs6138178 variant is located in the first intron of SNRPB on chromosome 20p13 and encodes a nuclear component of small ribonucleoprotein particles (snRNPs) which are essential for pre-mRNA splicing. The SNP is in high LD with rs3746687 (r2 > 0.8), a variant in the 3′-UTR of ZNF434, encoding a zinc finger protein of unknown function. Interestingly, rs6138178 was selected for this study based on an association with breast cancer in premenopausal women (P2df = 7 × 10−4) in a Mayo Clinic breast cancer case–control study, which is consistent with the association with risk in BRCA1 carriers who develop breast cancer at a younger age. SNP rs6602595, identified through the UK1 GWAS, is located in the first intron of CAMK1D, which encodes a calcium-/calmodulin-dependent serine/threonine protein kinase. CAMK1D is frequently amplified and overexpressed in basal-like breast tumors, and has been shown to promote an epithelial-to-mesenchymal transition of breast epithelial cells, consistent with the basal-like tumor phenotype (22). Since BRCA1 mutant breast tumors are predominantly basal-like (23), it is possible that common genetic variation in CAMK1D may contribute to this tumor phenotype. The rs9393597 SNP associated with risk in BRCA2 carriers was also initially associated with breast cancer in the UK1 GWAS. The closest gene to rs9393597 is FAM65B, encoding a potential myogenic factor in cell differentiation, cytoskeletal rearrangements and filopodia formation. The rs12652447 SNP associated with risk in BRCA2 carriers was selected following near-genome wide significant validation by CGEMS of a UK1 GWAS candidate SNP. This variant is located within an intron of FBXL7, which encodes an F-box containing protein that constitutes ubiquitin-dependent ligase complexes (SKP1-Cullin-F-box) essential in phosphorylation-dependent ubiquitination and proteosome-dependent degradation.
As noted earlier, none of the 8 SNPs in BRCA1 carriers and 12 SNPs in BRCA2 carriers displayed significant associations in both BRCA1 and BRCA2 carriers. The absence of SNPs displaying significance in both populations (Supplementary Material, Tables S1 and S2) is consistent with evidence that BRCA1 and BRCA2 carriers represent pathologically distinct diseases (23). Analysis of genetic risk factors for breast cancer identified through GWAS suggests that more of these loci are significantly associated with breast cancer risk in BRCA2 carriers than BRCA1 carriers (18,19), which is consistent with the outcome of our own study. The ER-negative nature of BRCA1 breast cancer also raises the possibility of the existence of a distinct set of SNPs that modify risk in BRCA1 carriers but may not necessarily display strong associations in the general population.
In an effort to demonstrate the potential utility for breast cancer risk modifiers in cancer risk assessment for BRCA1 and BRCA2 carriers, we conducted preliminary analyses in which the effects of the SNPs with strongest associations and combinations of the SNPs on the age-related cumulative risk of breast cancer in carriers were estimated (Fig. 2). The average cumulative risk at the age of 50 years was estimated at 43% among BRCA1 carriers and almost 20% among BRCA2 carriers, which is consistent with previous assessments (1). In BRCA1 carriers, subjects with the combination of the minor alleles of rs6602595 and the major alleles of rs6138178 showed an increase in risk at age 50 from 43 to 58%, whereas individuals with the opposite alleles showed a decrease in risk at age 50–27%. Overall, significantly more carriers were predicted to have elevated breast cancer risk than reduced risk (40.8 versus 20.9%). In BRCA2 carriers, the minor alleles of the SNPs were associated with an increase in risk from an average of 20 up to 50% by age 50. These observations serve to demonstrate the potential utility of SNPs for cancer risk assessment in BRCA1 and BRCA2 carriers. The results suggest that combinations of several modifier loci, when identified, may allow identification of subsets of carriers at high risk of cancer at young age who can benefit from early intervention, and carriers at low or even population-based levels of risk who may delay or avoid current intervention methods such as prophylactic oophorectomy and mastectomy. However, in the context of the current study, it should be recognized that the influence of the SNPs on age-related risk of breast cancer may well be attenuated upon further validation with larger numbers of BRCA1 and BRCA2 carriers. Similarly, the incorporation of family history and environmental risk factors that also influence the lifetime risk of breast cancer may substantially modify the age-related risk models shown here. Despite the need for more refined estimation of risk in the future, our results support the notion that the addition of fully characterized modifier loci to risk models will lead to improved risk assessment for patients carrying deleterious mutations in BRCA1 and BRCA2. Thus, in time modifier loci may significantly influence the approach to genetic counseling, prevention and routine care of these high-risk patients.
MATERIALS AND METHODS
BRCA1 and BRCA2 mutation carriers
Nine large studies from eight countries (Australia, Austria, Britain, Czech Republic, France, Germany, the Netherlands and the USA) provided DNA samples, clinical and risk factor data from BRCA1 and BRCA2 mutation carriers for this study (Table 2). The majority of carriers were recruited through cancer genetics clinics offering genetic testing, and were enrolled into site-specific studies (Mayo and UPENN) or national studies. Eligibility was restricted to individuals with pathogenic BRCA1 or BRCA2 mutations (10) over 18 years of age, with available genomic DNA from blood samples, and phenotypic information including country of residence, year of birth, mutation description, age at last follow-up, diagnosis date of invasive breast and/or ovarian cancer, age of bilateral mastectomy or oophorectomy, and family identifiers (Table 4). All carriers were recruited to studies at the host institutions under IRB-approved protocols. Only self-reporting Caucasian women were included in the analysis. Carriers with both BRCA1 and BRCA2 mutations were excluded. Duplicate samples were identified based on the year of birth, exact mutation description, age at onset of disease and SNP genotyping data from the present and past CIMBA studies and were included only once in the analysis. The date of blood draw was identified as the date of ascertainment for each carrier.
Table 4.
Summary characteristics for the 5457 eligible BRCA1 and BRCA2 carriers used in the analysis
| BRCA1 |
BRCA2 |
|||
|---|---|---|---|---|
| Characteristic | Unaffecteda | Affectedb | Unaffecteda | Affectedb |
| Number | 1540 | 1911 | 887 | 1119 |
| Person-years follow-up | 65 612 | 77 316 | 39 322 | 49 428 |
| Median age (IQR) | 41 (34, 50) | 39 (34, 46) | 43 (35, 52) | 43 (37, 50) |
| Age at censure, n (%) | ||||
| <30 | 232 (15.1) | 186 (9.7) | 101 (11.4) | 42 (3.8) |
| 30–39 | 473 (30.7) | 823 (43.1) | 267 (30.1) | 356 (31.8) |
| 40–49 | 445 (28.9) | 605 (31.2) | 253 (28.5) | 441 (39.4) |
| 50–59 | 251 (16.3) | 223 (11.7) | 164 (18.5) | 198 (17.7) |
| 60–69 | 93 (6.0) | 67 (3.5) | 71 (8.0) | 67 (6.0) |
| 70+ | 46 (3.0) | 7 (0.4) | 31 (3.5) | 15 (1.3) |
| Year of birth, n (%) | ||||
| <1920 | 7 (0.5) | 15 (0.8) | 6 (0.7) | 5 (0.5) |
| 1920–29 | 34 (2.2) | 74 (3.9) | 23 (2.6) | 47 (4.2) |
| 1930–39 | 115 (7.5) | 180 (9.4) | 65 (7.3) | 146 (13.1) |
| 1940–49 | 221 (14.4) | 427 (22.3) | 130 (14.7) | 292 (26.1) |
| 1950–59 | 371 (24.1) | 606 (31.7) | 225 (25.4) | 366 (32.7) |
| 1960+ | 792 (51.4) | 609 (31.9) | 438 (49.4) | 263 (23.5) |
| Mutation description, n (%) | ||||
| Ashkenazi Jewishc | 130 (8.5) | 144 (7.6) | 41 (4.6) | 49 (4.4) |
| Other | 1410 (91.5) | 1767 (92.4) | 846 (95.4) | 1070 (95.6) |
| After ovarian case exclusion | ||||
| Number | 1226 | 1911 | 784 | 1119 |
| Person-years follow-up | 50 240 | 77 316 | 33 678 | 49 428 |
| Median age (IQR) | 39 (32, 48) | 39 (34, 46) | 41 (34, 50) | 43 (37, 50) |
| After prevalent cases exclusion | ||||
| Number | 1540 | 894 | 887 | 544 |
| Person-years follow-up | 65 580 | 36 730 | 39 331 | 24 491 |
| Median age (IQR) | 41 (34, 50) | 40 (34, 46) | 43 (35, 52) | 44 (38, 50) |
IQR, interquartile range.
aUnaffected (earliest of age at last follow up, age at ovarian cancer diagnosis, age at prophylactic mastectomy; age 80).
bAffected refers to carriers diagnosed with breast cancer.
cAshkenazi Jewish includes 185delAG and 5382insC for BRCA1 and 6174delT for BRCA2.
SNP selection and genotyping
The exact concentration of DNA in each sample was measured using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). The quality of the DNA was assessed by electrophoresis on E-Gel-96 1% agarose gels (Invitrogen). Samples with severely degraded DNA were excluded from the study. A total of 5685 samples from mutation carriers and 110 duplicates (2%) at a concentration of 50 ng/µl (250 ng total per sample) were selected for genotyping. A single Illumina custom-designed 384-plex OPA containing 350 non-overlapping SNPs from two breast cancer GWAS and a Mayo Clinic candidate SNP study were generated. This included 142 SNPs displaying the most significant associations (P <1 × 10−3) with breast cancer in stage 2 of the CGEMS breast cancer GWAS (17), 54 SNPs from the UK1 GWAS (11) displaying the most significant associations (P < 1 × 10−3) with breast cancer when combining data from SEARCH and CGEMS, 99 SNPs displaying the most significant associations (P < 1 × 10−3) with breast cancer in the combined SEARCH stage 1 and 2 data and 55 SNPs displaying the most significant associations (P < 1 × 10−2) with breast cancer in associations studies using the Mayo Clinic Breast Cancer Study (MCBCS) (24–26). DNA samples were arranged as a random mixture of breast cancer affected and unaffected carriers in 63 × 96 well plates with four DNA controls consisting of a trio of CEPH controls plus a random genomic control per plate. Following genotyping, SNPs with a minor allele frequency (MAF) <0.05 or with a call rate <95% were excluded from the analysis (n = 13). An additional four SNPs which deviated significantly from Hardy–Weinberg equilibrium (HWE) (P < 1 × 10−7) were excluded. A total of 154 samples were excluded due to missing phenotypic data, and 74 samples were excluded due to SNP call rates <95%. Using the genotypes available, three duplicate individuals from different study centers were identified and removed. The concordance rate between duplicates was 100%. After all quality control was completed, a total of 5457 unique mutation carriers (3451 BRCA1 and 2006 BRCA2) with genotyping data for 333 SNPs were analyzed (Table 2).
Statistical analyses
Associations between SNPs and breast cancer risk in BRCA1 and BRCA2 mutation carriers were evaluated using a Cox proportional hazards model. Each subject was followed from birth to breast cancer, or when unaffected by breast cancer to the earliest of bilateral mastectomy, ovarian cancer, last follow-up or age 80. To adjust for the non-random sampling of mutation carriers, analyses were carried out within a weighted cohort framework (27). Individuals were assigned weights based upon mutation status, breast cancer affected/unaffected status and age at event or age at censoring, such that the observed breast cancer incidence rates in the study sample were consistent with established breast cancer risk estimates for BRCA1 and BRCA2 mutation carriers (27). Analyses were stratified by year of birth (based on quartiles of unaffected), ethnicity, country of residence, study site and mutation status. A robust variance approach was used to estimate the standard errors of the parameters to allow for the dependence of individuals from the same family. Each of the 333 SNPs that passed quality control was formally evaluated for associations with risk under a log-additive model. In addition, estimates were separately generated for one and two copies of the minor allele versus two copies of the major allele. Between-study heterogeneity for each SNP was assessed by fitting a model that estimated study-specific log-hazard ratios (HR), and comparing this against the model that estimated a single-pooled log-hazard ratio via a likelihood ratio test. Forest plots were used to assess consistency of study specific effects for rs6138178 (SNRPB), rs6602595 (CAMK1D), rs9393597 (LOC134997) and rs12652447 (FBXL7).
To determine the degree to which the inclusion of ovarian cancer cases may have influenced the risk association, analyses were performed after excluding all individuals diagnosed with ovarian cancer. To further explore the influence of inclusion of prevalent cases in the study, analyses were performed after exclusion of subjects who were diagnosed with breast cancer more than 5 years prior to age of mutation diagnosis. Because of the good 5-year survival rate for breast cancer, this avoids substantial survival bias in the collection of BRCA1 carriers. In addition, a further analysis treating prophylactic oophorectomy as a time-dependent covariate was conducted.
Cumulative risk assessment in BRCA1 and BRCA2 mutation carriers
The proportion of genetic modifying variance explained by each of the SNPs was estimated by ln(c)/σ2, as previously described (18,28,29), where c was the estimated coefficient of variation in incidence rates due to each SNP, obtained from the SNP genotype frequencies and the ordinal hazard ratio estimates, and σ2 was the total polygenic variance component for breast cancer risk, estimated at 1.32 for BRCA1 and 1.73 for BRCA2 mutation carriers (5). In addition, the HR estimates of candidate SNPs were integrated with estimates of either BRCA1 or BRCA2 penetrance (1) to further estimate the influence of candidate SNPs on the absolute risk of breast cancer. This was achieved by constraining breast cancer incidence to match estimates of BRCA1 or BRCA2 penetrance by year of age and by estimating the cumulative incidence of breast cancer that corresponded to each genotype, or genotype combination, by applying the estimated HRs such that the appropriate baseline breast cancer incidence was retained while reflecting the relative risk differences among the genotypes, or genotype combinations. Plots of the resulting breast cancer cumulative incidence estimates corresponding to each genotype of each candidate SNP were generated.
A series of three models were fit to the data for the top two SNPs, separately for BRCA1 and BRCA2 carriers, in order to assess the combined effects of two candidate BRCA1 modifiers and also two candidate BRCA2 modifiers on breast cancer risk. The first model estimated a separate HR for each combination of genotypes, with those simultaneously homozygous for the major alleles serving as the reference group. The second model estimated an ordinal (log-additive) trend for each SNP, together with a multiplicative interaction between the ordinal effects for the two SNPs. The third model simply estimated an ordinal effect for each of the two SNPs. Score tests that accounted for familial clustering were used to test if increasing model complexity significantly improved model fit. Estimates from the models that most parsimoniously fit the data from the two SNPs were integrated with estimates of BRCA1 and BRCA2 penetrance (1) to calculate the cumulative risk of breast cancer for each combination of SNP genotypes in BRCA1 and BRCA2 carriers separately.
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by R01 grants (CA122340, CA128978) and a Breast Cancer Specialized Program of Research Excellence grant [P50 CA116201 (SPORE)] from the National Cancer Institute at the National Institutes of Health, an award from the Breast Cancer Research Foundation and an award from the Komen Race for the Cure Foundation to F.J.C. The UK1 GWAS was funded by a Cancer Research UK award to D.F.E. The CGEMS GWAS was funded by intramural funds from the National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the CGEMS and UK1 GWAS for providing candidate SNPs.
Conflict of Interest statement. None declared.
Appendix
EPIDEMIOLOGICAL STUDY OF BRCA1 & BRCA2 MUTATION CARRIERS (EMBRACE)
EMBRACE Collaborating Centers are: Coordinating Centre, Cambridge: Susan Peock, Margaret Cook, Clare Oliver, Debra Frost. North of Scotland Regional Genetics Service, Aberdeen: Helen Gregory, Zosia Miedzybrodzka. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Carole McKeown, Amy Taylor. South West Regional Genetics Service, Bristol: Alan Donaldson. East Anglian Regional Genetics Service, Cambridge: Joan Paterson. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark Rogers, Emma McCann. St James's Hospital, Dublin, and National Centre for Medical Genetics, Dublin: John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt. South East Thames Regional Genetics Service, Guys Hospital London: Louise Izatt, Gabriella Pichert, Caroline Langman. North West Thames Regional Genetics Service, Harrow: Huw Dorkins. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell. Yorkshire Regional Genetics Service, Leeds: Carol Chu, Tim Bishop, Julie Miller. Merseyside and Cheshire Clinical Genetics Service, Liverpool: Ian Ellis. Manchester Regional Genetics Service, Manchester: D. Gareth Evans, Fiona Lalloo, Felicity Holt. North East Thames Regional Genetics Service, NE Thames: Alison Male, Anne Robinson. Nottingham Centre for Medical Genetics, Nottingham: Carol Gardiner. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Lucia D'Mello, Elizabeth Page, Audrey Ardern-Jones, Anita Mitra. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Emma Tyler, Donna McBride. The EMBRACE study was funded by Cancer Research UK. S.P. is funded by Cancer Research-UK Grants C1287/A10118 and C1287/A8874. D.G.E. and F.L. are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. R.E./E.B./L.D'M. are also supported by Cancer Research UK Grant C5047/A8385. A.C.A. is a Cancer Research UK Senior Cancer Research Fellow. D.F.E. is a Principal Research Fellow of Cancer Research UK.
GERMAN CONSORTIUM OF HEREDITARY BREAST AND OVARIAN CANCER (GC-HBOC)
We thank all families for providing samples for this study. This work was supported by grants from the German Cancer Aid (Grant numbers 107054 and 107353).
THE GEMO STUDY (GENETIC MODIFIERS OF CANCER RISK IN BRCA1/2 MUTATION CARRIERS: CANCER GENETICS NETWORK ‘GROUPE GÉNÉTIQUE ET CANCER’, FÉDÉRATION NATIONALE DES CENTRES DE LUTTE CONTRE LE CANCER, FRANCE)
Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon/Centre Léon Bérard, and UMR5201 CNRS, Université de Lyon, Lyon: Olga Sinilnikova, Laure Barjhoux, Sophie Giraud, Mélanie Léone, Sylvie Mazoyer. INSERM U509, Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Antoine de Pauw. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Audrey Remenieras, Véronique Byrde, Olivier Caron, Gilbert Lenoir. Centre Jean Perrin, Clermont-Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, François Eisinger. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier. CHU de Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis. Centre René Huguenin, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Michel Longy, Nicolas Sevenet. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU de Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung. CHU de Dijon: Cécile Cassini, Laurence Olivier-Faivre. CHU de St-Etienne: Fabienne Prieur. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU de Limoges: Laurence Vénat-Bouvet. Creighton University, Omaha, USA: Henry T. Lynch. We wish to thank all the GEMO collaborating members for their contribution to this study. The GEMO study was supported by the Ligue National Contre le Cancer and the Association ‘Le cancer du sein, parlons-en!’ Award.
HEBON (HEREDITARY BREAST AND OVARIAN CANCER RESEARCH GROUP NETHERLANDS)
Coordinating center: Netherlands Cancer Institute, Amsterdam: Frans Hogervorst, Senno Verhoef, Anouk Pijpe, Laura van 't Veer, Flora van Leeuwen, Matti Rookus. Erasmus Medical Center, Rotterdam: Margriet Collée, Ans van den Ouweland, Mieke Kriege, Mieke Schutte, Maartje Hooning, Caroline Seynaeve. Leiden University Medical Center, Leiden: Christi van Asperen, Juul Wijnen, Maaike Vreeswijk, Rob Tollenaar, Peter Devilee. Radboud University Nijmegen Medical Center, Nijmegen: Marjolijn Ligtenberg, Nicoline Hoogerbrugge. University Medical Center Utrecht, Utrecht: Margreet Ausems, Rob van der Luijt. Amsterdam Medical Center: Cora Aalfs, Theo van Os. VU University Medical Center, Amsterdam: Hans Gille, Quinten Waisfisz, Hanne Meijers-Heijboer. University Hospital Maastricht, Maastricht: Encarna Gomez-Garcia, Kees van Roozendaal, Marinus Blok. University Medical Center Groningen University: Jan Oosterwijk, Annemieke van der Hout, Marian Mourits. The Netherlands Foundation for the detection of hereditary tumours, Leiden, the Netherlands: Hans Vasen. The HEBON study is supported by the Dutch Cancer Society grants NKI 1998-1854, NKI 2004-3088 and NKI 2007-3756.
KCONFAB (KATHLEEN CUNNINGHAM FOUNDATION CONSORTIUM FOR RESEARCH INTO FAMILIAL BREAST CANCER)
We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow Up Study (funded by NHMRC grants 145684, 288704 and 454508) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. A.B.S. and G.C.T. are Senior and Senior Principal Research Fellows, respectively, of the NHMRC.
MODIFIER STUDY OF QUANTITATIVE EFFECTS ON DISEASE (MODSQUAD)
Csilla Szabo (Mayo Clinic College of Medicine, Rochester, MN); Michal Zikan, Petr Pohlreich, Zdenek Kleibl (First Faculty of Medicine, Charles University, Prague, Czech Republic); Lenka Foretova, Eva Machackova, Miroslava Lukesova (Masaryk Memorial Cancer Institute, Brno, Czech Republic); Kathleen Claes, Kim De Leeneer, Bruce Poppe, Anne De Paepe (Ghent University, Ghent, Belgium). Csilla Szabo was supported by Susan G. Komen Foundation Basic, Clinical, and Translational Research grant (BCTR0402923) and the Mayo Rochester Early Career Development Award for Non-Clinician Scientists; Michal Zikan by the Grant Agency of the Czech republic grant No. 301/08/P103. Lenka Forebova through the Ministry of Health of the CR grant MZ0 MOU 2005, Kathleen Claes by grant 1.5.150.07 from the Fund for Scientific Research Flanders (FWO), Anne De Peepe by grant 12051203 from the Ghent University. Bruce Poppe is Senior Clinical Investigator of the Fund for Scientific Research of Flanders (FWO—Vlaanderen).
REFERENCES
- 1.Antoniou A., Pharoah P.D., Narod S., Risch H.A., Eyfjord J.E., Hopper J.L., Loman N., Olsson H., Johannsson O., Borg A., et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 2003;72:1117–1130. doi: 10.1086/375033. doi:10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simchoni S., Friedman E., Kaufman B., Gershoni-Baruch R., Orr-Urtreger A., Kedar-Barnes I., Shiri-Sverdlov R., Dagan E., Tsabari S., Shohat M., et al. Familial clustering of site-specific cancer risks associated with BRCA1 and BRCA2 mutations in the Ashkenazi Jewish population. Proc. Natl Acad. Sci. USA. 2006;103:3770–3774. doi: 10.1073/pnas.0511301103. doi:10.1073/pnas.0511301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebbeck T.R. Inherited predisposition and breast cancer: modifiers of BRCA1/2-associated breast cancer risk. Environ. Mol. Mutagen. 2002;39:228–234. doi: 10.1002/em.10050. doi:10.1002/em.10050. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou A.C., Pharoah P.D., McMullan G., Day N.E., Stratton M.R., Peto J., Ponder B.J., Easton D.F. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br. J. Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. doi:10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniou A.C., Cunningham A.P., Peto J., Evans D.G., Lalloo F., Narod S.A., Risch H.A., Eyfjord J.E., Hopper J.L., Southey M.C., et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br. J. Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. doi:10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begg C.B., Haile R.W., Borg A., Malone K.E., Concannon P., Thomas D.C., Langholz B., Bernstein L., Olsen J.H., Lynch C.F., et al. Variation of breast cancer risk among BRCA1/2 carriers. J. Am. Med. Assoc. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. doi:10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenevix-Trench G., Milne R.L., Antoniou A.C., Couch F.J., Easton D.F., Goldgar D.E. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. doi:10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couch F.J., Sinilnikova O., Vierkant R.A., Pankratz V.S., Fredericksen Z.S., Stoppa-Lyonnet D., Coupier I., Hughes D., Hardouin A., Berthet P., et al. AURKA F31I polymorphism and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a consortium of investigators of modifiers of BRCA1/2 study. Cancer Epidemiol. Biomarkers Prev. 2007;16:1416–1421. doi: 10.1158/1055-9965.EPI-07-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao B., Xie X.J., Huang C., Shames D.S., Chen T.T., Lewis C.M., Bian A., Zhang B., Olopade O.I., Garber J.E., et al. RASSF1A polymorphism A133S is associated with early onset breast cancer in BRCA1/2 mutation carriers. Cancer Res. 2008;68:22–25. doi: 10.1158/0008-5472.CAN-07-5183. doi:10.1158/0008-5472.CAN-07-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou A.C., Sinilnikova O.M., Simard J., Leone M., Dumont M., Neuhausen S.L., Struewing J.P., Stoppa-Lyonnet D., Barjhoux L., Hughes D.J., et al. RAD51 135G– > C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am. J. Hum. Genet. 2007;81:1186–1200. doi: 10.1086/522611. doi:10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 14.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. doi:10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. doi:10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniou A.C., Spurdle A.B., Sinilnikova O.M., Healey S., Pooley K.A., Schmutzler R.K., Versmold B., Engel C., Meindl A., Arnold N., et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am. J. Hum. Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. doi:10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antoniou A.C., Sinilnikova O.M., McGuffog L., Healey S., Nevanlinna H., Heikkinen T., Simard J., Spurdle A.B., Beesley J., Chen X., et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. doi:10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Closas M., Hall P., Nevanlinna H., Pooley K., Morrison J., Richesson D.A., Bojesen S.E., Nordestgaard B.G., Axelsson C.K., Arias J.I., et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. doi:10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebbeck T.R., Lynch H.T., Neuhausen S.L., Narod S.A., Van't Veer L., Garber J.E., Evans G., Isaacs C., Daly M.B., Matloff E., et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 2002;346:1616–1622. doi: 10.1056/NEJMoa012158. doi:10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 22.Bergamaschi A., Kim Y.H., Kwei K.A., La Choi Y., Bocanegra M., Langerod A., Han W., Noh D.Y., Huntsman D.G., Jeffrey S.S., et al. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol. Oncol. 2008;2:327–339. doi: 10.1016/j.molonc.2008.09.004. doi:10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakhani S.R., Reis-Filho J.S., Fulford L., Penault-Llorca F., van der Vijver M., Parry S., Bishop T., Benitez J., Rivas C., Bignon Y.J., et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin. Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. doi:10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Goode E.L., Fredericksen Z.S., Vierkant R.A., Pankratz V.S., Liu-Mares W., Rider D.N., Vachon C.M., Cerhan J.R., Olson J.E., et al. Association of genetic variation in genes implicated in the beta-catenin destruction complex with risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 2008;17:2101–2108. doi: 10.1158/1055-9965.EPI-08-0134. doi:10.1158/1055-9965.EPI-08-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelemen L.E., Couch F.J., Ahmed S., Dunning A.M., Pharoah P.D., Easton D.F., Fredericksen Z.S., Vierkant R.A., Pankratz V.S., Goode E.L., et al. Genetic variation in stromal proteins decorin and lumican with breast cancer: investigations in two case–control studies. Breast Cancer Res. 2008;10:R98. doi: 10.1186/bcr2201. doi:10.1186/bcr2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson J.E., Wang X., Goode E.L., Pankratz V.S., Fredericksen Z.S., Vierkant R.A., Pharoah P.D., Cerhan J.R., Couch F.J. Variation in genes required for normal mitosis and risk of breast cancer. Breast Cancer Res. Treat. 2010;119:453–462. doi: 10.1007/s10549-009-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniou A.C., Goldgar D.E., Andrieu N., Chang-Claude J., Brohet R., Rookus M.A., Easton D.F. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet. Epidemiol. 2005;29:1–11. doi: 10.1002/gepi.20074. doi:10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- 28.Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am. J. Hum. Genet. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- 29.Antoniou A.C., Easton D.F. Polygenic inheritance of breast cancer: implications for design of association studies. Genet. Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. doi:10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.