Abstract
Down syndrome (DS) is caused by the presence of an extra copy of human chromosome 21 (Hsa21) and is the most common genetic cause for developmental cognitive disability. The regions on Hsa21 are syntenically conserved with three regions located on mouse chromosome 10 (Mmu10), Mmu16 and Mmu17. In this report, we describe a new mouse model for DS that carries duplications spanning the entire Hsa21 syntenic regions on all three mouse chromosomes. This mouse mutant exhibits DS-related neurological defects, including impaired cognitive behaviors, reduced hippocampal long-term potentiation and hydrocephalus. These results suggest that when all the mouse orthologs of the Hsa21 genes are triplicated, an abnormal cognitively relevant phenotype is the final outcome of the elevated expressions of these orthologs as well as all the possible functional interactions among themselves and/or with other mouse genes. Because of its desirable genotype and phenotype, this mutant may have the potential to serve as one of the reference models for further understanding the developmental cognitive disability associated with DS and may also be used for developing novel therapeutic interventions for this clinical manifestation of the disorder.
INTRODUCTION
Down syndrome (DS), caused by human trisomy 21, is the most frequent live-born human chromosomal disorder, occurring in one in 700–800 newborns (1–3). Owing to its polygenic basis, the phenotypes of this genomic disorder are involved with several major organ systems. Among them, abnormalities of the nervous system are one of the most important burdens for patients. DS is the most common genetic cause of mental retardation (4,5). The average IQ of individuals with DS is significantly lower than normal (5,6). Cognitive deficits include impairment in spatial memory and long-term memory as well as difficulties in acquiring new skills (4,7,8). Neuropsychological tests have shown that adolescents with DS exhibit deficits in hippocampal functions (4,9). Other neurological abnormalities include hydrocephalus (10) and Alzheimer-type neuropathological alterations (11,12). Relatively little is known of the molecular mechanisms underlying cognitively relevant phenotypes in DS, and no treatments have yet proven effective.
To understand the molecular mechanisms of cognitive dysfunction in DS, the mouse has been used extensively as a model organism, which is based on the syntenic conservation between human and mouse genomes (Fig. 1). One group of mouse models was generated to carry Hsa21 or a fragment of it (13,14). Tc1 is currently the most desirable strain from this trans-species group, carrying Hsa21 with only two small deletions in which ∼8% of Hsa21 genes are deleted. However, the random loss of this human chromosome during mouse development resulted in variable levels of mosaicism of the human chromosome in different tissues (13). Nevertheless, the presence of the human chromosome in mice causes impaired cognitive behaviors and hippocampal long-term potentiation (LTP) (13), an activity-dependent, sustained increase in the efficacy of synaptic transmission in the hippocampus, which is viewed as an important cellular manifestation of learning and memory (15–18). The other group of models carries three exact copies of mouse syntenic regions of Hsa21 in various sizes. Ts65Dn is the most widely used model from this group (19). Ts65Dn mice are trisomic for ∼13.4 Mb of the Hsa21 syntenic region on Mmu16, which contains approximately 99 orthologs of Hsa21 genes (20,21). However, the entire Hsa21 syntenic region on Mmu16 spans 22.9 Mb and contains 115 Hsa21 gene orthologs (Fig. 1) (Supplementary Material, Table S2) (http://www.ensembl.org http://genome.ucsc.edu). Furthermore, the mouse genome carries approximately 60 other Hsa21 gene orthologs located in two additional Hsa21 syntenic regions on Mmu10 and Mmu17 (Fig. 1) (Supplementary Material, Tables S1 and S3). For the Hsa21 gene orthologs that are not triplicated in Ts65Dn, some are known to play roles in regulating neuronal functions, such as S100b (22) and Trpm2 (23,24), and others have unknown functions, so their effects on the central nervous system cannot be excluded. More importantly, a recent study showed that the triplication of 12 genes located within the Hsa21 syntenic region on Mmu17 results in a significant increase in hippocampal LTP (25). Therefore, the Hsa21 gene orthologs that are not triplicated in Ts65Dn may contribute significantly to the cognitively relevant phenotypes related to DS.
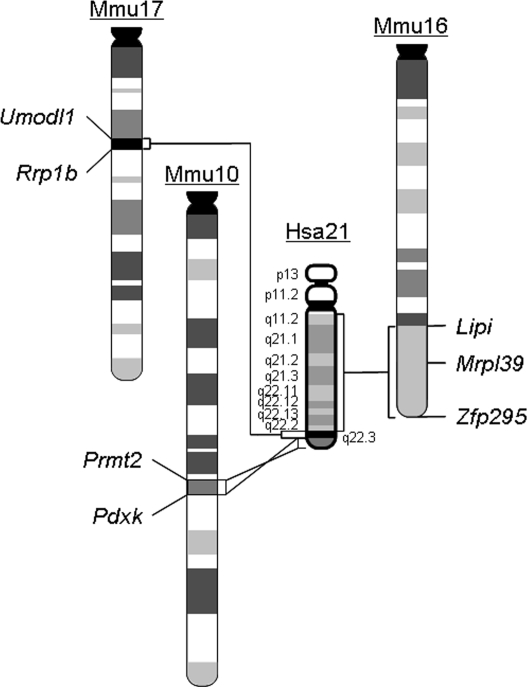
Figure 1.
Schematic representation of Hsa21 and the syntenic regions on Mmu10, Mmu16 and Mmu17. The endpoints of the syntenic regions are indicated. Mrpl39 is the proximal endpoint for the segmental trisomy in Ts65Dn.
Because of the potential species-specific differences between Hsa21 and the mouse syntenic regions, the transcriptional regulations of the Hsa21 gene orthologs and the interactions of the proteins encoded by these orthologs in a trans-species mouse mutant may be different from those in a mouse mutant carrying three copies of a mouse syntenic region, even if the triplicated regions carry the same orthologous genes in both mutants. Therefore, we decided, in this study, to generate a mouse model trisomic for all three Hsa21 syntenic regions and to analyze the impact of the simultaneous presence of the three segmental trisomies on the cognitively relevant phenotype.
RESULTS
Generation of a mouse model of DS trisomic for all the Hsa21 syntenic regions
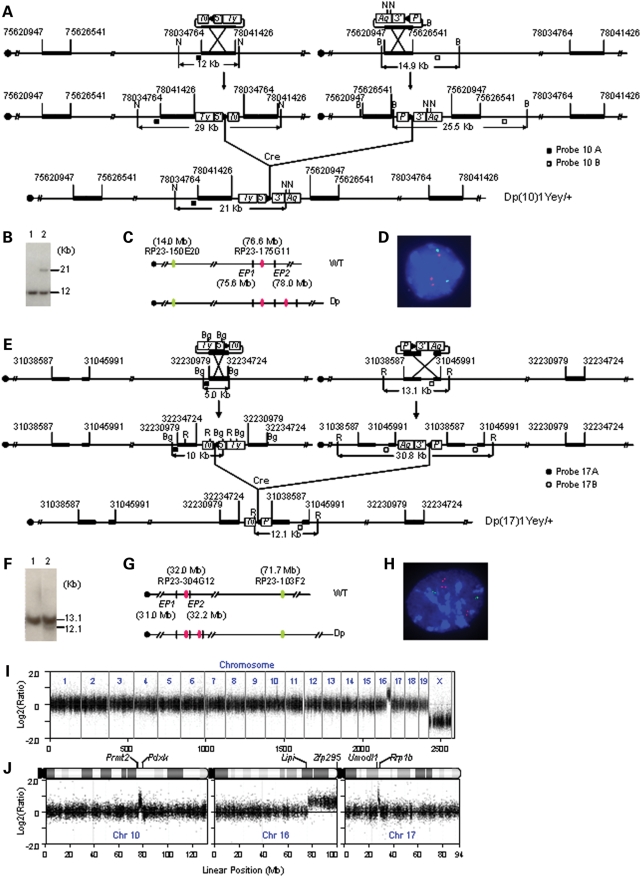
The Prmt2 and Pdxk genes are located at the proximal and distal ends of an ∼2.3 Mb region on Mmu10, respectively, which is syntenic to the distal part of human 21q22.3 (Fig. 1). This mouse syntenic region contains 41 genes orthologous to their counterparts on Hsa21 (Supplementary Material, Table S1) (http://www.ensembl.org; http://genome.ucsc.edu). The Abcg1 and Rrp1b genes are located at the proximal and distal ends of an ∼1.1 Mb region on Mmu17, respectively, which is syntenic to the proximal part of human 21q22.3. This mouse syntenic region contains approximately 19 genes orthologous to their counterparts on Hsa21 (Supplementary Material, Table S3) (http://www.ensembl.org; http://genome.ucsc.edu). To generate a duplication of the syntenic region on Mmu10, we targeted loxP to regions proximal to Prmt2 and distal to Pdxk in the genome of AB2.2 mouse embryonic stem (ES) cells (Fig. 2A). Dp(Prmt2-Pdxk)1Yey, abbreviated as Dp(10)1Yey or Ts2Yey, was generated by transfecting a Cre-expression vector into double-targeted ES cells (Fig. 2A; see Materials and Methods). A similar strategy was used to generate Dp(Abcg1-Rrp1b)1Yey, abbreviated as Dp(17)1Yey or Ts3Yey, by targeting loxP to regions proximal to Abcg1 and distal to Rrp1b, in AB2.2 ES cells (Fig. 2E; see Materials and Methods). The mutant ES cells carrying Dp(10)1Yey or Dp(17)1Yey were used to generate Dp(Prmt2-Pdxk)1Yey or Dp(Abcg1-Rrp1b) germ-line chimeras, respectively. The constitutional mouse mutants carrying the desired chromosomal duplications were generated by crossing the chimeric males with C57BL/6J females, and their genotypes were confirmed by Southern blot analysis (Fig. 2B and F) and fluorescent in situ hybridization (FISH) (Fig. 2C, D, G and H). Dp(10)1Yey/+, Dp(17)1Yey/+ as well as Dp(16)1Yey/+ (i.e. Ts1Yey) (26) mice were backcrossed to wild-type C57BL/6J mice for five generations.
Figure 2.
Development of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice. (A) Strategy to generate Dp(10)1Yey. B, BamHI; N, NheI; 5′, 5′HPRT fragment; 3′, 3′HPRT fragment; N, neomycin-resistance gene; P, puromycin-resistance gene; Ty, Tyrosinase transgene; Ag, Agouti transgene; arrowhead, loxP site. (B) Southern blot analysis of NheI-digested mouse-tail DNA using Probe 10B. Lane 1, the wild-type mouse; lane 2, Dp(10)1Yey/+ mouse. (C) Schematic of the genomic locations of BAC probes for FISH analysis. EP1 and EP2, endpoint 1 and endpoint 2, which were targeted with pTV(10)1EP1 and pTV(10)1EP2, respectively. (D) FISH analysis of interphase nuclei prepared from the embryonic fibroblasts carrying Dp(10)1Yey/+. (E) Strategy to generate Dp(17)1Yey. R, EcoRI; Bg, BglII. (F) Southern blot analysis of EcoRI-digested mouse-tail DNA using Probe 17A. Lane 1, the wild-type mouse; lane 2, Dp(17)1Yey/+ mouse. (G) Schematic of the genomic locations of BAC probes for FISH analysis. EP1 and EP2, endpoint 1 and endpoint 2, which were targeted with pTV(17)1EP1 and pTV(17)1EP2, respectively. (H) FISH analysis of interphase nuclei prepared from the embryonic fibroblasts carrying Dp(17)1Yey/+. (I) Whole-genome Agilent microarray CGH profile of DNA isolated from a Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mouse. (J) CGH profile of the mouse chromosomes 10, 16 and 17 of the Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mouse. Plotted are log2-transformed hybridization ratios of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mouse DNA versus wild-type mouse DNA.
After Dp(10)1Yey/+, Dp(16)1Yey/+ and Dp(17)1Yey/+ mice were established, we used the standard breeding strategy to generate compound mutant mice carrying two different duplications and then used these compound mutants to cross with the mutant carrying the third duplication to generate Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ (i.e. Ts1Yey;Ts2Yey;Ts3Yey) mice. Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ embryos were present at a normal Mendelian ratio at E18.5. However, ∼26% of them died soon after birth, which is similar to the neonatal mortality rate of the Dp(16)1Yey/+ model. Our preliminary result showed that the Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ model has DS-associated heart defects with a frequency similar to that of the Dp(16)1Yey/+ model (26) (Yu et al., unpublished data). The neonatal survival rates are similar among Dp(16)1Yey/+, Dp(10)1Yey/+;Dp(16)1Yey/+, Dp(16)1Yey/+;Dp(17)1Yey/+ and Dp(10)1Yey/+;Dp(16)1Yey/+Dp(17)1Yey/+ models. Dp(10)1Yey/+, Dp(17)1Yey/+ and Dp(10)1Yey/+;Dp(17)1Yey/+ models have a neonatal survival rate similar to that of the wild-type littermates. From the 172 progeny resulting from such a breeding scheme, ∼74% of the expected number of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice survived at weaning. The individual duplications in Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ embryos or mice were identified by Southern blot analysis (Fig. 2B and F) (26). The genotype was confirmed by Agilent microarray-based comparative genomic hybridization (CGH) using DNA isolated from a male Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mouse. As shown in Fig. 2I and J, mouse array CGH accurately detected single-copy duplications located in the Hsa21 syntenic regions on Mmu10, Mmu16 and Mmu17 by 343, 2, 141 and 166 oligonucleotide probes, respectively.
Using Taqman real-time quantitative PCR, we analyzed the mRNA levels for five and two genes duplicated in Dp(10)1Yey/+ mice and Dp(17)1Yey/+ mice, respectively, and showed, similar to Dp(16)1Yey/+ mice (26), that the duplications led to elevated mRNA levels in the brain for the genes located in the rearranged intervals (Table 1), reflecting gene-dosage increases associated with the duplications.
Table 1.
Normalized relative values (RQ) of expression in the brains
| Gene name | RQ ± S.E.M. | |
|---|---|---|
| Dp(10)1Yey/+ over +/+ | Adar2 | 1.61 ± 0.04 |
| Dnmt3l | 1.89 ± 0.04 | |
| Itgb2 | 1.86 ± 0.08 | |
| S100b | 1.50 ± 0.01 | |
| Trpc7 | 1.54 ± 0.03 | |
| Dp(17)1Yey/+ over +/+ | Cbs | 1.72 ± 0.04 |
| Pknox1 | 1.64 ± 0.02 |
The values represent the means of triplicated samples. Gapdh was used as an internal control and is disomic in all strains.
Impairment of hippocampal-mediated learning and memory
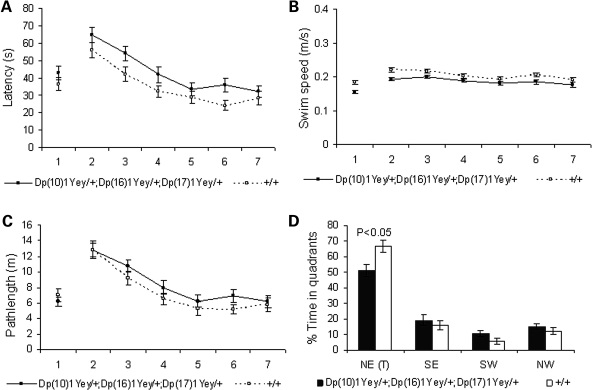
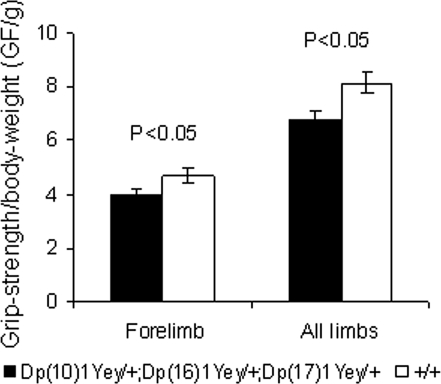
To examine the effect of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ on hippocampal-mediated learning and memory, we examined the mutant mice in the Morris water maze tasks. In the hidden platform version, while latency in finding the platform significantly decreased with training for both the mutant mice and the wild-type littermates (P < 0.01), the mutant mice had a significantly longer average latency (P < 0.01) (Fig. 3A). The difference in latency may be partially explained by the slower swimming speed of the mutant mice (P < 0.01) (Fig. 3B). Importantly, our results showed that the mutant mice took a longer path-length to locate the platform (P < 0.05) (Fig. 3C). Furthermore, in the probe test on the day after the training period, the mutant mice spent a significantly shorter time in the target quadrant (northeast, NE) than did the wild-type littermates (P < 0.05) (Fig. 3D). These results provide evidence that the mutant mice are impaired in spatial learning and memory. The slower swimming speed of the mutant mice may be caused by a muscle-strength deficit, a characteristic of patients with DS (27–29). To examine this possibility, we analyzed the grip strength of the mutant mice. Our results show that Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice are impaired in the grip strength of the forelimbs and all limbs (P < 0.05) (Fig. 4).
Figure 3.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice are impaired in Morris water maze tasks. The mutant mice (n = 15) and the wild-type littermates (n = 15) were examined in the Morris water maze, as described in Materials and Methods. (A) Latency to locate the platform (s, second). (B) Swimming speed during the learning trials (m/s, meter/second). (C) Path length to locate the platform (m, meter). (D) In the probe test on the day after the end of the training trials, the relative amount of time spent in different quadrants.
Figure 4.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice are impaired in grip strength. The muscle strength of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice (n = 10) and the wild-type littermates (n = 15) was analyzed using a grip-strength meter, as described in Materials and Methods. The grip-strength measurements (gram force/gram of body weight, GF/g) of the forelimbs and all limbs of the mice with different genotypes are shown.
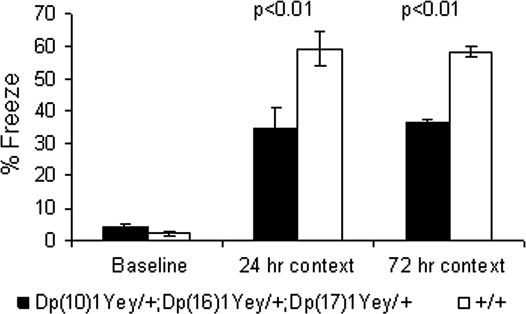
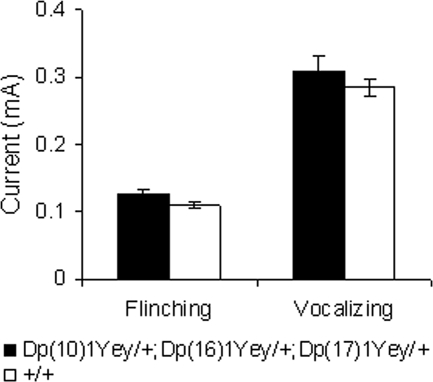
We performed additional tests to assess hippocampal-mediated cognitive behaviors. In the contextual fear-conditioning test, the mutant mice had a normal baseline freezing level prior to presentation of the foot shock (P > 0.05; Fig. 5). Both the mutant mice and the wild-type littermates increased their freezing behavior after being returned to the test chamber 24 h after the initial training and foot shock, but the mutant mice froze significantly less (P < 0.01, Fig. 5). After the 24 h context test, the context of the test chamber was altered, as described in Materials and Methods. The freezing levels of all strains in the altered context were similar (P > 0.05) and were significantly lower than their freezing levels earlier when they were exposed to the original context. This result suggests that none of the mouse strains exhibited generalized freezing in all conditions (30). Seventy-two hours after the initial training, the mice were returned to the original test chamber for the additional context test, and the mutant mice again exhibited a decreased freezing level (P < 0.01, Fig. 5). The foot-shock sensitivity test showed that there was no difference in the mean threshold of the current to elicit flinching or vocalizing between the mutant mice and the wild-type littermates (P > 0.05) (Fig. 6). These results indicate that the mutant mice were impaired in context-associated learning.
Figure 5.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice are impaired in the contextual fear-conditioning test. The mutant mice (n = 15) and the wild-type littermates (n = 15) were examined in the contextual fear-conditioning test, as described in Materials and Methods. The percentages of time spent freezing before the foot shock (baseline) as well as during the 24 and 72 h contextual tests are shown.
Figure 6.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice exhibit normal sensitivity to the electric foot shock. The minimal levels of currents (mA) needed to elicit a response, either flinching or vocalizing, from the mutant mice (n = 15) and the wild-type littermates (n = 15) are shown.
Impairment of hippocampal LTP
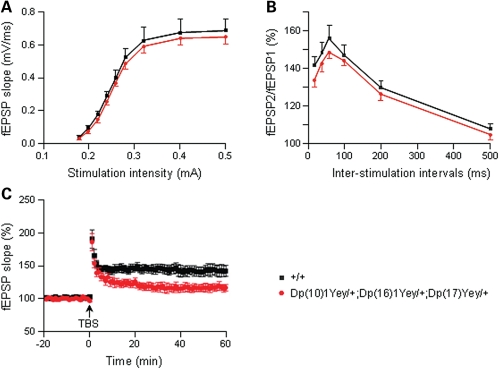
To explore the physiological basis of cognitive dysfunction in Dp(10)1Yey/+;(16)1Yey/+;Dp(17)1Yey/+ mice, we performed in vitro analysis of hippocampal synaptic transmission and plasticity by carrying out electrophysiological recordings in the CA1 region of the hippocampus in brain slices. First, we assessed the basal synaptic transmission by analyzing field excitatory postsynaptic potentials (fEPSPs) evoked by stimuli of various intensities and found no differences in the stimulus-intensity/response curve (i.e. input/output curve) between the mutant mice and the wild-type littermates (P > 0.05; Fig. 7A). To examine presynaptic function, we compared paired-pulse facilitation exhibited by Dp(10)1Yey/+; (16)1Yey/+;Dp(17)1Yey/+ mice and the wild-type littermates and found the genotype has no significant effect on this short-lasting form of synaptic plasticity (P > 0.05; Fig. 7B). We then investigated hippocampal LTP, which is considered a major physiological mechanism of learning and memory (15,31,32). The fEPSP of the CA1 region of the hippocampus in brain slices was induced by theta-burst stimulation (TBS). The baseline fEPSPs as well as evoked potentials after the TBS induction were recorded for each brain slice. The comparative analysis of the time course revealed that the mutant mice had a significant deficit in hippocampal LTP (P < 0.05) (Fig. 7C).
Figure 7.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice are impaired in hippocampal LTP. The electrophysiological recordings were carried out using hippocampal slices. (A) Input–output curves generated by applying stimuli of increasing intensity and measuring the initial slopes of the fEPSPs for the mutant mice (n = 13) and the wild-type littermates (n = 14). (B) Paired-pulse facilitation was measured by applying two closely spaced stimuli for the mutant mice (n = 18) and the wild-type littermates (n = 17), which was expressed as the ratio of the second synaptic response to the first synaptic response as a function of interpulse interval. (C) For analyzing hippocampal LTP, the fEPSP was induced by TBS. Recordings were carried out before and after the TBS inductions for the mutant mice (n = 7) and the wild-type littermates (n = 7). Evoked potentials were normalized to the fEPSP recorded prior to TBS induction (baseline = 100%). The data are presented as the percentage of fEPSP as a function of time.
Hydrocephalus
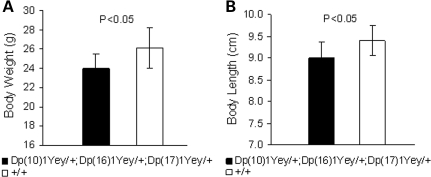
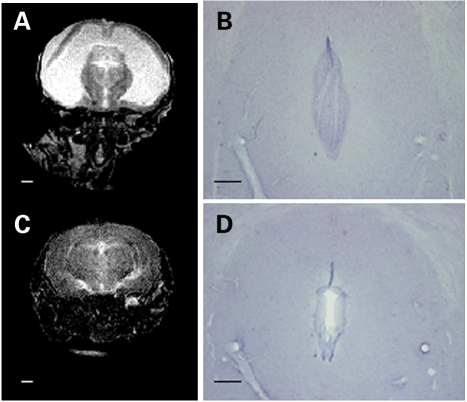
Approximately 93.5% of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice have a normal appearance, with only small reductions in body weight and body length (nose to anus) (Fig. 8). However, ∼6.5% of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice exhibit rounded and enlarged craniums at ∼6–8 weeks of age. The mutant mice exhibiting this phenotype usually died at ∼8–10 weeks of age. Further examinations revealed that these mutant mice had hydrocephalus, with lateral ventricles of the brains abnormally dilated (Fig. 9A). In these mutant mice, we also detected aqueductal stenosis as an associated defect (Fig. 9B). No hydrocephalus has been detected in any wild-type littermates.
Figure 8.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)Yey/+ causes small reductions in body weight and body length. Body weights (A) and body lengths (nose to anus) (B) of the male Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)Yey/+ mice (n = 7) and the male wild-type littermates (n = 9) are shown.
Figure 9.
Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)Yey/+ mice exhibit hydrocephalus. The brains of a Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(16)1Yey/+ mouse with a rounded and enlarged cranium at 1 month of age (A and B) and a wild-type littermate (C and D) were analyzed. Coronal views of T2-weighted MR images (A and C) show the lateral ventricles are severely dilated in the mutant mouse (A). Scale bar, 1 mm. The hematoxylin-stained histology sections of the same brains (B and D) reveal aqueductal stenosis in the mutant mouse (B). Scale bar, 0.2 mm.
DISCUSSION
Because DS is caused by the triplication of Hsa21, and the orthologs of the Hsa21 genes are located in three syntenic regions in the mouse genome, an ideal mouse model of DS should harbor Hsa21 (13) or segmental trisomies for all three mouse syntenic regions. These types of models have been considered important genetic tools for advancing DS research (33–38). Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice generated from this study carry a genotype of such desired models.
Recent studies suggest that the alteration of a biological pathway and, in turn, a DS phenotype could be the consequence of a functional interaction of two or more triplicated Hsa21 gene orthologs (39). Dyrk1a, one of the gene orthologs triplicated in Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice, has been proposed as a causative ortholog for the abnormal cognitively relevant phenotype in DS (39,40). However, the triplication of the Dyrk1a ortholog alone by BAC transgenics causes enhanced hippocampal LTP in mutant mice (41), even though Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice exhibited reduced hippocampal LTP. Similarly, the triplication of 12 genes in the Hsa21 syntenic region on Mmu17 was found to be associated with enhanced hippocampal LTP in Ts1Yah mice (25). The latter suggests that the cognitively relevant phenotypes in DS are the consequence of interactions among Hsa21 genes whose orthologs are located in different syntenic regions in mice. Therefore, Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ is a more desirable genotype for modeling human trisomy 21. Furthermore, our results show that when we recreated the evolutionarily conserved genotype, the final outcome at the phenotypic level also appears to be evolutionarily conserved: the DS-related abnormalities in the central nervous system, including impairments in learning, memory and hippocampal LTP as well as hydrocephalus (10). These results strengthen the possibility that the Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice could potentially be used as one of the reference models for understanding developmental cognitive disability in DS.
Another popular mouse model for DS is Ts65Dn mice. Ts65Dn is a derivative chromosome of a reciprocal translocation, and the studies on other mutant mice have shown that the presence of translocation chromosomes may be associated with impaired gametogenesis (42,43). Ts65Dn mice are maintained in the B6C3HF1 background, and backcrossing Ts65Dn mice to inbred wild-type mice has led to drastic reduction in fertility, probably because the adverse effects of the Ts65Dn chromosome on spermatogenesis and oogenesis are more severe in an inbred background. On the other hand, no impairment of fertility has been observed after Dp(10)1Yey/+, Dp(16)1Yey/+ and Dp(17)1Yey/+ mice were backcrossed to C57BL/6J mice for five generations, probably because these mutants do not carry a translocation chromosome and the third copies of the Hsa21 syntenic regions in these mutants are present as genomic duplications embedded within the original chromosomes. Ts65Dn mice and Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice shared the similarities in impairments in hippocampal-mediated behaviors and hippocampal LTP (19,44–49), suggesting the critical genes associated with these phenotypes may reside within the Mrpl39-Zfp295 genomic segment (Fig. 1). However, the result from Ts1Yah mice suggests that the orthologs of Hsa21 genes in other syntenic regions also contribute significantly to the DS-related phenotypes in patients with trisomy 21 and in Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice (25).
Hydrocephalus and associated aqueductal stenosis has been observed in patients with DS (50–53). On the basis of the analysis of 2894 cases of DS, Torfs and Christianson found a 10.1-fold increase in the incidence of hydrocephalus in DS patients compared with the non-DS population (P < 10−6) (10). Among mouse models of DS, enlargement of brain ventricles has been observed in Ts2Cje mice (54) that have the same triplicated region as the Ts65Dn mice (55). However, the hydrocephalus observed in Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice has not been reported in any other mouse models of DS. There are no known genes on Hsa21 at present that are associated with hydrocephalus. Our result suggests that hydrocephalus may be caused by the triplication of an Hsa21 gene ortholog that is not triplicated in Ts2Cje or by the functional interactions between triplicated genes located in the Mrpl39-Zfp295 region (Fig. 1) and in another Hsa21 syntenic region(s). These possibilities can now be examined in animal models for the first time.
Beta-amyloid plaques and neurofibrillary tangles are the most commonly observed Alzheimer-type neuropathology in patients with DS after ∼40 years of age. But the analysis of Ts65Dn mice and Tc1 mice revealed no amyloid plaques or neurofibrillary tangles in these models (13,19,56). We also performed a preliminary examination of the brains of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice at 12, 15, 18 and 26 months of age using immunohistochemistry and thioflavin-S staining, as described in Materials and Methods. Our result shows no evidence of beta-amyloid plaques or neurofibrillary tangles in our mutant mice, although the transcriptional level of App was increased by 50% in the mutant mice (26). The reason for this difference between patients with DS and Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice is currently unknown.
Although more than one gene may contribute to a DS phenotype, recent studies also suggest that, for some specific phenotypes, individual genes could be identified for playing the conspicuous roles by the gene-subtraction strategy (57–60). In those experiments, a compound mutant was generated to carry a null allele of the gene and a triplicated Hsa21 syntenic region that often was Ts65Dn. The contribution of the gene to the phenotype was established based on elimination or significant alleviation of a DS-related phenotype observed in a segmentally trisomic mouse strain (57–60). Because of the difference between Ts65Dn and Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice, introducing the genotype of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ into the compound mutants in future gene-subtraction experiments would enable us to assess the role of an Hsa21 gene ortholog that is not triplicated in Ts65Dn.
The advance of chromosomal manipulation technologies has made it possible to establish the desired trans-species and segmental trisomic mouse models. These models should facilitate better understanding of the mechanisms underlying developmental cognitive disability in DS as well as the development of new therapies for this clinical phenotype of the disorder.
MATERIALS AND METHODS
Generation of mouse mutants carrying the desired duplications using Cre/loxP-mediated chromosome engineering
To generate Dp(10)1Yey in mouse ES cells, the pTV(10)1EP1-targeting vector (Fig. 2A) was isolated from the 3′HPRT vector genomic library (61) using PCR primers 5′-GCTGGCCTCTGACTGTCTTGTTCC-3′ (forward) and 5′-GGGGCCATGTCACTGTCCTGCTA-3′ (reverse). The pTV(10)1EP2-targeting vector (Fig. 2A) was isolated from the 5′HPRT vector genomic library using PCR primers 5′-TCTGGAGCCATCTTCATAAAGCTG-3′ (forward) and 5′-AACTCACCGGTGATTGTGAAGA-3′ (reverse). pTV(10)1EP1 and pTV(10)1EP2 were used for inserting loxP into the regions proximal to Prmt2 and distal to Pdxk, respectively (Figs 1 and 2A). To generate Dp(17)1Yey, a loxP site with the desired orientation was inserted into a region proximal to Abcg1. For this specific targeting, the pTV(17)1EP1-targeting vector was constructed by reversing the orientation of the mouse genomic insert in MICER clone MHPP309a23 using the restriction enzyme site AscI that flanks the insert (Fig. 2E) (62). In addition, to facilitate the generation of a gap probe (Probe 17B) for Southern blot analysis, a 2.7 kb AflII–AflII fragment was deleted from the mouse genomic insert in the pTV(17)1EP1. MHPN353c19 was utilized as the pTV(17)1EP2-targeting vector for inserting loxP into the region distal to Rrp1b (Fig. 2E) (62).
The AB2.2 ES cells (63) were cultured for electroporation, as described previously (64). Prior to electroporation, the targeting vectors were linearized in the homology regions by digestion with the desired restriction enzymes. The linearized targeting vectors were electroporated into ES cells, which were then selected with G418 or puromycin. Single-targeted ES cell clones were identified by Southern blot analysis using PCR products as probes (Fig. 2A and E). A Cre-expression vector, pOG231 (65), was transfected into double-targeted ES cell clones by electroporation to induce Cre/loxP-mediated chromosomal rearrangements, as described previously (66,67). The clones of ES cells carrying the duplications on Mmu10 or Mmu17 were identified by Southern blot analyses of NheI-digested ES cell DNA with Probe 10B or EcoRI-digested ES cell DNA with Probe 17A, respectively. Germ-line transmitting chimeras were generated from the ES cell lines carrying the duplications by microinjecting them into blastocysts that were isolated from albino C57B6/J-Tyrc-Brd females, as described previously (68). Dp(10)1Yey/+ and Dp(17)1Yey/+ mice were identified by Southern blot analyses of NheI-digested tail DNA with Probe 10B or EcoRI-digested tail DNA with Probe 17A, respectively (Fig. 2B and F) and confirmed by FISH (see below).
Fluorescent in situ hybridization
The interphase nuclei of embryonic fibroblasts were prepared, as described previously (69). BAC clones were used as probes for FISH. To detect the chromosomal duplication between Prmt2 and Pdxk, BAC clone RP23-175G11 was labeled with digoxigenin and detected with anti-digoxigenin-rhodamine antibody. BAC clone RP23-150E20 was labeled with biotin and detected with fluorescein isothiocyanate–avidin. Chromosomes were counter-stained with DAPI (4′,6′-diamidino-2-2phenylindole) (Fig. 2D). To detect the chromosomal duplication between Abcg1 and Rrp1b, BAC clone RP23-304G12 was labeled with digoxigenin and detected with anti-digoxigenin-rhodamine antibody. BAC clone RP23-103F2 was labeled with biotin and detected with fluorescein isothiocyanate–avidin (Fig. 2H).
Mice
The mutant mice and littermates were maintained at a temperature- and humidity-controlled animal facility. Unless otherwise noted, mice used in the experiments were 2–4 months old. Before behavioral experiments, each mouse was prehandled for 2 min every day for a week. The experimental procedures were approved by the Institutional Animal Care and Use Committee.
Array-based CGH
To confirm the genotype of Dp(10)1Yey/+;Dp(16)1Yey/+;Dp(17)1Yey/+ mice, an oligonucleotide array containing 244 000 probes designed for mouse CGH was utilized (Agilent Technologies), which is composed of 60-mer oligonucleotides at an average spatial resolution of 6.4 Kb. The genome coordinates of the probes in the array were predetermined by Agilent Technologies. Genomic DNA was prepared from the tail tissue of a male mutant mouse using a DNeasy Tissue Kit (Qiagen Inc., Valencia, CA, USA). Genomic DNA isolated from a wild-type female littermate was used as the reference control. The genomic DNA from the mutant mouse and the wild-type littermate (1 µg each) was fluorescently labeled using an Agilent Genomic DNA Labeling Kit. Hybridization to the Agilent mouse 244K CGH array was performed for 40 h at 65°C. After hybridization, the slide was washed and scanned in an Agilent microarray scanner to generate high-resolution images for both Cy3 (the mutant mouse) and Cy5 (the wild-type littermate) channels. Image analysis was performed using Feature Extraction version 9.1 and CGH Analytics version 3.4.27 (Agilent Technologies Inc., Santa Clara, CA, USA).
Real-time quantitative PCR
Real-time quantitative PCR was used to analyze RNA levels of the genes located within the duplicated intervals in mutant mice. Gapdh is located on Mmu 6 and served as a reference gene of the disomic state for all the mice examined. Total RNAs were isolated from mouse brains using TRIzol Reagent (Invitrogen Corp., Carlsbad, CA, USA). An amount of 1 µg of the pooled RNA from three mice with the same genotype was used to generate cDNA by using Superscript version III reverse transcriptase (Invitrogen Corp.). The specific primers and probes for the genes were obtained from the TagMan® Gene Expression Assays System of Applied Biosystems, Inc. A 0.5 µg of cDNA from each genotype was analyzed by ABI 7900HT Real-Time Thermocycler (Applied Biosystems, Foster City, CA, USA) with the following amplification conditions: an initial activation and denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and primer annealing and extension at 60°C for 1 min.
Morris water maze test
A standard Morris water maze test was carried out in a circular pool (152 cm in diameter) of water at 25 ± 1°C (70–73). The experimental data were collected and analyzed using HVS Water 2020, an imaging-tracking and analysis system (HVS Image Ltd, Twickenham, Middlesex, UK). Each mouse had four trials each day. Visible-platform and hidden-platform training trials were carried out on day 1 and days 2–7, respectively. The amount of time spent finding the platform (latency), the distance traveled (path length) and the swimming speed were recorded. On day 8, a probe test was performed in which the platform was removed from the water and each mouse was allowed 60 s to search the pool. The time spent in each quadrant was measured.
Grip-strength measurement
The muscle strength of the mice was evaluated by a grip-strength meter (Columbus Instruments, Columbus, OH, USA). To measure the forelimb grip strength, a mouse was led to grip the grid of the apparatus using its forepaws. The mouse was then gently pulled away horizontally until its grasp was broken. The peak tension was measured by the grip-strength meter. The test was repeated five times for each mouse, and the highest and lowest values were removed. The forelimb grip strength of the mouse was calculated based on the average of the remaining three values and normalized to its body weight. A similar procedure was used to measure the combined forelimb and hindlimb grip strength with the exception that the mouse was allowed to grip the grid by both the fore and hind paws.
Contextual fear-conditioning test
The contextual fear-conditioning test was performed, as described previously (70,74), using the Fear-Conditioning Video Tracking System (Med Associates Inc., St Albans, VT, USA). The test chamber floor was a grid of stainless steel rods connected to an electric shock generator. A video camera was mounted on the front wall and a ceiling light illuminated the chamber interior through the transparent ceiling. Prior to conditioning, each mouse had 2 min to explore the test chamber (baseline activity). A foot shock (1 mA scrambled) was then administered for 2 s, which was controlled by a computer program (Video Freeze Software V.1.8, Med Associates Inc.). The mouse was removed from the chamber 30 s later. Approximately 24 h later, each mouse was returned to the chamber and monitored for freezing behavior for 3 min during which no foot shock was delivered. Freezing behavior was recorded automatically by the Video Freeze Software. Mean freezing activity during the contextual exposure was calculated as a measure of contextual learning. Two hours after the 24 h context test, the context was altered by cleaning the chamber with 1% acetic acid, covering the grid floor with a sheet of Perspex and inserting two sheets of Perspex into the chamber to give it a prism shape. Each subject was placed into the altered chamber and allowed 2 min for exploration. The freezing activity in the altered context was used to assess simply suppressed activity during all conditions (generalized freezing) (30). Seventy-two hours after the initial training, each mouse was again placed in the original test chamber for a final 3 min of observation without any foot shock being delivered.
Foot-shock sensitivity test
To facilitate accurate interpretation of the data from the fear-conditioning tests, we performed a foot-shock sensitivity test using the fear-conditioning test chamber. A foot shock was delivered every 10 s starting at 0.05 mA, with a 0.05 mA increment between each shock (75). The minimal level of current needed to elicit flinching or vocalizing was recorded.
Electrophysiology
The procedures for electrophysiological recordings were described previously (76,77). Briefly, hippocampal slices (400 µm) from the mice were prepared as described (78) and allowed to recover in a holding chamber for at least 1 h. For electrophysiological measurements, a single slice was then submerged in artificial cerebrospinal fluid (ACSF) within a temperature-controlled tissue chamber and superfused with oxygenated ACSF. The ACSF contained 120 mm NaCl, 2.5 mm KCl, 1.3 mm MgSO4, 1.0 mm NaH2PO4, 26 mm NaHCO3, 2.5 mm CaCl2 and 11 mm d-glucose. To record fEPSPs, the stimulating and recording electrodes were placed in the Schaffer collaterals and stratum radiatum of CA1, respectively, separated by a distance of 200–300 µm. Basal synaptic transmission was examined based on the relationship between the evoked presynaptic volley and the initial fEPSP slope (i.e. input–output curve). Then paired-pulse facilitation, a short-lasting form of synaptic plasticity indicative of presynaptic release probability, was assessed by measuring two closely spaced synaptic responses (fEPSP2/fEPSP1) at various interstimulus intervals. LTP was induced by TBS (15 bursts of four pulses at 100 Hz, delivered at an interburst interval of 200 ms) through the stimulating electrode. Recordings were carried out for 80 min for each brain slice (20 min of baseline recording and 60 min after the TBS induction). Traces were obtained by pClamp 9.2 and analyzed using the Clampfit 9.2 program (Molecular Devices, Sunnyvale, CA, USA).
Magnetic resonance imaging
The mice were transcardially perfused with 20 ml of PBS followed by 30 ml of 4% paraformaldehyde. Skin from the heads was removed, and the heads were then immersed in 4% paraformaldehyde at 4°C overnight. Magnetic resonance (MR) imaging scans were performed using a General Electric (GE) CSI 4.7 T/33 cm horizontal bore magnet (GE NMR Instruments, Fremont, CA, USA) with upgraded radio frequency (RF) and computer systems incorporating AVANCE digital electronics (Bruker BioSpec platform with Para-VisionR Version 2.1 Operating System, Bruker Medical, Billerica, MA, USA). MR data were acquired using a G060 removable gradient coil insert generating a maximum field strength of 950 mT/m and a custom-designed 35 mm RF transceiver coil. High-resolution T2-weighted rapid acquisitions with relaxation enhancement MR imaging pulse sequences were used to obtain multi-slice images. MR data analyses were carried out using Analyze PC (Version 7.0, AnalyzeDirect, Overland Park, KS, USA) and MATLAB (Version 7.0; Mathworks Inc., Natick, MA, USA).
Tissue processing, histology, immunohistochemistry and thioflavin-S staining
To collect brain tissues for analysis, the mice were perfused intracardially with 20 ml of PBS (pH 7.2), followed by 30 ml of 4% paraformaldehyde. The brains were removed and post-fixed in 4% paraformaldehyde at 4°C overnight. The tissues were further processed by the Roswell Park Cancer Institute Pathology Core Facility. Tissue processing and immunohistochemistry were performed as described (79). For detecting aqueducts, 100 µm thick brain sections were obtained by vibratome sectioning and stained with hematoxylin. Antibody 14F1 (Immuno-Biological Laboratories, Minneapolis, MN, USA), which recognizes both mouse and human APP proteins (80), was used to detect amyloid plaques. Thioflavin-S staining was performed according to the standard procedure (81). To detect Alzheimer-type neuropathological changes, Tg(APPswe,PSEN1dE9)85Dbo/J mice (82) of the same ages were used as a control. Five mice per strain were used for examining amyloid plaques and neurofibrillary tangles at 12, 15 and 18 months of age. Three mice per strain were used for the same examination at 26 months of age.
Statistical methods
The data from the Morris water maze probe test, grip-strength measurement, contextual fear-conditioning test, foot-shock sensitivity test and body-weight and body-length measurement were subjected to a one-way ANOVA between genotypes. ANOVA did not detect any effects from gender in all the behavioral tests, so the data from males and females were pooled and analyzed together for these experiments. Data from the 6-day training trials of the Morris water maze hidden-platform version were analyzed using a two-way (genotype × day) ANOVA, with the genotype as a between-subject factor and the day as a repeated-measures factor. The electrophysiology data were analyzed with a Student's t-test. All values reported in the text and figures were expressed as means ± S.E.M.
FUNDING
This project was supported in part by grants to Y.E.Y. from the Roswell Park Alliance Foundation, the Louis Sklarow Memorial Fund, the Fondation Jerome Lejeune, the Children's Guild Foundation and the NIH (R01HL091519).
SUPPLEMENTARY MATERIAL
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Jeffery Conroy, Devin McQuaid, Paul Szurek, Jeffrey LaDuca, Joseph Spernyak and Mukund Seshadri for their assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.CDC. Improved national prevalence estimates for 18 selected major birth defects—United States, 1999–2001. MMWR Morb. Mortal. Wkly Rep. 2006;54:1301–1305. [PubMed] [Google Scholar]
- 2.Hook E.B., Cross P.K., Schreinemachers D.M. Chromosomal abnormality rates at amniocentesis and in live-born infants. JAMA. 1983;249:2034–2038. doi:10.1001/jama.249.15.2034. [PubMed] [Google Scholar]
- 3.Hook E.G. Epidemiology of Down syndrome. In: Pueschel S.M., Rynders J.E., editors. Down Syndrome. Advances in Biomedicine and the Behavioral Sciences. Cambridge: Ware Press; 1982. p. 11. [Google Scholar]
- 4.Pennington B.F., Moon J., Edgin J., Stedron J., Nadel L. The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child Dev. 2003;74:75–93. doi: 10.1111/1467-8624.00522. doi:10.1111/1467-8624.00522. [DOI] [PubMed] [Google Scholar]
- 5.Pulsifer M.B. The neuropsychology of mental retardation. J. Int. Neuropsych. Soc. 1996;2:159–176. doi: 10.1017/s1355617700001016. doi:10.1017/S1355617700001016. [DOI] [PubMed] [Google Scholar]
- 6.Chapman R.S., Hesketh L.J. Behavioral phenotype of individuals with Down syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2000;6:84–95. doi: 10.1002/1098-2779(2000)6:2<84::AID-MRDD2>3.0.CO;2-P. doi:10.1002/1098-2779(2000)6:2<84::AID-MRDD2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 7.Haxby J.V. Neuropsychological evaluation of adults with Down's syndrome: patterns of selective impairment in non-demented old adults. J. Ment. Defic. Res. 1989;33:193–210. doi: 10.1111/j.1365-2788.1989.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 8.Nadel L. Down syndrome in cognitive neuroscience perspective. In: Tager-Flusberg H., editor. Neurodeveolpmental Disorders. Cambridge, MA: MIT Press; 1999. pp. 197–221. [Google Scholar]
- 9.Uecker A., Mangan P.A., Obrzut J.E., Nadel L. Down syndrome in neurobiological perspective: an emphasis on spatial cognition. J. Clin. Child. Psychol. 1993;22:266–276. doi:10.1207/s15374424jccp2202_12. [Google Scholar]
- 10.Torfs C.P., Christianson R.E. Anomalies in Down syndrome individuals in a large population-based registry. Am. J. Med. Genet. 1998;77:431–438. doi:10.1002/(SICI)1096-8628(19980605)77:5<431::AID-AJMG15>3.0.CO;2-J. [PubMed] [Google Scholar]
- 11.Mann D.M., Esiri M.M. The pattern of acquisition of plaques and tangles in the brains of patients under 50 years of age with Down's syndrome. J. Neurol. Sci. 1989;89:169–179. doi: 10.1016/0022-510x(89)90019-1. doi:10.1016/0022-510X(89)90019-1. [DOI] [PubMed] [Google Scholar]
- 12.Wisniewski K.E., Wisniewski H.M., Wen G.Y. Occurrence of neuropathological changes and dementia of Alzheimer's disease in Down's syndrome. Ann. Neurol. 1985;17:278–282. doi: 10.1002/ana.410170310. doi:10.1002/ana.410170310. [DOI] [PubMed] [Google Scholar]
- 13.O'Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M.L., Cooke S., Sesay A., Modino S., Vanes L., Hernandez D., et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. doi:10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara T., Tomizuka K., Miyabara S., Takehara S., Kazuki Y., Inoue J., Katoh M., Nakane H., Iino A., Ohguma A., et al. Mice containing a human chromosome 21 model behavioral impairment and cardiac anomalies of Down's syndrome. Hum. Mol. Genet. 2001;10:1163–1175. doi: 10.1093/hmg/10.11.1163. doi:10.1093/hmg/10.11.1163. [DOI] [PubMed] [Google Scholar]
- 15.Bliss T.V., Collingridge G.L. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. doi:10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 16.Lynch M.A. Long-term potentiation and memory. Physiol. Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. doi:10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 17.Martin S.J., Grimwood P.D., Morris R.G. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu. Rev. Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. doi:10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 18.O'Keefe J. Hippocampus, theta, and spatial memory. Curr. Opin. Neurobiol. 1993;3:917–924. doi: 10.1016/0959-4388(93)90163-s. doi:10.1016/0959-4388(93)90163-S. [DOI] [PubMed] [Google Scholar]
- 19.Reeves R.H., Irving N.G., Moran T.H., Wohn A., Kitt C., Sisodia S.S., Schmidt C., Bronson R.T., Davisson M.T. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. doi:10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 20.Akeson E.C., Lambert J.P., Narayanswami S., Gardiner K., Bechtel L.J., Davisson M.T. Ts65Dn—localization of the translocation breakpoint and trisomic gene content in a mouse model for Down syndrome. Cytogenet. Cell Genet. 2001;93:270–276. doi: 10.1159/000056997. doi:10.1159/000056997. [DOI] [PubMed] [Google Scholar]
- 21.Kahlem P., Sultan M., Herwig R., Steinfath M., Balzereit D., Eppens B., Saran N.G., Pletcher M.T., South S.T., Stetten G., et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. doi:10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. doi:10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 23.Eisfeld J., Luckhoff A. Trpm2. Handb. Exp. Pharmacol. 2007;179:237–252. doi: 10.1007/978-3-540-34891-7_14. doi:10.1007/978-3-540-34891-7_14. [DOI] [PubMed] [Google Scholar]
- 24.Olah M.E., Jackson M.F., Li H., Perez Y., Sun H.S., Kiyonaka S., Mori Y., Tymianski M., MacDonald J.F. Ca2+-dependent induction of TRPM2 currents in hippocampal neurons. J. Physiol. 2009;587:965–979. doi: 10.1113/jphysiol.2008.162289. doi:10.1113/jphysiol.2008.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira P.L., Magnol L., Sahun I., Brault V., Duchon A., Prandini P., Gruart A., Bizot J.C., Chadefaux-Vekemans B., Deutsch S., et al. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of Down syndrome. Hum. Mol. Genet. 2009;18:4756–4769. doi: 10.1093/hmg/ddp438. doi:10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Yu T., Morishima M., Pao A., LaDuca J., Conroy J., Nowak N., Matsui S., Shiraishi I., Yu Y. Duplication of the entire 22.9-Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum. Mol. Genet. 2007;16:1359–1366. doi: 10.1093/hmg/ddm086. doi:10.1093/hmg/ddm086. [DOI] [PubMed] [Google Scholar]
- 27.Morris A.F., Vaughan S.E., Vaccaro P. Measurements of neuromuscular tone and strength in Down's syndrome children. J. Ment. Defic. Res. 1982;26:41–46. doi: 10.1111/j.1365-2788.1982.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 28.Pitetti K.H., Climstein M., Mays M.J., Barrett P.J. Isokinetic arm and leg strength of adults with Down syndrome: a comparative study. Arch. Phys. Med. Rehabil. 1992;73:847–850. [PubMed] [Google Scholar]
- 29.Cioni M., Cocilovo A., Di Pasquale F., Araujo M.B., Siqueira C.R., Bianco M. Strength deficit of knee extensor muscles of individuals with Down syndrome from childhood to adolescence. Am. J. Ment. Retard. 1994;99:166–174. [PubMed] [Google Scholar]
- 30.Balogh S.A., Radcliffe R.A., Logue S.F., Wehner J.M. Contextual and cued fear conditioning in C57BL/6J and DBA/2J mice: context discrimination and the effects of retention interval. Behav. Neurosci. 2002;116:947–957. doi: 10.1037//0735-7044.116.6.947. doi:10.1037/0735-7044.116.6.947. [DOI] [PubMed] [Google Scholar]
- 31.Bear M.F., Abraham W.C. Long-term depression in hippocampus. Annu. Rev. Neurosci. 1996;19:437–462. doi: 10.1146/annurev.ne.19.030196.002253. doi:10.1146/annurev.ne.19.030196.002253. [DOI] [PubMed] [Google Scholar]
- 32.Malenka R.C., Nicoll R.A. Long-term potentiation—a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. doi:10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 33.Antonarakis S.E. Chromosome 21: from sequence to applications. Curr. Opin. Genet. Dev. 2001;11:241–246. doi: 10.1016/s0959-437x(00)00185-4. doi:10.1016/S0959-437X(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 34.Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 2004;5:725–738. doi: 10.1038/nrg1448. doi:10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 35.Davisson M.T., Bechtel L.J., Akeson E.C., Fortna A., Slavov D., Gardiner K. Evolutionary breakpoints on human chromosome 21. Genomics. 2001;78:99–106. doi: 10.1006/geno.2001.6639. doi:10.1006/geno.2001.6639. [DOI] [PubMed] [Google Scholar]
- 36.Patterson D., Costa A.C. Down syndrome and genetics—a case of linked histories. Nat. Rev. Genet. 2005;6:137–147. doi: 10.1038/nrg1525. doi:10.1038/nrg1525. [DOI] [PubMed] [Google Scholar]
- 37.Reeves R.H. Down syndrome mouse models are looking up. Trends Mol. Med. 2006;12:237–240. doi: 10.1016/j.molmed.2006.04.005. doi:10.1016/j.molmed.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Wiseman F.K., Alford K.A., Tybulewicz V.L., Fisher E.M. Down syndrome—recent progress and future prospects. Hum. Mol. Genet. 2009;18:R75–R83. doi: 10.1093/hmg/ddp010. doi:10.1093/hmg/ddp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arron J.R., Winslow M.M., Polleri A., Chang C.P., Wu H., Gao X., Neilson J.R., Chen L., Heit J.J., Kim S.K., et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. doi:10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 40.Lepagnol-Bestel A.M., Zvara A., Maussion G., Quignon F., Ngimbous B., Ramoz N., Imbeaud S., Loe-Mie Y., Benihoud K., Agier N., et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum. Mol. Genet. 2009;18:1405–1414. doi: 10.1093/hmg/ddp047. doi:10.1093/hmg/ddp047. [DOI] [PubMed] [Google Scholar]
- 41.Ahn K.J., Jeong H.K., Choi H.S., Ryoo S.R., Kim Y.J., Goo J.S., Choi S.Y., Han J.S., Ha I., Song W.J. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 2006;22:463–472. doi: 10.1016/j.nbd.2005.12.006. doi:10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Burgoyne P.S., Mahadevaiah S., Mittwoch U. A reciprocal autosomal translocation which causes male sterility in the mouse also impairs oogenesis. J. Reprod. Fertil. 1985;75:647–652. doi: 10.1530/jrf.0.0750647. [DOI] [PubMed] [Google Scholar]
- 43.de Boer P., Searle A.G., van der Hoeven F.A., de Rooij D.G., Beechey C.V. Male pachytene pairing in single and double translocation heterozygotes and spermatogenic impairment in the mouse. Chromosoma. 1986;93:326–336. doi: 10.1007/BF00327591. [DOI] [PubMed] [Google Scholar]
- 44.Costa A.C., Grybko M.J. Deficits in hippocampal CA1 LTP induced by TBS but not HFS in the Ts65Dn mouse: a model of Down syndrome. Neurosci. Lett. 2005;382:317–322. doi: 10.1016/j.neulet.2005.03.031. doi:10.1016/j.neulet.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 45.Costa A.C., Scott-McKean J.J., Stasko M.R. Acute injections of the NMDA receptor antagonist memantine rescue performance deficits of the Ts65Dn mouse model of Down syndrome on a fear conditioning test. Neuropsychopharmacology. 2008;33:1624–1632. doi: 10.1038/sj.npp.1301535. doi:10.1038/sj.npp.1301535. [DOI] [PubMed] [Google Scholar]
- 46.Escorihuela R.M., Fernandez-Teruel A., Vallina I.F., Baamonde C., Lumbreras M.A., Dierssen M., Tobena A., Florez J. A behavioral assessment of Ts65Dn mice: a putative Down syndrome model. Neurosci. Lett. 1995;199:143–146. doi: 10.1016/0304-3940(95)12052-6. doi:10.1016/0304-3940(95)12052-6. [DOI] [PubMed] [Google Scholar]
- 47.Kleschevnikov A.M., Belichenko P.V., Villar A.J., Epstein C.J., Malenka R.C., Mobley W.C. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J. Neurosci. 2004;24:8153–8160. doi: 10.1523/JNEUROSCI.1766-04.2004. doi:10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sago H., Carlson E.J., Smith D.J., Rubin E.M., Crnic L.S., Huang T.T., Epstein C.J. Genetic dissection of region associated with behavioral abnormalities in mouse models for Down syndrome. Pediatr. Res. 2000;48:606–613. doi: 10.1203/00006450-200011000-00009. doi:10.1203/00006450-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Siarey R.J., Stoll J., Rapoport S.I., Galdzicki Z. Altered long-term potentiation in the young and old Ts65Dn mouse, a model for Down syndrome. Neuropharmacology. 1997;36:1549–1554. doi: 10.1016/s0028-3908(97)00157-3. doi:10.1016/S0028-3908(97)00157-3. [DOI] [PubMed] [Google Scholar]
- 50.Dumont M., Weyland M. Apropos of a case of combined mongolism and hydrocephalus. Discussion of the etiological factor. Mises Point Accouch Pediatre. 1961;627:3–34. [PubMed] [Google Scholar]
- 51.Forcelini C.M., Mallmann A.B., Crusius P.S., Seibert C.A., Crusius M.U., Zandona D.I., Carazzo C., Crusius C.U., Goellner E., Ragnini J., et al. Down syndrome with congenital hydrocephalus: case report. Arq. Neuropsiquiatr. 2006;64:869–871. doi: 10.1590/s0004-282x2006000500031. [DOI] [PubMed] [Google Scholar]
- 52.Jayaraman A., Ballweg G.P., Donnenfeld H., Chusid J.G. Hydrocephalus in Down's syndrome. Childs Brain. 1976;2:202–207. [PubMed] [Google Scholar]
- 53.Zadikoff C. Down's syndrome with hydrocephalus treated by compressive head binding. S. Afr. Med. J. 1977;51:353–355. [PubMed] [Google Scholar]
- 54.Ishihara K., Amano K., Takaki E., Shimohata A., Sago H.C., Epstein C.J., Yamakawa K. Enlarged brain ventricles and impaired neurogenesis in the Ts1Cje and Ts2Cje mouse models of Down syndrome. Cereb. Cortex. 2009;20:1131–1143. doi: 10.1093/cercor/bhp176. doi:10.1093/cercor/bhp176. [DOI] [PubMed] [Google Scholar]
- 55.Villar A.J., Belichenko P.V., Gillespie A.M., Kozy H.M., Mobley W.C., Epstein C.J. Identification and characterization of a new Down syndrome model, Ts[Rb(12.1716)]2Cje, resulting from a spontaneous Robertsonian fusion between T(171)65Dn and mouse chromosome 12. Mamm. Genome. 2005;16:79–90. doi: 10.1007/s00335-004-2428-7. doi:10.1007/s00335-004-2428-7. [DOI] [PubMed] [Google Scholar]
- 56.Cooper J.D., Salehi A., Delcroix J.D., Howe C.L., Belichenko P.V., Chua-Couzens J., Kilbridge J.F., Carlson E.J., Epstein C.J., Mobley W.C. Failed retrograde transport of NGF in a mouse model of Down's syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc. Natl Acad. Sci. USA. 2001;98:10439–10444. doi: 10.1073/pnas.181219298. doi:10.1073/pnas.181219298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cataldo A.M., Petanceska S., Peterhoff C.M., Terio N.B., Epstein C.J., Villar A., Carlson E.J., Staufenbiel M., Nixon R.A. App gene dosage modulates endosomal abnormalities of Alzheimer's disease in a segmental trisomy 16 mouse model of Down syndrome. J. Neurosci. 2003;23:6788–6792. doi: 10.1523/JNEUROSCI.23-17-06788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salehi A., Delcroix J.D., Belichenko P.V., Zhan K., Wu C., Valletta J.S., Takimoto-Kimura R., Kleschevnikov A.M., Sambamurti K., Chung P.P., et al. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. doi:10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Sussan T.E., Yang A., Li F., Ostrowski M.C., Reeves R.H. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. doi:10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 60.Baek K.H., Zaslavsky A., Lynch R.C., Britt C., Okada Y., Siarey R.J., Lensch M.W., Park I.H., Yoon S.S., Minami T., et al. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126–1130. doi: 10.1038/nature08062. doi:10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng B., Mills A.A., Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. doi:10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams D.J., Biggs P.J., Cox T., Davies R., van der Weyden L., Jonkers J., Smith J., Plumb B., Taylor R., Nishijima I., et al. Mutagenic insertion and chromosome engineering resource (MICER) Nat. Genet. 2004;36:867–871. doi: 10.1038/ng1388. doi:10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 63.Bradley A., Zheng B., Liu P. 13 years of manipulating the mouse genome: a personal history. Int. J. Dev. Biol. 1998;42:943–950. [PubMed] [Google Scholar]
- 64.Ramirez-Solis R., Davis A.C., Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 65.O'Gorman S., Dagenais N.A., Qian M., Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. doi:10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu P., Zhang H., McLellan A., Vogel H., Bradley A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramirez-Solis R., Liu P., Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. doi:10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 68.Bradley A. Production and analysis of chimeric mice. In: Robertson E., editor. Teratocarcinomas and Embryonic Stem Cells—a Practical Approach. IRL Press; 1987. pp. 113–151. [Google Scholar]
- 69.Robertson E. Embryo-derived stem cell lines. In: Robertson E., editor. Teratocarcinomas and Embryonic Stem Cells—a Practical Approach. IRL Press; 1987. pp. 77–112. [Google Scholar]
- 70.Clapcote S.J., Lazar N.L., Bechard A.R., Roder J.C. Effects of the rd1 mutation and host strain on hippocampal learning in mice. Behav. Genet. 2005;35:591–601. doi: 10.1007/s10519-005-5634-5. doi:10.1007/s10519-005-5634-5. [DOI] [PubMed] [Google Scholar]
- 71.Clapcote S.J., Roder J.C. Survey of embryonic stem cell line source strains in the water maze reveals superior reversal learning of 129S6/SvEvTac mice. Behav. Brain Res. 2004;152:35–48. doi: 10.1016/j.bbr.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 72.D'Hooge R., Nagels G., Franck F., Bakker C.E., Reyniers E., Storm K., Kooy R.F., Oostra B.A., Willems P.J., De Deyn P.P. Mildly impaired water maze performance in male Fmr1 knockout mice. Neuroscience. 1997;76:367–376. doi: 10.1016/s0306-4522(96)00224-2. [DOI] [PubMed] [Google Scholar]
- 73.McIlwain K.L., Merriweather M.Y., Yuva-Paylor L.A., Paylor R. The use of behavioral test batteries: effects of training history. Physiol. Behav. 2001;73:705–717. doi: 10.1016/s0031-9384(01)00528-5. doi:10.1016/S0031-9384(01)00528-5. [DOI] [PubMed] [Google Scholar]
- 74.Lu Y.M., Jia Z., Janus C., Henderson J.T., Gerlai R., Wojtowicz J.M., Roder J.C. Mice lacking metabotropic glutamate receptor 5 show impaired learning and reduced CA1 long-term potentiation (LTP) but normal CA3 LTP. J. Neurosci. 1997;17:5196–5205. doi: 10.1523/JNEUROSCI.17-13-05196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosa E.F., Takahashi S., Aboulafia J., Nouailhetas V.L., Oliveira M.G. Oxidative stress induced by intense and exhaustive exercise impairs murine cognitive function. J. Neurophysiol. 2007;98:1820–1826. doi: 10.1152/jn.01158.2006. doi:10.1152/jn.01158.2006. [DOI] [PubMed] [Google Scholar]
- 76.Jia Z., Agopyan N., Miu P., Xiong Z., Henderson J., Gerlai R., Taverna F.A., Velumian A., MacDonald J., Carlen P., et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron. 1996;17:945–956. doi: 10.1016/s0896-6273(00)80225-1. doi:10.1016/S0896-6273(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 77.Meng Y., Zhang Y., Tregoubov V., Janus C., Cruz L., Jackson M., Lu W.Y., MacDonald J.F., Wang J.Y., Falls D.L., et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. doi:10.1016/S0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 78.Henderson J.T., Georgiou J., Jia Z., Robertson J., Elowe S., Roder J.C., Pawson T. The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron. 2001;32:1041–1056. doi: 10.1016/s0896-6273(01)00553-0. doi:10.1016/S0896-6273(01)00553-0. [DOI] [PubMed] [Google Scholar]
- 79.Li Z., Szurek P.F., Jiang C., Pao A., Bundy B., Le W.D., Bradley A., Yu Y.E. Neuronal differentiation of NTE-deficient embryonic stem cells. Biochem. Biophys. Res. Commun. 2005;330:1103–1109. doi: 10.1016/j.bbrc.2005.03.090. doi:10.1016/j.bbrc.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 80.Nishitomi K., Sakaguchi G., Horikoshi Y., Gray A.J., Maeda M., Hirata-Fukae C., Becker A.G., Hosono M., Sakaguchi I., Minami S.S., et al. BACE1 inhibition reduces endogenous Abeta and alters APP processing in wild-type mice. J. Neurochem. 2006;99:1555–1563. doi: 10.1111/j.1471-4159.2006.04178.x. doi:10.1111/j.1471-4159.2006.04178.x. [DOI] [PubMed] [Google Scholar]
- 81.Sun A., Nguyen X.V., Bing G. Comparative analysis of an improved thioflavin-S stain, Gallyas silver stain, and immunohistochemistry for neurofibrillary tangle demonstration on the same sections. J. Histochem. Cytochem. 2002;50:463–472. doi: 10.1177/002215540205000403. [DOI] [PubMed] [Google Scholar]
- 82.Jankowsky J.L., Slunt H.H., Gonzales V., Jenkins N.A., Copeland N.G., Borchelt D.R. APP processing and amyloid deposition in mice haplo-insufficient for presenilin 1. Neurobiol. Aging. 2004;25:885–892. doi: 10.1016/j.neurobiolaging.2003.09.008. doi:10.1016/j.neurobiolaging.2003.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.