Abstract
We previously showed that mutations in LIS1 and DCX account for ∼85% of patients with the classic form of lissencephaly (LIS). Some rare forms of LIS are associated with a disproportionately small cerebellum, referred to as lissencephaly with cerebellar hypoplasia (LCH). Tubulin alpha1A (TUBA1A), encoding a critical structural subunit of microtubules, has recently been implicated in LIS. Here, we screen the largest cohort of unexplained LIS patients examined to date to determine: (i) the frequency of TUBA1A mutations in patients with lissencephaly, (ii) the spectrum of phenotypes associated with TUBA1A mutations and (iii) the functional consequences of different TUBA1A mutations on microtubule function. We identified novel and recurrent TUBA1A mutations in ∼1% of children with classic LIS and in ∼30% of children with LCH, making this the first major gene associated with the rare LCH phenotype. We also unexpectedly found a TUBA1A mutation in one child with agenesis of the corpus callosum and cerebellar hypoplasia without LIS. Thus, our data demonstrate a wider spectrum of phenotypes than previously reported and allow us to propose new recommendations for clinical testing. We also provide cellular and structural data suggesting that LIS-associated mutations of TUBA1A operate via diverse mechanisms that include disruption of binding sites for microtubule-associated proteins (MAPs).
INTRODUCTION
Development of the human brain requires an exquisitely orchestrated program of neuronal migration and positioning. Lissencephaly (LIS, smooth brain) comprises a group of severe brain malformations associated with deficient neuronal migration that results in mental retardation, epilepsy and when severe a shortened lifespan (1–3). In the most severe form, the surface of the cerebral hemispheres is completely smooth (agyria), whereas less severe forms of LIS are characterized by simplified folding patterns with abnormally broad gyri (pachygyria). Classic LIS (agyria–pachygyria spectrum) is typically characterized by a loosely organized and markedly thickened four-layer cortex (compared with the normal six-layered cortical architecture) and apparently normal cerebellum, although some patients have mild vermis hypoplasia (4). However, some rare forms of LIS are associated with a disproportionately small cerebellum, referred to as LIS with cerebellar hypoplasia (LCH). Several subtypes are known including LCH group b associated with Reelin pathway defects and another type characterized by a very poorly organized two-layer cortex (4,5). Classic LIS and LCH most often occur as isolated malformations, but rarely occur as part of malformation syndromes.
LIS has a strong genetic basis. Several genes that underlie LIS are directly or indirectly associated with modulation of microtubules, which are cytoskeletal elements involved in key cellular processes including mitosis, cytokinesis, vesicle transport, and neuronal migration. Heterozygous or hemizygous mutations in the microtubule-associated genes LIS1 on 17p13.3 and DCX on Xq22.3q23 account for ∼85% of classic LIS (2,3,6,7). Co-deletion of the nearby LIS1 and YWHAE genes underlies Miller–Dieker syndrome (8–10), which consists of severe classic LIS and characteristic facial dysmorphism. Mutations in several other genes including ARX, RELN and VLDLR have been identified in other forms of LIS but collectively these account for only a small proportion of patients with LIS.
The most recent gene implicated in LIS encodes tubulin alpha1A (TUBA1A) and resides on chromosome 12q13.12 (11–14). TUBA1A is a critical structural subunit of microtubules that is transiently expressed during neuronal development (15). Here, we report screening of the largest cohort of LIS patients examined to date to determine the frequency of TUBA1A mutations in patients with LIS, the spectrum of phenotypes associated with TUBA1A mutations, and the functional consequences of different TUBA1A mutations on microtubule function.
We identified novel and recurrent TUBA1A mutations in ∼1% of children with classic LIS and surprisingly in ∼30% of children with LCH spanning a wide spectrum of severity. We also detected a novel TUBA1A mutation in one child with agenesis of the corpus callosum (ACC) and cerebellar hypoplasia (CBLH) without LIS. We provide structural and functional data suggesting that LIS-associated mutations of TUBA1A operate via diverse mechanisms, perturbing tubulin α–β dimerization and most commonly affecting binding sites for microtubule-associated proteins (MAPs).
RESULTS
TUABA1A mutations identified in 7% of unexplained classic LIS and 32% of LCH
We screened the complete TUBA1A coding region and flanking 5′- and 3′-untranslated regions using bi-directional Sanger sequencing in 125 patients with LIS, including 72 patients with classic LIS, 22 with subcortical band heterotopia (SBH), 29 with LCH and two with LIS and ACC. Mutations of DCX and LIS1 were excluded by sequencing or by review of brain imaging, so that patients with posterior-predominant LIS (the LIS1 pattern) had normal sequencing results for LIS1, but may or may not have had DCX testing, and vice versa. We identified 5 missense mutations in 72 patients with classic LIS (∼7%) and 10 missense mutations in 29 patients with LCH (∼34%) (Table 1). We subsequently added mutations in two further patients with TUBA1A mutations found by clinical testing (p.M377V and p.R390C), but did not include them in our cohort analysis. Eight mutations (p.L92V, p.V137D, p.D218Y, p.A270T, p.N329S, p.M377V, p.R390C and p.M425K) were novel, while three (p.R402C, p.R402H and p.R422H) were recurrent mutations previously reported in other patients. We identified p.R402C in five patients, p.R402H in two patients, and p.R422H in two patients.
Table 1.
TUBA1A mutations identified in LIS
| Subject ID | DNA ID | Diagnosis | Status | Amino acid | Inheritancea | Group assignment |
|---|---|---|---|---|---|---|
| CM-66 | CDF-0005 | LCH | Novel | L92V (c.274C > G) | De novo | LCH×Sev group 4 |
| CM-107 | CDF-0023 | LCH | Novel | V137D | Unknown | LCH×mild group 3 |
| LR07-213 | 07.3123 | LCH | Novel | D218Y (c.652G > T) | De novo | LCH×Sev group 4 |
| LR07-244 | 06.1048 | LCH | Novel | A270T (c.808G > A) | Unknown | LCH×Mod group 3 |
| LR05-388 | 06.1242 | LCH | Novel | N329S (c.986A > G) | De novo | LCH×Sev group 4 |
| LP95-073 | 00.0107 | LIS | Reported (13,14) | R402C (c.1204C > T) | Unknown | LIS×Mod group 1 |
| LR07-008 | 05.2831 | LIS | Reported (13,14) | R402C (c.1204C > T) | De novo | LIS×Mod group 1 |
| LR06-210 | 07.1424 | LIS | Reported (13,14) | R402C (c.1204C > T) | De novo | LIS×Mod group 1 |
| LR08-035 | 08.0750 | LIS | Reported (13,14) | R402C (c.1204C > T) | De novo | LIS×Mod group 1 |
| LR06-064 | 06.1029 | LIS | Reported (13,14) | R402C (c.1204C > T) | Unknown | LIS×Mod group 1 |
| LP97-039 | 99.1608 | LCH | Reported (12,14) | R402H (c.1205G > A) | De novo | LIS×Sev group 2 |
| LP97-041 | 01.2278 | LCH | Reported (12,14) | R402H (c.1205G > A) | De novo | LIS×Sev group 2 |
| LR05-052 | CDF-0014 | LCH | Reported (11,13) | R422H (c.1265G > A) | De novo | LCH×Mod group 3 |
| LR08-340 | CDF-0016 | LCH | Reported (11,13) | R422H (c.1265G > A) | De novo | LCH×Mod group 3 |
| LR08-388 | 08.3908 | LCH | Novel | M425K | Unknown | LCH×Sev group 4 |
Sev, severe; Mod, moderate.
aParental DNA unavailable for testing.
Defining the phenotypic spectrum of TUBA1A mutations
Detailed brain imaging studies were available for all 17 patients with TUBA1A mutations. We immediately recognized that mutations involving the arginine residue at codon 402 resulted in phenotypes indistinguishable from those associated with mutations of the LIS1 gene, while mutations at other codons produced more complex forms of LIS. From our detailed review of these imaging studies, we were able to delineate five distinct phenotype groups that are most easily appreciated with multiple views in several planes, as shown in Figs 1–4 and Supplementary Material, Figure S1.
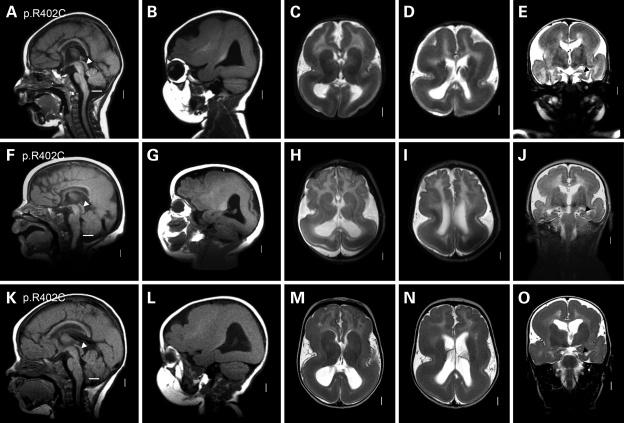
Figure 1.
Intermediate severity LIS identical to the LIS1-associated phenotype in group 1. In this and other brain imaging figures, each row shows multiple images from the same patient. The columns contain midline sagittal (far left column) and parasagittal (second column) images, axial images through the deep nuclei (center column) and bodies of the lateral ventricles (fourth column) and coronal images through the hippocampus (far right column). When coronal images were not available, low axial images through the cerebellum are shown instead. The horizontal white lines in the low posterior fossa in the midline sagittal images (left column) indicate the expected level of the lower border of the vermis at the level of the obex. A set of normal control images is shown in the top row of Supplementary Material, Figure S1. This figure shows three group 1 patients with the recurrent p.R402C TUBA1A mutation. All three have frontal pachygyria and posterior agyria consistent with classic LIS grade 3 and a posterior more severe than anterior (p > a) gradient. Other features include dysmorphic corpus callosum with small rostrum and genu plus a flattened anterior body, poorly myelinated and so unseen internal capsules and round hippocampi with thick leaves [black arrowheads in (E), (J) and (O)]. One patient also has a mildly small cerebellar vermis [space between horizontal white line and lower border of vermis in (A)]. This appearance is essentially identical to patients with heterozygous LIS1 null mutations and deletions. In addition, the tectum appears mildly enlarged (arrowheads in (A), (F) and (K), which differs from other types of LIS. These images come from subjects LP95-073 (A–E), LR07-008 (F–J) and LR08-035 (K–O).
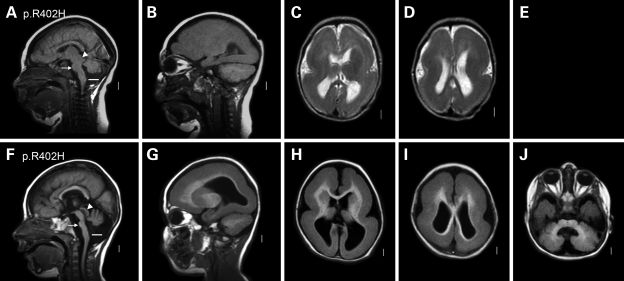
Figure 2.
Severe LIS resembling MDS in group 2. This figure shows two group 2 patients with the recurrent p.R402H TUBA1A mutation. Both have nearly complete agyria consistent with classic LIS grade 1. Both also have dysmorphic corpus callosum with small rostrum–genu and flat anterior body and poorly myelinated and so unseen internal capsules. The tectum again appears mildly enlarged [arrowheads in (A) and (F)], and both have a small cerebellar vermis [note horizontal white line well below the lower border of the vermis in (A) and (F)]. The gyral malformation is essentially identical to MDS (due to deletion 17p13.3 that includes both LIS1 and YWHAE), while the cerebellar hypoplasia is more severe than most—but not all—children with MDS. These images come from subjects LP97-039 (A–D) and LP97-041 (F–J).
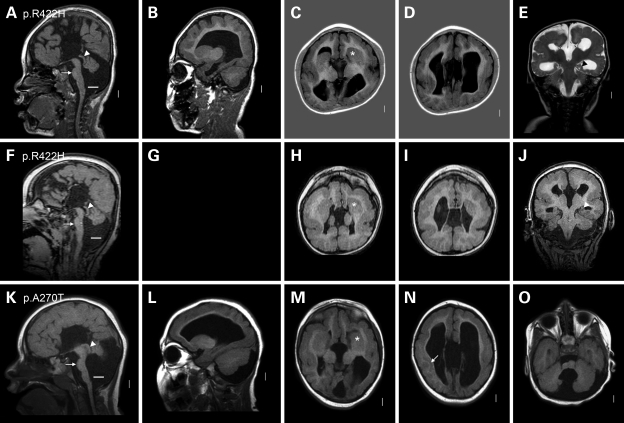
Figure 3.
A novel and distinctive pattern of LCH in group 3. This figure shows three group 3 patients with TUBA1A missense mutations and a novel pattern of malformations. All three have diffuse pachygyria or LIS grade 4 that appears mildly asymmetric and most severe over the central (mid- and posterior frontal, perisylvian and anterior parietal) regions rather than over the posterior pole. One patient has a prominent layer of gray matter beneath the right perisylvian region [arrow in (N)] that resembles SBH, but the overlying cortex appears thick so this is not true SBH. The hippocampi appear small and abnormally open and round with thick leaves [arrowheads in (E) and (J)]. The basal ganglia are malformed, appearing as large round structures in which the caudate, putamen and globus pallidus cannot be distinguished [asterisks in (C), (H) and (M)]. In the top row, the left basal ganglia appear to be located lateral rather than medial to the frontal horn [asterisk in (C)]. Associated malformations include complete (A and K) or nearly complete (F) ACC, thin brainstem with flat pons [arrows in (A), (F) and (K)], enlarged tectum [arrowheads in (A), (F) and (K)] and small cerebellar vermis [large space between the lower vermis and white lines in (A), (F) and (K)]. The cerebellar hemispheres are mildly small as well (O). These images come from subjects LR05-052 (A–E), LR08-340 (F–J) and LR07-244 (K–O).
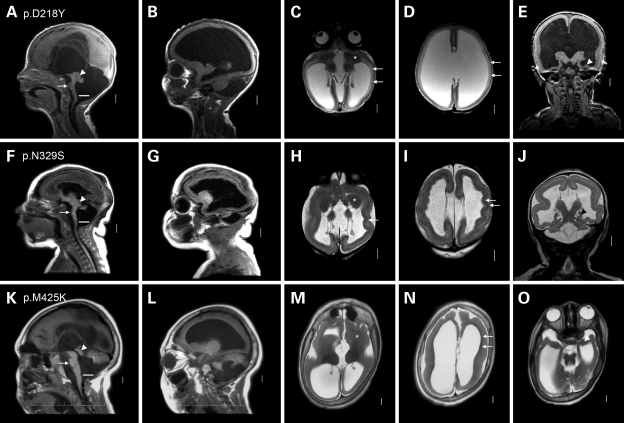
Figure 4.
Group 4 consists of severe LCH. This figure shows three group 4 patients with heterogeneous TUBA1A missense mutations and the most severe form of LCH. All three have a low forehead indicating microcephaly, marked thinning of the cortex and white matter with severe ventriculomegaly and nearly complete agyria except for very limited pachygyria over the frontal poles. However, the appearance of the cerebral cortex differs between the three patients. In the first patient, the ventriculomegaly is severe and the cerebral wall including the cortex is abnormally thin. In the first and third, the cortex is thick with a smooth lower border [arrows in (C), (D), (N)]. In the second, the cortex is thick with an undulating almost sinusoidal lower border [arrows in (H) and (I)]. Compare this pattern with subject LP99-059 shown in Fig. 6F in a prior publication (5). The hippocampi are very small and globular [arrowheads in (E), (J) and (O)]. The basal ganglia are very small and dysplastic with no differentiation between caudate, putamen and globus pallidus [asterisks in (C), (H) and (M)]. Other changes are also severe, including complete ACC, very thin brainstem with flat pons [arrows in (A), (F) and (K)], enlarged tectum [arrowheads in (A), (F) and (K)], severe diffuse cerebellar hypoplasia [large space between lower border of the vermis and white line in (A), (F) and (K)] and relatively enlarged posterior fossa. These overlapping patterns correspond to our prior LCH groups c and f (5). These images come from subjects LR05-388 (A–E), LR07-213 (F–J), and LR08-388 (K–O).
The first group consists of five patients with the recurrent p.R402C mutation (Fig. 1). All had classic LIS with frontal pachygyria and posterior agyria corresponding to classic LIS grade 3 with posterior more severe than anterior gradient, rounded hippocampi with a thick outer rim (Ammon's horn), intact but dysmorphic corpus callosum (missing rostrum plus flat genu and anterior body) and most often normal cerebellar structure. We observed a slightly more severe gyral malformation intermediate between grades 2 and 3, and mild cerebellar vermis hypoplasia in one patient each. This appearance is identical to the most common pattern found with mutations or deletions of the LIS1 gene.
Our second group consists of two patients with the recurrent p.R402H mutation (Fig. 2). Both had severe, classic LIS with nearly complete agyria, the same hippocampal and callosal defects as in the first group and moderate cerebellar vermis hypoplasia. This pattern resembles the brain imaging appearance found in patients with the Miller–Dieker syndrome (MDS) and co-deletion of LIS1 and YWHAE, except the cerebellar vermis hypoplasia is more severe in our two patients.
The third group consists of five patients with mild or moderate LCH and heterogeneous missense mutations (p.V137D, p.A270T, p.M377V, p.R422H) throughout the gene (Fig. 3). The brain imaging appearance is novel, consisting of diffuse pachgyria with no areas of agyria, mild-to-moderate asymmetry of the gyral malformation that could appear most severe over either the central convexity or the posterior pole and a thick cortex that is, however, not as thick as seen in patients with LIS1 (or TUBA1A p.R402C or p.R402H) mutations. The asymmetry is unusual for pachygyria; we considered whether this could be polymicrogyria, but the cortex is too thick and cortical infolding was not seen so we think this is a variant of pachgyria and not polymicrogyria. The posterior frontal, perisylvian and parietal regions were more severely affected than the occipital poles, another striking difference from the LIS1 mutation pattern in which the entire posterior brain including the occipital poles is most severely involved. One patient had a prominent deep cellular layer in the subcortical perisylvian region that resembles a short band heterotopia, but the overlying cortex is not normal indicating pachgyria and not SBH (Fig. 3N). The hippocampus is malformed as in the previous groups. Our three patients with severe pachygyria also had absent or nearly absent corpus callosum, thin brainstem and severe cerebellar hypoplasia. The cerebellar malformation consisted of small and upwardly rotated vermis, cystic dilatation of the 4th ventricle, less severe cerebellar hemisphere hypoplasia and mild to moderately enlarged posterior fossa, thus meeting the criteria for Dandy–Walker malformation, although the shape of the vermis differed from the typical form of this disorder. The least severely affected subjects had only moderate pachgyryia, mildly dysmorphic corpus callosum with absent spenium and mild cerebellar vermis hypoplasia.
Our fourth group consists of four patients with novel heterozygous missense mutations associated with severe LCH that corresponds to our previous LCH groups c, d and f (5) by brain imaging (p.D218Y, p.N329S; p.M425K; Fig. 4) or fetal neuropathology at 21 weeks gestation (p.L92V). In all, the walls of the cerebral hemispheres are thin, and the cerebral surface completely smooth except for a few abortive sulci at the frontal pole in the third patient. The cortex demonstrates a slightly different pattern in each patient. In the first, the cerebral wall is very thin, but the cortex and white matter can be distinguished (LR07-213; p.D218Y; Fig. 4A–E). In the second, the cortex fills the entire mantle and creates an undulating pattern along the walls of the lateral ventricles, a rare LIS pattern (LR05-388; p.N329S; Fig. 4F–J). The same pattern was seen in an LCH patient with no sample available to test (patient LP99-059 shown in Fig. 6D–F in 5). In the third patient, the cortex and white matter can barely be distinguished. The amygdala and hippocampus are seen as small thickenings of the temporal wall (LR08-388, p.M425K; Fig. 4K–O). The corpus callosum is absent in all three patients. In the first two, the brainstem is remarkably thin and the cerebellum consists of a nubbin with the vermis <1 cm in size. The posterior fossa is mildly enlarged, but the overall appearance is atypical for DWM because of the minute cerebella. In the third patient, the brainstem is moderately small, and the cerebellar defects meet the criteria for mild DWM.
The remaining subject was identified following fetal termination for ventriculomegaly and other anomalies at ∼21 weeks gestation (CM–66, p.L92V). The brain was small (weight 31 g, expected 45 g) and had a generalized deficiency of neuronal migration with neurons seen diffusely through the mantle (thus no layers), a small amorphous group of neurons in the area of the hippocampus, severe dilatation of the third and lateral ventricles and absent corpus callosum. The brainstem was very small with absent descending corticospinal tracts and inferior olives and small basis pontis. The cerebellum was also very small with absent dentate nuclei and formation of only an indistinct cerebellar cortex.
Our last group consists of a single patient carrying a novel de novo heterozygous missense mutation p.R390C with mildly reduced number of gyri and shallow sulcal depth that we designate as a ‘simplified’ gyral pattern, plus complete ACC and moderate CBLH (Supplementary Material, Fig. S1). The cortical-white matter interface appeared mildly irregular in places, but the cortex was not thick and we found this unconvincing for pachygyria, polymicrogyria or another cortical dysplasia; however, the resolution of the study was low. This phenotype is closest to—but milder than—mild-moderate LCH.
Cellular studies of TUBA1A mutations
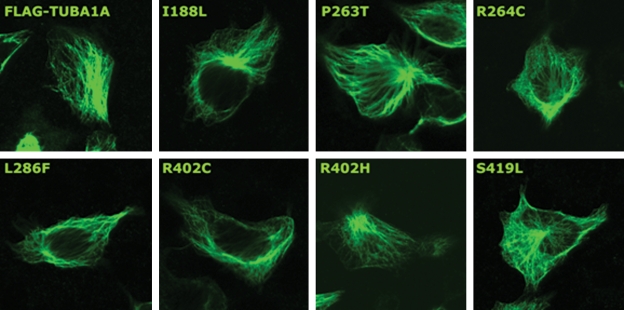
Since all TUBA1A mutations reported to date are de novo and affect a single allele of an autosomal gene, a dominant-negative mechanism of action is plausible. Consistent with this hypothesis, TUBA1A mutations reported to date do not profoundly affect the ability of mutant TUBA1A to incorporate into the cytoskeleton in recombinant systems (12,16). To confirm this finding for a range of TUBA1A mutants reported here and elsewhere, we introduced the mutations I188L, P263T, R264C, L286F, R402H, R402C and S419L into a construct expressing C-terminally FLAG-tagged TUBA1A and transfected these into P19 cells (Fig. 5). Confocal microscopy revealed that in each case, recombinant TUBA1A is clearly made and the typical tubulin architecture is observed. This suggests that these mutants readily incorporate into the cellular cytoskeletal network. This finding does not support a recent study that reported haploinsufficiency as a cause of the disease phenotype (16) and we chose to undertake detailed structural modeling to predict the functional consequences of existing and novel TUBA1A mutations.
Figure 5.
Recombinant wild-type and mutant FLAG-tagged TUBA1A incorporate into the normal interphase microtubule network. After transfection into P19 cells, wild-type and mutant recombinant TUBA1A were visualized using methanol fixation and immunostaining using anti-FLAG antibodies.
Structural modeling of TUBA1A mutations
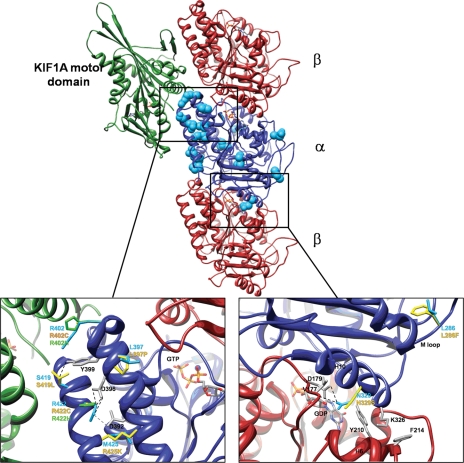
The N329S mutation is located on α-helix H10 of α-tubulin (residues 324–337) (17) at the interface between α- and β-tubulin monomers. H10 interacts with α-helix H6 (residues 206–214) and the adjacent loop (the B5–H5 loop, residues 171–183) on β-tubulin, both of which directly interact with GTP (Fig. 6). In the wild-type structure, the interaction of H10 with β-tubulin is stabilized by the side chains of two residues, K326 and N329. The K326 amino group forms an H bond with the backbone oxygen of Y210 and is also involved in a cation–π interaction with the aromatic ring of F214. The N329 carboxamide group forms two H bonds, one with the D179 carboxylate group and the other with the V177 backbone oxygen (on the B5–H5 loop). The N329S mutation will not allow the latter H bond interactions and therefore is likely to destabilize the interaction between α- and β-tubulin monomers around H6 and the B5–H5 loop, possibly affecting the conformation necessary for GTP binding.
Figure 6.
Mapping of TUBA1A mutations onto the 3D structure of kinesin KIF1A–microtubule complex. The KIF1A/microtubule complex: α-tubulin is shown in blue ribbons, β-tubulin in dark and KIF1A in green [PDB ID: 2HXF (33)]. Residues mutated in TUBA1A are shown as light blue spheres; ADP and ATP molecules in stick representation. Substitutions R402H/R402C, S419L, R422H/R422C, L397P and M425K on α-helices H11–H12 are likely to affect H11–H12 interactions, position, orientation and interactions with tubulin-binding proteins (e.g. KIF1A, DCX, MAP2c and Dynein). The guanidinium ion of R402 is involved in a cation–π interaction with the aromatic ring of TUBA1A residue Y399, which in turn forms an H bond with S419-Oγ. Both the R402H and R402C mutations will abolish the cation–π interaction, since neither histidine nor cysteine can be involved in this type of interaction. Because a leucine side chain cannot form an H bond, the S419L mutation cannot stabilize Y399 in the ideal position to form the cation–π interaction with R402. The guanidinium ion of R422 forms multiple H bonds with the carboxylate group of D396 and a salt bridge with the carboxylate group of D392. Neither the R422C nor R422H mutations can participate in these interactions (although histidine is able to form an H bond, it is too far from D396). The M425K mutation might also interrupt this network by competing with R422 for the interaction with D392 (due to a change from a neutral residue to a positively charged one). The L397P mutation is expected to introduce a kink in H11, which will affect the position of D396 and D392 relative to R422. C: The N329S mutation is located on α-helix H10 at the interface between α - and β-tubulin subunits, close to the GTP binding site. The interaction of H10 with α-tubulin is stabilized by the side chains of residues N329 and K326. The N329 forms two H bonds, one with D179 and the other with V177. The N329S mutation will not allow the forming of the H bonds with V177 and therefore is likely to destabilize interactions at the α–β interface. H bonds are marked in black and salt bridges in gray.
Six additional TUBA1A mutations cluster around α-helices H11 and H12 of α-tubulin (R402H, R402C, S419L, R422H, R422C and M425K) and they are likely to affect the interaction between these helices. Because H11–H12 lie on the interface between α-tubulin and various MAPs [e.g. kinesin KIF1A (18), DCX (19), MAP2c (20), and Dynein (21)], the mutations are also likely to affect the interaction between tubulin and MAPs. The interaction network between H11 and H12 is complex. First, the guanidinium ion of R402 is involved in a cation–π interaction with the aromatic ring of Y399, which in turn forms an H bond with S419-Oγ. Clearly, both the R402H and the R402C mutations will abolish the cation–π interaction, since neither histidine nor cysteine can be involved in this type of interaction. Furthermore, because a leucine side chain cannot form an H bond, the S419L mutation cannot stabilize Y399 in the ideal position to form a cation–π interaction with R402. Second, the guanidinium ion of R422 forms three H bonds with the carboxylate group of D396 and a salt bridge with the carboxylate group of D392. Neither the R422C nor the R422H mutations can form these interactions (although histidine is able to form an H bond, it is too far from D396). The M425K mutation might also interrupt this network by competing with R422 for the interaction with D392 (due to a change from a neutral residue to a positively charged one). Finally, the L397P mutation is expected to introduce a kink in H11 (since the cyclization of the proline side chain prevents the regular backbone H bond formation), which will affect the position of D396 and D392 relative to R422 and possibly their interaction.
DISCUSSION
The tubulin gene family in humans is extensive, comprising at least nine α, nine β, two γ and single δ and ε subunits, all expressed in different temporal, spatial and subcellular locations. TUBA1A is one of three α-tubulin genes that cluster on chromosome 12q13.12. The encoded protein is formed by a core of two β-sheets surrounded by α-helices and can be divided into three functional domains: (i) an N-terminal domain (residues 1–205) that contains the guanine nucleotide-binding region; (ii) an intermediate domain (residues 206–381) containing the Taxol drug-binding site and (iii) a C-terminal domain (resides 382–451) that likely constitutes the binding surface for MAPs and molecular motors such as kinesins and dyneins (17).
Mutation detection rates and testing recommendations in LIS-SBH subtypes
We identified TUBA1A mutations in 17 patients, most (67%) residing in the C-terminal domain. Five were detected in patients with unexplained classic LIS (∼7%), all at codon R402. Another 10 mutations were found in patients with LCH (∼34%) scattered through most of the coding region of the gene. In children with classic LIS resembling the LIS1 (posterior predominate) pattern, the expected mutation detection rate is ∼85% (7). Our data predict that another ∼1% of patients with classic LIS will have mutations of TUBA1A (i.e. 7% of the remaining 15% of LIS yet unexplained).
In children with LCH—a rare subtype of LIS—these results increase the mutation detection rate from essentially nil up to ∼30%, making TUBA1A the first gene to explain a significant proportion of this rare phenotype. A few patients with another LCH variant [LCH group b (5)] have mutations of RELN or VLDLR (22–24).
Although published reports (14,25) suggest that TUBA1A mutations are found in some patients with SBH, we found no mutations in a series of 22 patients with unexplained SBH. We did see small areas partly resembling SBH in two patients with the mild–moderate form of LCH (as shown in Fig. 3N), but the brain imaging phenotype in these individuals was clearly dominated by pachygyria, not SBH.
Importantly, we found one mutation in a girl with ACC and CBLH but without LIS. This observation suggests that mutations of TUBA1A may be responsible for a subset of children with isolated ACC, isolated CBLH or—as in our patient—combined ACC and CBLH without obvious LIS. Further studies in these latter groups are clearly needed.
These results have significant implications for clinical molecular testing for LIS. We conclude that sequencing of TUBA1A is so far indicated in two groups of patients. First are those with classic LIS and negative deletion and sequencing analysis for LIS1 and DCX. On brain imaging, we recognize classic LIS based on the presence of extensive agyria–pachgyria, 12–20 mm thick cortex and no more than mild CBLH. Second are those with LIS and moderate-to-severe CBLH, the so-called LCH, whether or not the corpus callosum is small. Experts in brain imaging in this group of disorders could exclude patients with a frontal gradient (LIS most severe in the anterior frontal lobes) and patients with the rare pattern associated with RELN or VLDLR mutations (mild frontal pachgyria, hippocampal hypoplasia and very small and near-afoliar cerebellum), but we do not recommend excluding these patterns generally. Although we did find one subject with combined ACC and CBLH without LIS, we do not have sufficient data to justify clinical testing of TUBA1A in children without LIS at this time.
Genotype–phenotype analysis
Our genotype–phenotype analysis revealed two major phenotype subsets, each with varying degrees of severity. The first subset (groups 1 and 2) involves all seven patients with mutations of codon R402, where the brain phenotype matches the well-known phenotype associated with mutations in LIS1 or combined LIS1 and YWHAE mutations. Specifically, the recurrent p.R402C phenotype resembles the LIS1 deletion or intragenic mutation phenotype, whereas the p.R402H phenotype resembles MDS caused by the co-deletion of LIS1 and YWHAE. The second subset (groups 3 and 4) involves patients with mutations of any other codon that differ dramatically from the LIS1-associated pattern. Mutations of other codons have much more variable phenotypes with more variable severity and (anterior–posterior) gradient of LIS and more consistent hypoplasia of the corpus callosum and cerebellum.
The second subset includes children with the rare LCH phenotype. In group 3, the gyral pattern often appeared most severe in the middle (posterior frontal or perisylvian-parietal regions), rather than at the frontal or occipital pole. Also, the callosal and cerebellar defects varied substantially in severity. Finally, the LCH seen in our group 4 is far more severe than ever seen in patients with LIS1 or codon R402 mutations. Taken together, our genotype–phenotype analysis suggests that mutations involving R402 specifically disrupt the LIS1-associated neuronal migration pathway, whereas other mutations are likely to disrupt multiple pathways and functions.
We reviewed the descriptions and available brain images for other published patients with TUBA1A mutations and found them generally compatible with our analysis of the phenotype spectrum (11–14,25). This was particularly true for the high-resolution images provided in Fig. 1 in the most recent paper (13).
Mechanisms of action for TUBA1A mutations
The mechanism(s) leading to the LIS or LCH phenotypes with heterozygous mutations is not clear, and we considered both haploinsufficiency and a dominant-negative effect. Several lines of evidence may support a dominant-negative mechanism of action. First, our data and those of others (12,16) show that mutant TUBA1A protein readily incorporates into the cellular cytoskeletal network, rather than failing to incorporate as might be expected with loss of function. Second, the observation of a nonsense mutation, frameshift mutation or gene deletion in even a single patient would support haploinsufficiency, but only missense mutations of TUBA1A have been identified to date. To extend this observation, we searched several large cytogenetic databases including Decipher (https://decipher.sanger.ac.uk/), DGAP (http://www.bwhpathology.org/dgap/) and ECARUCA (http://agserver01.azn.nl:8080/ecaruca/ecaruca.jsp), as well as PubMed, but found no reports of individuals with TUBA1A deletions. Further, no 12q13.12 deletions that contain TUBA1A were found in approximately 15 000 subjects with developmental disorders screened by microarrays (Swaroop Aradhya, GeneDx; Lisa Schaffer and Jill Rosenfeld, Signature Genomics, personal communications, 2009). We cannot exclude the possibility that heterozygous null mutations of TUBA1A cause early embryonic lethality, which would support haploinsufficiency. However, we think that the pathogenic mechanisms are more complex than simple haploinsufficiency given that LIS-associated TUBA1A mutations incorporate and maintain typical microtubule structure. These mutations could disrupt the binding sites for MAPs, leading to altered function or possibly dominant-negative effects.
Recurrent R402 mutations
Mutations involving the R402 residue have been reported in a total of eleven patients to date [seven described here and four elsewhere (13,14)], demonstrating that this codon is a mutational hotspot in TUBA1A. Our modeling data show that the R402 residue is located on α-helix H11, which lies at the outer surface of microtubules at the interface between α-tubulin and various MAPs and molecular motor proteins, including kinesin KIF1A (18). Our cellular assays clearly demonstrated that mutant R402 proteins are translated, localize to the cytoplasm and incorporate properly into the cytoskeletal network. Collectively, our data indicate that mutations involving the R402 residue are more likely to affect the interaction between tubulin and tubulin-binding proteins than to affect overall tubulin architecture. However, further biochemical experiments would be needed to substantiate these predictions. Given that R402 mutations result in a LIS1-like phenotype, we hypothesize that mutations involving this residue specifically disrupt the LIS1 signaling pathway. We propose that this may occur in a kinesin-dependent manner that involves a LIS1-dynein-transportable microtubule (tMT) complex, based on a recent observation that LIS1 and cytoplasmic dynein co-migrate towards the plus end of microtubules in a complex with tMT (26). In this model, LIS1 fixes dynein on tMT (freighter tubulins), which are then transported to the plus end of cytoskeletal MTs in a kinesin-dependent manner. Thus, we hypothesize that the R402 mutation specifically interferes with kinesin-mediated anterograde transport of LIS1-coupled dynein, without disrupting other functions of the TUBA1A protein.
We further hypothesize that the LCH-associated mutations of TUBA1A disrupt two or more distinct neuronal migration pathways that converge on TUBA1A, such as LIS1-related and DCX-related pathways, resulting in the chaotic LIS gradient and severe callosal and cerebellar phenotypes. It follows that mutations at less critical residues might cause corpus callosum and cerebellar malformations without obvious LIS, as we now document in one patient.
In conclusion, we have identified numerous novel mutations in TUBA1A, including several recurrent mutations at two codons that result in homogenous phenotypes. We provide mutation frequency data that will contribute to clinical diagnostics and demonstrate a wider spectrum of phenotypes than previously reported that now includes severe and moderate classic LIS (groups 1 and 2), severe and moderate LCH (groups 3 and 4) and ACC and CBLH not associated with LIS (group 5). We also show that LIS-associated mutations are likely to act in a dominant-negative manner by incorporating into microtubules and disrupting tubulin-MAP interactions. Certainly, tubulin genes are emerging as strong candidates for analysis in neuronal migration disorders, given recent additional reports of mutations in TUBB2B in asymmetrical polymicrogyria (27) and TUBA8 in polymicrogyria with optic nerve hypoplasia (28).
MATERIALS AND METHODS
Patients
The Institutional Review Boards at the University of Chicago and the University Hospital of Wales approved this study. From our large cohort of subjects with unexplained LIS documented by review of brain imaging (available for all subjects and reviewed by W.B.D. or D.T.P.) and DNA samples available (n = 125), we selected 72 patients with classic LIS (29 females and 43 males), 22 with SBH (13 females and 9 males), 29 with LCH (17 females and 12 males) most of whom also had partial or complete ACC and two females with LIS and ACC but normal cerebellum on imaging. We combined the latter two groups for further analysis. All LIS subgroups were defined years before this study was conducted (5,29,30). Testing for nonsense mutations, missense mutations, deletions or duplications in LIS1 and DCX was negative in most patients (7). In the remainder, the most appropriate gene was tested with negative results. Specifically, we tested only LIS1 in some patients with posterior-predominant LIS and tested DCX only in some with anterior-predominant LIS. We subsequently added two patients in whom TUBA1A mutations were found in clinical laboratories (one unexpectedly given the lack of LIS) and referred to us.
DNA isolation, amplification and sequencing
DNA was extracted from leukocytes from EDTA-treated blood using either the Puregene kit (Gentra Systems, Inc., Minneapolis, MN, USA) or the MagNAPure Total Nucleic Acid Extraction system (Roche Diagnostics, Indianapolis, IN, USA) following the manufacturer's protocols. PCR-amplification primers were designed using Primer3 (http://frodo.wi.mit.edu/) with M13 forward and reverse tails added to each primer to facilitate high-throughput DNA sequencing (Table 1). DNA was amplified in a reaction comprising: 20 ng genomic DNA, 1× buffer I (1.5 mm MgCl2, Applied Biosystems, Foster City, CA, USA), 1 mm dNTPs (Applied Biosystems), 0.4 µm primer (each of forward and reverse; IDT, Coralville, IA, USA) and 0.25 U AmpliTaq Gold (Applied Biosystems) in a total volume of 10 µl. Thermocycling conditions were as follows: 94°C for 10 min; 35 cycles of 94°C for 30 s, annealing temperature (53–60°C) for 30 s and 72°C for 30 s and final extension of 72°C for 10 min. Variations in reaction composition and cycling conditions were required for a small number of amplicons. PCR products were purified in a 10 µl reaction comprising 6.6 U Exonuclease I and 0.66 U shrimp alkaline phosphatase that were incubated at 37°C for 30 min followed by 80°C for 15 min. Sequencing reactions were performed using BigDye terminators on an ABI 3730XL 96-capillary automated DNA sequencer (Applied Biosystems) at The University of Chicago DNA Sequencing and Genotyping Core Facility. Sequence data were imported as ABI files into Mutation Surveyor v3.10 (SoftGenetics, State College, PA, USA). Sequence contigs were assembled by aligning ABI files against GenBank reference sequence files obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Genbank/). To screen for putative mutations, the entire length of the sample trace was manually inspected for quality and variation from the reference trace. All detected variants were visually reviewed by two trained individuals and were confirmed using bi-directional sequencing.
Transfection experiments
Mutations were introduced into the pRK5TUBA1A-CFLAG expression construct using the Quickchange site-directed mutagenesis kit (Stratagene). This construct expresses C-terminally FLAG-tagged TUBA1A under the control of the CMV promoter. Wild-type and mutant constructs were transfected into P19 cells (ECACC, 95102107) using Lipofectamine LTX reagent (Invitrogen). After 24 h, cells were fixed in ice-cold methanol, permeabilized with 0.1% (v/v) Triton X-100 and stained with antibodies recognizing FLAG (Sigma) and secondary anti-mouse antibodies conjugated to Alexa 488 (Molecular Probes).
Structural modeling of TUBA1A mutations
Two atomic models of tubulin complexes were used for the analysis of the mutations: (a) protofilaments of α- and β-tubulin heterodimers and (b) the kinesin KIF1A-microtubule complex. The former model was based on docking of the α- and β-tubulin heterodimer crystal structure (PDB id: 1JFF) (31) into the cryo electron microscopy (cryoEM) map of the intact microtubules at 8 Å resolution (32) (coordinates kindly provided by Ken Downing, Life Sciences Division, Berkeley Lab), while the latter was based on docking of the same structure (PDB id: 1JFF) in addition to the kinesin KIF1A motor domain crystal structure (PDB id: 1VFV) into the cryoEM map of the complex in an AMPPNP (5′-adenylyl-β,γ-imidodiphosphate) form at 10 Å resolution (PDB id: 2HXF) (33). In the original models, the interfaces between the domains suffer from minor clashes, most likely due to conformational differences between the structures of the domains in their isolated form and in the context of their assemblies. To remove these clashes, we first added hydrogens using VMD (34) and then applied conjugate gradient energy minimization to the interface between two heterodimers of α- and β-tubulin in the first model and to the interface between KIF1A and α-tubulin in the second model. The minimization was performed using NAMD (35) with the CHARMM27 forcefield (36). The final models were visualized and analysed with chimera (37); mutations were introduced based on the Dunbrack backbone-dependent rotamer library (38) and H bonds were identified with the FindHBond method (39). Cation–π interactions were identified using the program CaPTURE (40).
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported in part by a grant from the NIH to W.B.D. (1P01-NS039404 and 1R01-NS050375), the Wales Office of Research and Development (M.I.R. and T.D.C.) and the Medical Research Council (K.H., R.J.H. and M.T.). We thank Autism Speaks for granting a Postdoctoral Fellowship Award to R.A.K. Funding to pay the Open Access Charge was provided by the National Institutes for Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the parents of the many children with LIS who have allowed us to include them in our studies over the past 25 years.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dobyns W.B., Truwit C.L., Ross M.E., Matsumoto N., Pilz D.T., Ledbetter D.H., Gleeson J.G., Walsh C.A., Barkovich A.J. Differences in the gyral pattern distinguish chromosome 17-linked and X-linked lissencephaly. Neurology. 1999;53:270–277. doi: 10.1212/wnl.53.2.270. [DOI] [PubMed] [Google Scholar]
- 2.Pilz D.T., Macha M.E., Precht K.S., Smith A.C., Dobyns W.B., Ledbetter D.H. Fluorescence in situ hybridization analysis with LIS1 specific probes reveals a high deletion mutation rate in isolated lissencephaly sequence. Genet. Med. 1998;1:29–33. doi: 10.1097/00125817-199811000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Pilz D.T., Matsumoto N., Minnerath S., Mills P., Gleeson J.G., Allen K.M., Walsh C.A., Barkovich A.J., Dobyns W.B., Ledbetter D.H., et al. LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum. Mol. Genet. 1998;7:2029–2037. doi: 10.1093/hmg/7.13.2029. doi:10.1093/hmg/7.13.2029. [DOI] [PubMed] [Google Scholar]
- 4.Forman M.S., Squier W., Dobyns W.B., Golden J.A. Genotypically defined lissencephalies show distinct pathologies. J. Neuropathol. Exp. Neurol. 2005;64:847–857. doi: 10.1097/01.jnen.0000182978.56612.41. doi:10.1097/01.jnen.0000182978.56612.41. [DOI] [PubMed] [Google Scholar]
- 5.Ross M.E., Swanson K., Dobyns W.B. Lissencephaly with cerebellar hypoplasia (LCH): a heterogeneous group of cortical malformations. Neuropediatrics. 2001;32:256–263. doi: 10.1055/s-2001-19120. doi:10.1055/s-2001-19120. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso C., Leventer R.J., Matsumoto N., Kuc J.A., Ramocki M.B., Mewborn S.K., Dudlicek L.L., May L.F., Mills P.L., Das S., et al. The location and type of mutation predict malformation severity in isolated lissencephaly caused by abnormalities within the LIS1 gene. Hum. Mol. Genet. 2000;9:3019–3028. doi: 10.1093/hmg/9.20.3019. doi:10.1093/hmg/9.20.3019. [DOI] [PubMed] [Google Scholar]
- 7.Haverfield E.V., Whited A.J., Petras K.S., Dobyns W.B., Das S. Intragenic deletions and duplications of the LIS1 and DCX genes: a major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur. J. Hum. Genet. 2008;7:911–918. doi: 10.1038/ejhg.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso C., Leventer R.J., Ward H.L., Toyo-Oka K., Chung J., Gross A., Martin C.L., Allanson J., Pilz D.T., Olney A.H., et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller–Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 2003;72:918–930. doi: 10.1086/374320. doi:10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori M., Adachi H., Tsujimoto M., Arai H., Inoue K. Miller–Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected] Nature. 1994;370:216–218. doi: 10.1038/370216a0. doi:10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 10.Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H. Isolation of a Miller–Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. doi:10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 11.Bahi-Buisson N., Poirier K., Boddaert N., Saillour Y., Castelnau L., Philip N., Buyse G., Villard L., Joriot S., Marret S., et al. Refinement of cortical dysgeneses spectrum associated with TUBA1A mutations. J. Med. Genet. 2008;45:647–653. doi: 10.1136/jmg.2008.058073. doi:10.1136/jmg.2008.058073. [DOI] [PubMed] [Google Scholar]
- 12.Keays D.A., Tian G., Poirier K., Huang G.J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnar Z., et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. doi:10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris-Rosendahl D.J., Najm J., Lachmeijer A.M., Sztriha L., Martins M., Kuechler A., Haug V., Zeschnigk C., Martin P., Santos M., et al. Refining the phenotype of alpha-1a Tubulin (TUBA1A) mutation in patients with classical lissencephaly. Clin. Genet. 2008;74:425–433. doi: 10.1111/j.1399-0004.2008.01093.x. doi:10.1111/j.1399-0004.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- 14.Poirier K., Keays D.A., Francis F., Saillour Y., Bahi N., Manouvrier S., Fallet-Bianco C., Pasquier L., Toutain A., Tuy F.P., et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum. Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. doi:10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- 15.Coksaygan T., Magnus T., Cai J., Mughal M., Lepore A., Xue H., Fischer I., Rao M.S. Neurogenesis in Talpha-1 tubulin transgenic mice during development and after injury. Exp. Neurol. 2006;197:475–485. doi: 10.1016/j.expneurol.2005.10.030. doi:10.1016/j.expneurol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Tian G., Kong X.P., Jaglin X.H., Chelly J., Keays D., Cowan N.J. A pachygyria-causing alpha-tubulin mutation results in inefficient cycling with CCT and a deficient interaction with TBCB. Mol. Biol. Cell. 2008;19:1152–1161. doi: 10.1091/mbc.E07-09-0861. doi:10.1091/mbc.E07-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogales E., Wolf S.G., Downing K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. doi:10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 18.Kikkawa M., Okada Y., Hirokawa N. 15 Å resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. doi:10.1016/S0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 19.Moores C.A., Perderiset M., Francis F., Chelly J., Houdusse A., Milligan R.A. Mechanism of microtubule stabilization by doublecortin. Mol. Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. doi:10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Al-Bassam J., Ozer R.S., Safer D., Halpain S., Milligan R.A. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 2002;157:1187–1196. doi: 10.1083/jcb.200201048. doi:10.1083/jcb.200201048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno N., Toba S., Edamatsu M., Watai-Nishii J., Hirokawa N., Toyoshima Y.Y., Kikkawa M. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 2004;23:2459–2467. doi: 10.1038/sj.emboj.7600240. doi:10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boycott K.M., Flavelle S., Bureau A., Glass H.C., Fujiwara T.M., Wirrell E., Davey K., Chudley A.E., Scott J.N., McLeod D.R., et al. Homozygous deletion of the very low density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am. J. Hum. Genet. 2005;77:477–483. doi: 10.1086/444400. doi:10.1086/444400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S.E., Shugart Y.Y., Huang D.T., Shahwan S.A., Grant P.E., Hourihane J.O., Martin N.D., Walsh C.A. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat. Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 24.Zaki M., Shehab M., El-Aleem A.A., Abdel-Salam G., Koeller H.B., Ilkin Y., Ross M.E., Dobyns W.B., Gleeson J.G. Identification of a novel recessive RELN mutation using a homozygous balanced reciprocal translocation. Am. J. Med. Genet. A. 2007;143A:939–944. doi: 10.1002/ajmg.a.31667. [DOI] [PubMed] [Google Scholar]
- 25.Fallet-Bianco C., Loeuillet L., Poirier K., Loget P., Chapon F., Pasquier L., Saillour Y., Beldjord C., Chelly J., Francis F. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain. 2008;131:2304–2320. doi: 10.1093/brain/awn155. doi:10.1093/brain/awn155. [DOI] [PubMed] [Google Scholar]
- 26.Yamada M., Toba S., Yoshida Y., Haratani K., Mori D., Yano Y., Mimori-Kiyosue Y., Nakamura T., Itoh K., Fushiki S., et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. doi:10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaglin X.H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X.P., Bomont P., et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdollahi M.R., Morrison E., Sirey T., Molnar Z., Hayward B.E., Carr I.M., Springell K., Woods C.G., Ahmed M., Hattingh L., et al. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am. J. Hum. Genet. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. doi:10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobyns W.B., Stratton R.F., Greenberg F. Syndromes with lissencephaly. I: Miller–Dieker and Norman–Roberts syndromes and isolated lissencephaly. Am. J. Med. Genet. 1984;18:509–526. doi: 10.1002/ajmg.1320180320. doi:10.1002/ajmg.1320180320. [DOI] [PubMed] [Google Scholar]
- 30.Barkovich A.J., Jackson D.E., Jr, Boyer R.S. Band heterotopias: a newly recognized neuronal migration anomaly. Radiology. 1989;171:455–458. doi: 10.1148/radiology.171.2.2468173. [DOI] [PubMed] [Google Scholar]
- 31.Lowe J., Li H., Downing K.H., Nogales E. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. doi:10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 32.Li H., DeRosier D.J., Nicholson W.V., Nogales E., Downing K.H. Microtubule structure at 8 Å resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. doi:10.1016/S0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 33.Kikkawa M., Hirokawa N. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 2006;25:4187–4194. doi: 10.1038/sj.emboj.7601299. doi:10.1038/sj.emboj.7601299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. doi:10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. doi:10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell A.D., Jr, Bashford D., Bellott M., Dunbrack J.R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. doi:10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Dunbrack R.L., Jr Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. doi:10.1016/S0959-440X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 39.Mills J.E., Dean P.M. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput. Aided Mol. Des. 1996;10:607–622. doi: 10.1007/BF00134183. doi:10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 40.Gallivan J.P., Dougherty D.A. Cation–pi interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. doi:10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.