Abstract
Lafora disease (LD) is an autosomal recessive, progressive myoclonus epilepsy, which is characterized by the accumulation of polyglucosan inclusion bodies, called Lafora bodies, in the cytoplasm of cells in the central nervous system and in many other organs. However, it is unclear at the moment whether Lafora bodies are the cause of the disease, or whether they are secondary consequences of a primary metabolic alteration. Here we describe that the major genetic lesion that causes LD, loss-of-function of the protein laforin, impairs autophagy. This phenomenon is confirmed in cell lines from human patients, mouse embryonic fibroblasts from laforin knockout mice and in tissues from such mice. Conversely, laforin expression stimulates autophagy. Laforin regulates autophagy via the mammalian target of rapamycin kinase-dependent pathway. The changes in autophagy mediated by laforin regulate the accumulation of diverse autophagy substrates and would be predicted to impact on the Lafora body accumulation and the cell stress seen in this disease that may eventually contribute to cell death.
INTRODUCTION
Lafora disease (LD) is an autosomal recessive, progressive myoclonus epilepsy that manifests during adolescence with generalized tonic–clonic seizures, myoclonus, absences, drop attacks and visual hallucinations. The disease results in progressive neurodegeneration, and death follows about 10 years after onset (1,2). The pathological hallmark of LD is the accumulation of polyglucosan inclusion bodies, called Lafora bodies, in the cytoplasm of cells in many organs. Lafora bodies contain around 90% of a poorly branched form of glycogen resembling amylopectin and 6% protein (3) and are also decorated by anti-ubiquitin antibodies (4,5). The extent of Lafora body deposition correlates with neuronal cell death and seizure frequency. Thus, it has been suggested that Lafora bodies may cause the pathology (6), but this has not yet been firmly established.
The great majority (90%) of mutations causing LD have been identified in two genes: EPM2A, which encodes laforin, a member of the dual-specificity protein phosphatase family, and EPM2B, which codes for malin, a protein with an NH2-terminal RING finger domain characteristic of an important group of E3-ubiquitin ligases (7–9). There is also evidence for a third minor locus (10). Patients carrying homozygous loss-of-function mutations in laforin or malin are indistinguishable suggesting that both proteins work together in the same physiological pathway. However, it is unclear how laforin and malin loss-of-function are related with the disease. Laforin interacts physically with malin and the complex causes specific ubiquitination and proteasome-dependent degradation of proteins involved in the regulation of glycogen biosynthesis (11–15). In addition, laforin acts as a glycogen phosphatase, and it appears that it can suppress excessive glycogen phosphorylation, thereby preventing the formation of the phosphorylated, poorly branched and aggregate-prone glycogen polymers that comprise Lafora bodies (16–19). While LD could be an error of carbohydrate metabolism, it has also been proposed to be a disorder of protein clearance (20).
Here we have considered the possibility that a deficiency in autophagy may be a feature of LD, and that it may contribute to the accumulation of Lafora bodies. Macroautophagy (which we will henceforth call autophagy) is a process that cells use to degrade intracytoplasmic proteins, protein complexes/oligomers, organelles and certain intracellular pathogens (21–24). It is initiated when cells form a double-layered vesicle, an autophagosome, around a portion of cytoplasm. Autophagosomes are trafficked to lysosomes where they ultimately fuse, allowing their contents to be degraded by the lysosomal hydrolases. We tested if autophagy was affected by the laforin mutation for three main reasons. First, autophagy is a crucial route for the removal of disease-associated, intracytoplasmic, aggregate-prone proteins, since it can engulf oligomeric and larger structures that cannot enter the narrow channel of the proteasome barrel (21–24). When autophagy is inhibited, then the clearance of such proteins is retarded, leading to increased aggregate formation. Second, when autophagy is inhibited in otherwise normal mice, then proteins accumulate in inclusions that are decorated by anti-ubiquitin antibodies (25). Third, autophagy is an important route for intracellular glycogen degradation (26,27). Our data in patient cells, laforin knockout mice and in cell culture systems show that laforin is a positive regulator of autophagy, via mammalian target of rapamycin kinase (mTOR), and therefore suggest that autophagy compromise may contribute to the pathology seen in LD.
RESULTS
Lack of laforin inhibits autophagosome formation
To assess macroautophagic activity, we first used the classical autophagosome marker LC3. During the formation of autophagic vacuoles, the cytoplasmic LC3-I isoform is converted by lipidation into LC3-II that has increased mobility in SDS–PAGE. LC3-II is the only known protein that associates specifically with autophagosomes and not with other organelles and is degraded in an autophagy-dependent fashion in lysosomes. Therefore, the levels of LC3-II (relative to actin or tubulin) correlate with the numbers of autophagosomes in the cells. The number of autophagosomes can be altered due to either changes in synthesis or in degradation. If one clamps LC3-II/autophagosome degradation by treating cells with lysosomal inhibitors (28), then changes in the levels of LC3-II will reflect alterations in the rate of LC3-II/autophagosome formation.
To further assess autophagic responses in our experiments, we have cultured cells in serum- and amino acid-free medium (Krebs–Henseleit medium) to stimulate autophagy via the well-described starvation response (H, for high proteolysis), or in full medium to assess basal autophagy (L, for low proteolysis). Human fibroblasts from LD patients lacking laforin (Supplementary Material, Fig. S1) had lower levels of LC3-II in the presence and absence of the lysosomal inhibitor bafilomycin A1, compared with control cells (Fig. 1A–C, and see also Supplementary Material, Fig. S2 for comparison with other control fibroblasts). Likewise, mouse embryonic fibroblasts (MEFs) from laforin null mice (Epm2a−/−, Supplementary Material, Fig. S1) had lower levels of LC3-II in the presence and absence of the lysosomal inhibitor bafilomycin A1, compared with control cells, in both full medium and starvation conditions (Fig. 1D–F). Thus, a lack of laforin impairs autophagosome synthesis both in human patient fibroblasts and in MEFs. This is consistent with a reduction in the numbers of autophagic vesicles in the LD patient cell lines (Laf-), as assessed by EGFP-LC3 vesicle numbers (Supplementary Material, Fig. S3). Note that LC3-I levels vary between different cell types, as does LC3-II induction in response to starvation. There may be some clonal variability in different cell lines, and that is why we have tested different control and patient human cells lines as well as MEFs. In all cases, we observed a reduction in autophagy in the laforin-defective cells.
Figure 1.
Loss of laforin decreases formation of autophagosomes. (A, B, D, E) Representative immunoblots, using antibodies which recognize LC3 or, as a loading control, actin, with extracts (75 µg protein) of human fibroblasts from a control individual (CTR-1) and from two different LD patients (Laf-1 and Laf-2) (A and B), or of MEFs from control (Epm2a+/+) and laforin-deficient (Epm2a−/−) mice (D and E), incubated for 2 h without (A and D) and with (B and E) bafilomycin A1 (400 nM) under conditions of high (H, Krebs-Henseleit medium) and low (L, full medium) proteolysis. The positions of LC3-I and LC3-II bands are indicated on the left. (C and F) The LC3-II bands from three independent experiments similar to those shown in (B) and (E), respectively, were quantified by densitometry and normalized to the corresponding actin bands. Data are expressed in percent relative to control cells.
Long-lived protein degradation is impaired in LD cells
Under basal conditions, autophagy only degrades a very small proportion of long-lived proteins. However, when cells are starved and autophagy is induced, then this process contributes to a substantial proportion of long-lived protein degradation. Consistent with our LC3 data above, long-lived protein degradation was impaired in LD cell lines, compared with normal fibroblasts, and also in MEFs from laforin null mice, compared with control wild-type MEFs (Fig. 2A and B). This was associated with a defect in autophagic protein degradation, which one can assess by measuring the amount of long-lived protein degradation that is sensitive to the autophagy inhibitor 3-methyladenine. When autophagy is impaired, there is an increase in ubiquitinated proteins (25). Conversely, the levels of ubiquitinated proteins are decreased in normal cells under starvation/high autophagy conditions. Levels of ubiquitinated proteins were elevated in LD fibroblasts, compared with control cells (Fig. 2C and D and Supplementary Material, Fig. S4A and B), and in laforin null MEFs, compared with control MEFs (Supplementary Material, Fig. S5), in both starvation and full media. These differences in the levels of ubiquitinated proteins were still present in cells treated with the proteasome inhibitor MG132 (Fig. 2D and Supplementary Material, Figs S4B and S5B and D), compatible with the observed differences among laforin and wild-type cells being proteasome-independent and likely due to autophagy.
Figure 2.
Loss of laforin decreases the degradation of long-lived proteins by macroautophagy and increases accumulation of polyubiquitinated proteins. (A and B) Intracellular protein degradation (total) and the amount of protein degraded by macroautophagy (MA) was calculated as described in Materials and Methods in control (CTR, black histograms) and laforin-deficient (Laf-, white histograms) human fibroblasts (A) and in MEFs from control (Epm2a+/+, black histograms) and laforin-deficient (Epm2a−/−, white histograms) mice (B). Each value represents the mean from three different experiments with duplicated samples, using in each case two different control cell lines and two different laforin-deficient cells. (C and D) Extracts (75 µg protein) of fibroblasts from a representative control individual (CTR-1) and from two different LD patients (Laf-1 and Laf-2), incubated for 2 h, without (C) or with (D) the proteasomal inhibitor MG-132 (20 µM) under conditions which produce high (H, Krebs–Henseleit medium) and low (L, full medium) proteolysis, were analysed on western blots using antibodies that recognize ubiquitinated proteins and actin, which serves as a loading control. The positions of molecular-mass markers and their size in kDa are indicated on the left. Upper gels: low exposure (exp.), lower gels: high exposure.
Autophagy is impaired in laforin knock-out mice
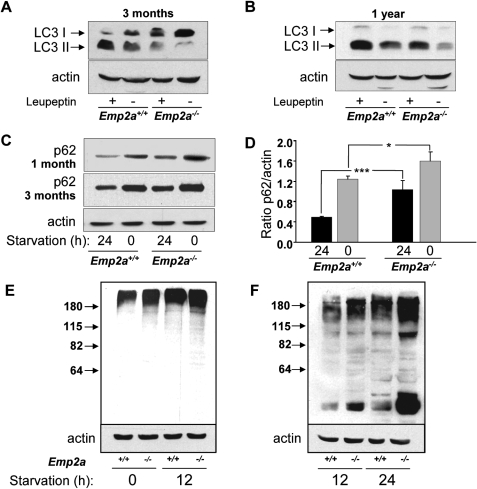
In order to test whether laforin regulates autophagy in vivo, we have studied laforin knockout mice (Epm2a−/−, Supplementary Material, Fig. S1). In these mice, we can assay autophagosome number in the liver and relate this to synthesis by measuring LC3-II in the presence or absence of the lysosomal inhibitor leupeptin. Leupeptin is a cysteine protease inhibitor which is probably incorporated into the cells by endocytosis and inhibits important lysosomal cathepsins. Therefore, it can be used as an equivalent to bafilomycin A1 in this context, but we have used leupeptin for the in vivo studies as it may have less organismal toxicity. Consistent with our data in the patient cell lines, liver extracts from laforin knockout mice have decreased numbers of autophagosomes that can be attributed to impaired autophagosome synthesis. This is apparent both in mice of 3 and 12 months of age (Fig. 3A and B).
Figure 3.
Loss of laforin slows macroautophagy in vivo. (A and B) Representative immunoblots using anti-LC3 or anti-actin of mouse liver lysates (100 µg protein) from control (Epm2a+/+) and laforin-deficient (Epm2a−/−) mice, 3 months (A) and 1 year (B) old, starved for 24 h and injected (+) or not (−) with 2 mg/100 g weight leupeptin. The positions of LC3-I and LC3-II bands are indicated. (C) Mouse liver lysates (50 µg protein) of 1-month and 3-month-old mice, starved for 24 h (high proteolysis) or fed (0, low proteolysis), were immunoblotted with p62 and actin antibodies. (D) The bands from four independent experiments (two from each 1-month and 3-month-old mice) similar to those shown in (C) were assessed by densitometry and normalized to the corresponding actin bands. (E and F) Control (Epm2a+/+) and laforin-deficient (Epm2a−/−) mice, 1 (E) and 3 (F) months old, were fed ad libitum (0) or fasted (starvation) for 12 or 24 h. Mouse liver lysates (100 µg protein) were analysed on western blots using antibodies that recognize ubiquitinated proteins and actin, which serves as a loading control. The positions of molecular-mass markers and their size in kDa are indicated on the left.
To confirm the observed autophagy impairment, we also assayed the levels of the endogenous autophagy substrate p62, which was increased in laforin-null mice (Fig. 3C and D). Likewise, the levels of ubiquitinated proteins were increased in both fed and starved laforin null mice compared with wild-type controls (Fig. 3E and F and Supplementary Material, Fig. S4C and D). While small differences were seen at 1 month of age and in 12 h starved mice at 3 months of age, these differences became very clear in 3-month-old mice starved for 24 h.
The mTOR pathway is upregulated by laforin deficiency
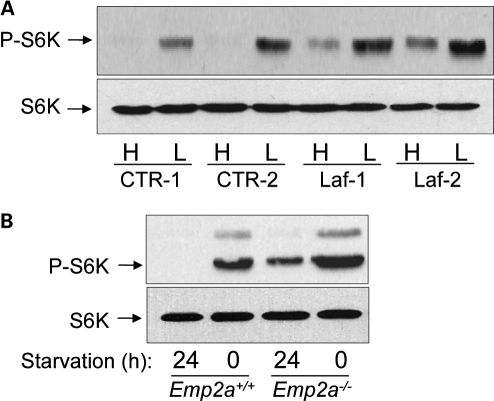
The mTOR is a major negative regulator of autophagy (29). mTOR can be inhibited, leading to autophagy induction, by starvation (30). Indeed, starvation of both control fibroblasts and wild-type mice leads to decreased phosphorylation (at the mTOR site) of the mTOR substrate p70S6 kinase (a conventional readout for mTOR kinase activity) (Fig. 4). Laforin deficiency in human fibroblasts (Fig. 4A) and in vivo (Fig. 4B) correlated with a marker of increased mTOR activity. This suggested that the autophagy defect in laforin null cells may be mTOR-dependent.
Figure 4.
mTOR signalling pathway is upregulated in laforin-deficient cells. (A) Human fibroblasts from two control individuals (CTR-1 and CTR-2) and from two different LD patients (Laf-1 and Laf-2) were incubated for 2 h under conditions of high (H, Krebs–Henseleit medium) and low (L, full medium) proteolysis in the cells. Then, extracts (75 µg) were prepared to determine the activation state of the down-stream target of mTOR p70S6 kinase (S6K) by immunoblot analysis, using antibodies which recognize phosphorylated Thr389 in p70S6 kinase (P-S6K) and total S6K. (B) mTOR activation was also analysed by the same procedures in liver extracts from 3-month-old control (Epm2a+/+) and laforin-deficient (Epm2a−/−) mice, starved for 24 h (high proteolysis conditions) or fed (0, low proteolysis conditions).
Laforin overexpression induces autophagy
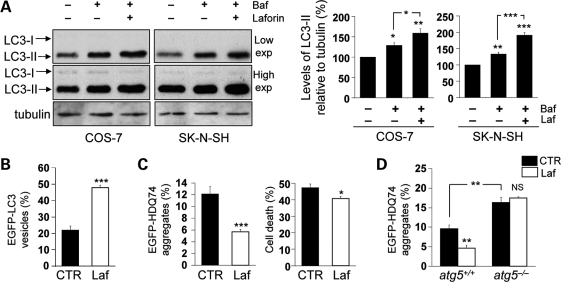
We also assessed whether laforin overexpression could induce autophagosome formation. Consistent with the laforin null data above, overexpression of laforin increased the levels of LC3-II in the presence of bafilomycin A1, suggesting an increase in autophagosome synthesis in both COS-7 and SK-N-SH (neuroblastoma) cell lines (Fig. 5A). Furthermore, laforin overexpression increased EGFP-LC3 vesicle numbers (Fig. 5B).
Figure 5.
Wild-type laforin induces autophagy and facilitates the clearance of autophagy substrates. (A) COS-7 or SK-N-SH cells, transfected with 2 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were treated with or without 400 nM bafilomycin A1 in the last 4 h of the 24 h post-transfection period. Overexpression of wild-type laforin increased autophagosome synthesis, as analysed by immunoblotting with anti-LC3 antibody (upper gels: low exposure (exp.), lower gels: high exposure) and densitometric analysis of LC3-II levels relative to tubulin. (B) COS-7 cells, transfected with 0.5 µg EGFP-LC3 and either 1.5 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were fixed and analysed for EGFP-LC3 vesicles at 24 h post-transfection. Overexpression of wild-type laforin increased the proportion of transfected (EGFP-positive) cells with EGFP-LC3 vesicles. (C) COS-7 cells, transfected with 0.5 µg EGFP-HDQ74 and either 1.5 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were fixed and analysed for EGFP-HDQ74 aggregates and cell death at 48 h post-transfection. Overexpression of wild-type laforin reduced the percentages of tranfected (EGFP-positive) cells with mutant huntingtin aggregates and cell death, assessed by apoptotic nuclear morphology (see Materials and Methods). (D) atg5+/+ and atg5−/− MEFs, transfected with 0.5 µg EGFP-HDQ74 and either 1.5 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were fixed and analysed for EGFP-HDQ74 aggregates at 48 h post-transfection. Overexpression of wild-type laforin reduced mutant huntingtin aggregates in atg5+/+ MEFs, but not in atg5−/− MEFs. atg5−/− MEFs had increased aggregates compared with atg5+/+ MEFs.
One way of assessing autophagic substrate clearance is to examine if a perturbation changes the percentage of cells with aggregates after expression of exogenous EGFP-HDQ74, an enhanced green fluorescent protein-tagged version of mutant huntingtin exon 1 with 74 polyglutamine (polyQ) repeats. Mutant huntingtin exon 1, which is associated with Huntington's disease, is an excellent autophagy substrate, and its levels and the percentage of cells with aggregates increases when autophagy is impaired. Likewise, the percentage of cells with aggregates is reduced when autophagy is induced (31). Laforin overexpression reduced the percentage of cells with mutant huntingtin aggregates, which was also associated with lower cell death (Fig. 5C). This was autophagy-dependent, as laforin only had these effects in autophagy-competent MEFs (atg5+/+), but not in Atg5 null, autophagy-incompetent cells (atg5−/−) (Fig. 5D).
Laforin induces autophagy via the mTOR pathway
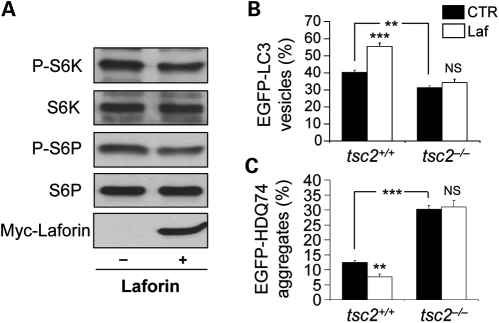
Consistent with the laforin knockout data above, laforin overexpression decreased mTOR activity, as assessed by phosphorylation of p70S6K (and the levels of phosphorylation of its substrate S6P) (Fig. 6A). To test whether laforin-mediated autophagy was mTOR-dependent, we tested its effects in cells where mTOR was constitutively activated due to loss of its upstream inhibitor TSC2 (32). While laforin overexpression together with mutant huntingtin exon 1 increased autophagosome numbers (Fig. 6B) and decreased the percentages of cells with polyQ aggregates (Fig. 6C) in wild-type MEFs (tsc2+/+), the mitigating effects of laforin overexpression were not seen in TSC2 null MEFs (tsc2−/−), which, as expected, had fewer autophagosomes and more aggregates due to impaired autophagy. These data suggest that laforin modulates autophagy either at the level of TSC2 or above this protein.
Figure 6.
Wild-type laforin reduces mTOR activity to regulate autophagy. (A) COS-7 cells, transfected with 2 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were analysed for mTOR activity at 24 h post-transfection by immunoblotting with anti-phospho-S6 kinase (P-S6K, Thr389) and anti-phospho-S6 ribosomal protein (P-S6P, Ser235/236) antibodies. Overexpression of wild-type laforin (detected with anti-myc antibody) reduced phosphorylation of S6K and S6P relative to the total proteins. (B) tsc2+/+ and tsc2−/− MEFs, transfected with 0.5 µg EGFP-LC3 and either 1.5 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were fixed and analysed for EGFP-LC3 vesicles at 24 h post-transfection. Overexpression of wild-type laforin increased the proportion of cells with EGFP-LC3 vesicles in tsc2+/+ MEFs, but not in tsc2−/− MEFs. tsc2−/− MEFs had a lower proportion of cells with EGFP-LC3 vesicles compared with tsc2+/+ MEFs. (C) tsc2+/+ and tsc2−/− MEFs, transfected with 0.5 µg EGFP-HDQ74 and either 1.5 µg pcDNA3.1 (empty vector) or Myc-Laforin for 4 h, were fixed and analysed for EGFP-HDQ74 aggregates at 48 h post-transfection. Overexpression of wild-type laforin reduced mutant huntingtin aggregates in tsc2+/+ MEFs, but not in tsc2−/− MEFs. tsc2−/− MEFs had increased aggregates compared with tsc2+/+ MEFs.
DISCUSSION
Our data show that a lack of laforin decreases autophagy. This was assessed using a range of assays. In patient cells, MEFs and mice, we observed decreased autophagosome formation when laforin activity was lost, as assessed by LC3-II blotting. This was associated with decreased clearance of long-lived proteins via autophagy and the accumulation of ubiquitinated proteins and the endogenous autophagy substrate, p62. The impaired autophagy associated with the lack of laforin correlated with increased activity of the autophagy inhibitor mTOR. In agreement with these observations, it has been recently shown that laforin-deficient mice have higher levels of the autophagy substrate tau (33) in their brains (34). Consistent with these data, we found that laforin overexpression induced autophagy and enhanced autophagy substrate clearance only in autophagy-competent cells. This was assessed by measuring the formation of autophagosomes by LC3-II western blotting, by immunofluorescence quantification of EGFP-LC3 vesicles (autophagosomes) and by assessing the accumulation of mutant huntingtin exon 1, an autophagy substrate, in wild-type and autophagy-incompetent (atg5−/−) cells. Thus, the ability of laforin to stimulate autophagy is not saturated in cells.
The accumulation of ubiquitinated proteins is a well-described phenomenon that occurs in autophagy-compromised tissues (25). This may occur if some proteins are specifically targeted to autophagy via ubiquitination and subsequent binding to adaptor proteins that link to the autophagic machinery (35,36). However, another reason for the accumulation of ubiquitinated proteins may be because autophagy inhibition results in a secondary reduction in flux through the ubiquitin-proteasome system. This phenomenon is driven by the accumulation of p62, which appears to impair the delivery of ubiquitinated proteins to the proteasome (37). Thus, it is possible that the reduced rates of autophagy in LD may have effects on the other major intracellular proteolytic system, the ubiquitin proteasome pathway.
The effects of laforin on autophagy are mTOR-dependent, as no effects are seen in tsc2−/− cells, and it is well known that TSC2 is a negative regulator of mTOR. Some studies have suggested that laforin dephosphorylates GSK3-β at Ser 9, thereby activating this enzyme (38). Lafora bodies are poorly branched polyglucosans, which could arise from an excess of glycogen synthase activity relative to glycogen branching enzyme activity. Glycogen synthase kinase 3 (GSK3) is the principal inhibitor of glycogen synthase. Thus, if laforin were a key GSK3 phosphatase, then one would expect that the loss of activity of this phosphatase would decrease GSK3 activity, increase glycogen synthase activity and thereby increase the formation of Lafora bodies. Unfortunately, this model is not supported by all studies, including analyses of Laforin null mice (34).
The exact mechanism for the laforin effect on autophagy is still elusive and may remain so for some time pending identification of authentic substrates. It is quite possible that laforin may have many substrates (such as other phosphatases), particularly since it is a dual specificity phosphatase that can hydrolyze phosphotyrosine as well as phosphoserine/threonine substrates in vitro (reviewed in 16). Thus the identification of the specific substrates relevant to autophagy will be the challenge. However, the consequences of its lack of activity in LD patients are easier to predict. Previous analyses of mice that lack laforin revealed progressive changes in the properties and structure of glycogen that paralleled the formation of Lafora bodies. One of the features observed was a progressive accumulation of glycogen, which also became more phosphorylated and insoluble (18). Since Lafora bodies are composed of poorly branched, water-insoluble, glycogen-like polymers that are also decorated with antiubiquitin antibodies, we believe that it is likely that decreased autophagy caused by laforin deficiency will enhance their accumulation, given that autophagy compromise leads to the accumulation of glycogen, ubiquitinated proteins and aggregate-prone proteins (27,39) (Fig. 7). Indeed, it appears that glycogen autophagy is regulated via the mTOR signalling pathway, which is perturbed by the loss of laforin activity. In addition to enhancing formation of Lafora bodies, which may be a toxic entity in LD, autophagy deficiency also increases cellular susceptibility to various toxic insults, which may further exacerbate the problem (40).
Figure 7.
Schematic representation of regulation of autophagy by laforin and the disease consequences. Wild-type laforin induces autophagy by inhibiting mTOR in a TSC2-dependent manner. Loss of laforin in LD activates mTOR and inhibits autophagy. An impairment of autophagy in LD may contribute to the disease pathogenesis (indicated in dashed line).
The situation we have described has similarities and differences when compared with lysosomal storage diseases associated with hydrolase deficiencies. Both sets of conditions exhibit impaired autophagic flux and the accumulation of various substrates in the affected tissues. However, there are important differences. In lysosomal storage diseases, autophagy is impaired not by reduced autophagosome synthesis, as occurs in laforin deficiency, but due to a partial block in the delivery of autophagosomes to lysosomes (22). Indeed, lysosomal storage diseases are frequently characterized by the accumulation of autophagosomes (22). Also, in lysosomal storage diseases, it is likely that there is an initial loss of lysosomal enzyme function that then leads to impaired autophagic flux, while in laforin deficiency the impaired autophagic flux may be a direct consequence of enzyme activity, since we saw increased autophagy with laforin overexpression. Currently, we do not know how early the autophagy defect is in the pathogenic cascade of LD. While it is likely that the Lafora bodies contribute to pathology, our data do raise the possibility that autophagy compromise may be an important driver of pathology in its own right (independent of the Lafora bodies), but also that the formation of these inclusions may be enhanced as a consequence of impaired clearance.
MATERIALS AND METHODS
Plasmids
EGFP-HDQ74 construct was described previously (41). Constructs received as kind gifts were EGFP-LC3 (42) and Myc-Laforin (43).
Mammalian cell culture and transfection
Human fibroblasts were obtained from two patients with two different mutations (Y86X (Laf-1) and R241X (Laf-2)) in the laforin gene and with the clinical features of LD. Control fibroblasts were obtained from healthy subjects matched by sex and age. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere in MEM supplemented with 15% foetal bovine serum, 1% MEM amino acids, 0.5% MEM non-essential amino acids, 1% glutamine, 1% vitamins, 100 units/ml penicillin and 100 µg/ml streptomycin (all from Invitrogen Life Technologies). For starvation conditions (high proteolysis medium, H), cells were switched, after washing, from full growth medium (low proteolysis medium, L) to Krebs–Henseleit medium (118.4 mm NaCl, 4.75 mm KCl, 1.19 mm KH2PO4, 2.54 mm MgSO4, 2.44 mm CaCl2·2H2O, 28.6 mm NaHCO3, 10 mm glucose) containing 10 mm Hepes, pH 7.4 and incubated for 2 h at 37°C.
COS-7 cells, SK-N-SH cells, atg5+/+ and atg5−/− MEFs (44) and tsc2+/+ and tsc2−/− MEFs (32) were maintained in DMEM supplemented with 10% foetal bovine serum, 100 units/ml penicillin/streptomycin and 2 mm l-glutamine (all from Sigma-Aldrich) in 37°C, 5% CO2 humidified incubator. Cells were transfected with DNA constructs for 4 h using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Laforin-deficient mice
Laforin-deficient Epm2a−/− mice (4) were kindly provided by Dr Antonio V. Delgado-Escueta. Male C57BL/6 wild-type and isogenic Epm2a−/− mice were maintained at the Centro de Investigaciones Biológicas (CSIC, Madrid) on a light–dark 12:12 cycle under constant temperature (23°C) with free access to food and water. In some experiments, mice were treated with leupeptin (0.2 mg/10 g body weight, intraperitoneally) 1 h before being sacrificed. MEFs were isolated from 14-day-old mouse embryos. Epm2a+/+ and Epm2a−/− MEFs were grown in DMEM supplemented with 15% inactivated foetal bovine serum, 1% MEM non-essential amino acids, 1% glutamine, 1% vitamins, 1% sodium pyruvate, 10−4 M mercaptoethanol, 100 units/ml penicillin and 100 µg/ml streptomycin. One-, 3- and 12-month-old knock-out, or control mice of the same age were sacrificed by cervical dislocation and livers were dissected for preparation of tissue extracts.
Western blot
Cell pellets were lysed on ice in Laemmli's buffer (62.5 mm Tris–HCl pH 6.8, 2% sodium dodecyl sulphate, 5% β-mercaptoethanol, 10% glycerol and 0.01% bromophenol blue) for 30 min in the presence of complete protease inhibitor cocktail (Roche Diagnostics), boiled for 5–7 min at 100°C and subjected to western blot analysis, as previously described (45,46). Mouse monoclonal antibodies which recognize human laforin have been described previously (12). Other primary antibodies include anti-LC3 (1:4,000 dilution; NB100-2220, Novus Biologicals and 1:500 dilution; clone 5F10, Nanotools), anti-p62 (1:1000 dilution; ab-56416, Abcam), anti-ubiquitinated proteins (1:5000 dilution; PW-8805 and PW-8810, BIOMOL), anti-p70S6 kinase (9202), anti-phospho-p70S6 kinase (Thr389) (9206), anti-S6 ribosomal protein (2217) and anti-phospho-S6 ribosomal protein (Ser235/236) (2211) (all 1:1000 dilutions from Cell Signalling Technology), anti-Myc (1:3000 dilution; 11667203001, Roche Diagnostics), anti-tubulin (1:4,000 dilution; T-6199) and anti-actin (1:5000 dilution; A-2066) (Sigma-Aldrich). Blots were probed with anti-mouse or anti-rabbit IgG-HRP secondary antibody (1:4000 dilutions) and visualized using ECL detection kit (GE Healthcare). Phospho-specific antibodies were always used in the first round of immunoblotting. After treating the membrane with stripping buffer (0.1 m glycine, pH 2.3), it was probed using the antibodies that recognize the total amount of the specific protein of interest.
Autophagy analysis in mammalian cell culture
Pulse-chase experiments and inhibition of autophagy
Human control and LD patient fibroblasts or Epm2a+/+ and Epm2a−/− MEFs were incubated for 48 h in fresh full medium with 2 µCi/ml [3H]valine (Hartmann Analytic Gmbh), followed by a 24 h chase in fresh full medium containing 10 mm L-valine to degrade short-lived proteins (47). Then, all cultures were incubated for 4 h under high proteolysis conditions in Krebs–Henseleit medium with 10 mm Hepes, pH 7.4, containing 10 mm valine and the indicated additions. Protein degradation, which was analysed 1 h later to ensure maximal effects of the various additions and for only 3 h to avoid possible secondary effects, was calculated at intervals of 1.5 h by measuring the net release of trichloroacetic acid-soluble radioactivity from the labelled cells into the culture medium and expressed as percentage of protein degraded in 1 h. The contribution of macroautophagy was calculated using 10 mm 3-methyladenine (Sigma-Aldrich) as previously described (47).
Assessment of autophagic flux by LC3-II levels with bafilomycin A1
Endogenous LC3-II levels, which directly correlate with autophagosome numbers (42), were detected with anti-LC3 antibody and densitometric analysis relative to actin. To assess autophagic flux, LC3-II was measured in the presence of 400 nm bafilomycin A1 (treated in the last 4 h), which clamped LC3-II/autophagosome degradation (48). This assay has been established previously with various autophagy modulators (46,49–51).
Assessment of autophagy by EGFP-LC3 vesicles
The percentage of EGFP-positive cells with >5 EGFP-LC3 vesicles were assessed with a fluorescence microscope, as previously described (50). Alternatively, the number of fluorescent dots per cell was counted.
Quantification of mutant huntingtin aggregates and cell death
The percentage of EGFP-positive cells with EGFP-HDQ74 aggregates was assessed with a Nikon Eclipse E600 fluorescence microscope (plan-apo 60X/1.4 oil immersion lens at room temperature) (Nikon, Inc.), as previously described (46,50). Cells were counted as aggregate positive if one or several aggregates of any size were visible within a cell. Cell death was assessed as we have done previously (40,50,51) by determining if there was apoptotic nuclear morphology using DAPI staining and was considered abnormal if the nucleus was fragmented or condensed to a small size resulting in a much stronger fluorescence signal compared with normal nuclei.
Statistical analyses
Densitometric analysis on the immunoblots was done by Image J software or with an Image Quant ECL (GE Healthcare), and the P-values were determined by factorial ANOVA test using STATVIEW v4.53 (Abacus Concepts), where the control condition was set to 100%. The y-axis values are shown in percentage (%) and the error bars denote standard error of mean. The P-values for assessing EGFP-HDQ74 aggregation or EGFP-LC3 vesicles were determined by unconditional logistical regression analysis, using the general log-linear analysis option of SPSS 9 software (SPSS, Chicago), as previously described (46,50,51). The convention we have used for P values in all figures is: ***P < 0.001; **P < 0.01; *P < 0.05; NS, non-significant.
SUPPLEMENTARY MATERIAL
FUNDING
Work in E.K.'s laboratory was supported by grants from the Spanish Ministry of Science and Innovation (BFU2008-00186BMC) and the Instituto de Salud Carlos III (Intramural Project, CIBERER). We are also grateful for Hughes Hall Research Fellowship (S.S. and V.I.K.), Wellcome Trust Senior Fellowship in Clinical Science (D.C.R.), Medical Research Council, EU Framework VI (EUROSCA) and the National Institute for Health Research Biomedical Research Centre at Addenbrooke's Hospital for funding. Funding to pay the Open Access Charge was provided by The Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A.V. Delgado-Escueta for Epm2a−/− mice; T. Yoshimori for EGFP-LC3 construct; P. Zheng for Myc-Laforin construct; N. Mizushima for atg5+/+ and atg5−/− MEFs; D.J. Kwiatkowski and A.R. Tee for tsc2+/+ and tsc2−/− MEFs. We also thank Nuria Mas for expert technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lafora G.R., Glueck B. Beitrag zur Histopathologie der myoklonischen Epilepsie. Z. Gesammte Neurol. Psychiatr. 1911;6:1–14. doi:10.1007/BF02863929. [Google Scholar]
- 2.Van Hoof F., Hageman-Bal M. Progressive familial myoclonic epilepsy with Lafora bodies. Electron microscopic and histochemical study of a cerebral biopsy. Acta Neuropathol. 1967;7:315–336. doi: 10.1007/BF00688087. [DOI] [PubMed] [Google Scholar]
- 3.Yokoi S., Nakayama H., Negishi T. Biochemical studies on tissues from a patient with Lafora disease. Clin. Chim. Acta. 1975;62:415–423. doi: 10.1016/0009-8981(75)90093-5. [DOI] [PubMed] [Google Scholar]
- 4.Ganesh S., Delgado-Escueta A.V., Suzuki T., Francheschetti S., Riggio C., Avanzini G., Rabinowicz A., Bohlega S., Bailey J., Alonso M.E., et al. Genotype-phenotype correlations for EPM2A mutations in Lafora's progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum. Mol. Genet. 2002;11:1263–1271. doi: 10.1093/hmg/11.11.1263. doi:10.1093/hmg/11.11.1263. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S., Satishchandra P., Gayathri N., Yasha T.C., Shankar S.K. Progressive myoclonic epilepsy: a clinical, electrophysiological and pathological study from South India. J. Neurol. Sci. 2007;252:16–23. doi: 10.1016/j.jns.2006.09.021. doi:10.1016/j.jns.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Yokoi S., Austin J., Witmer F., Sakai M. Studies in myoclonus epilepsy (Lafora body form). Isolation and preliminary characterization of Lafora bodies in two cases. Arch. Neurol. 1968;19:15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 7.Minassian B.A., Lee J.R., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 1998;20:171–174. doi: 10.1038/2470. doi:10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 8.Serratosa J.M., Gomez-Garre P., Gallardo M.E., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum. Mol. Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. doi:10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 9.Chan E.M., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. doi:10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 10.Chan E.M., Omer S., Ahmed M., Bridges L.R., Bennett C., Scherer S.W., Minassian B.A. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004;63:565–567. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- 11.Gentry M.S., Worby C.A., Dixon J.E. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl Acad. Sci. USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. doi:10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solaz-Fuster M.C., Gimeno-Alcaniz J.V., Ros S., Fernández-Sánchez M.E., García-Fojeda B., Criado-García O., Vilchez D., Domínguez J., Garcia-Rocha M., Sánchez-Piris M., et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum. Mol. Genet. 2008;17:667–678. doi: 10.1093/hmg/ddm339. doi:10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 13.Vilchez D., Ros S., Cifuentes D., Pujadas L., Valles J., García-Fojeda B., Criado-García O., Fernández-Sánchez E., Medrano-Fernández I., Domínguez J., et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat. Neurosci. 2007;10:1407–1413. doi: 10.1038/nn1998. doi:10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 14.Cheng A., Zhang M., Gentry M.S., Worby C.A., Dixon J.E., Saltiel A.R. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori's disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worby C.A., Gentry M.S., Dixon J.E. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J. Biol. Chem. 2008;283:4069–4076. doi: 10.1074/jbc.M708712200. doi:10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worby C.A., Gentry M.S., Dixon J.E. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. doi:10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tagliabracci V.S., Turnbull J., Wang W., Girard J.M., Zhao X., Skurat A.V., Delgado-Escueta A.V., Minassian B.A., Depaoli-Roach A.A., Roach P.J. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl Acad. Sci. USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. doi:10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tagliabracci V.S., Girard J.M., Segvich D., Meyer C., Turnbull J., Zhao X., Minassian B.A., Depaoli-Roach A.A., Roach P.J. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J. Biol. Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. doi:10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentry M.S., Dowen R.H., 3rd, Worby C.A., Mattoo S., Ecker J.R., Dixon J.E. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J. Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. doi:10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado-Escueta A.V. Advances in Lafora progressive myoclonus epilepsy. Curr. Neurol. Neurosci. Rep. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. doi:10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 21.Klionsky D.J., Cuervo A.M., Dunn W.A., Jr, Levine B., van der Klei I., Seglen P.O. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 22.García-Arencibia M., Hochfeld W.E., Toh P.P., Rubinsztein D.C. Semin. Cell Dev. Biol. 2010 doi: 10.1016/j.semcdb.2010.02.008. February 24, 2010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knecht E., Aguado C., Cárcel J., Esteban I., Esteve J.M., Ghislat G., Moruno J.F., Vidal J.M., Sáez R. Intracellular protein degradation in mammalian cells: recent developments. Cell. Mol. Life Sci. 2009;66:2427–2443. doi: 10.1007/s00018-009-0030-6. doi:10.1007/s00018-009-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravikumar B., Futter M., Jahreiss L., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Narayanan U., Renna M., et al. Mammalian macroautophagy at a glance. J. Cell Sci. 2009;122:1707–1711. doi: 10.1242/jcs.031773. doi:10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 26.Kotoulas O.B., Kalamidas S.A., Kondomerkos D.J. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. doi:10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Shea L., Raben N. Autophagy in skeletal muscle: implications for Pompe disease. Int. J. Clin. Pharmacol. Ther. 2009;47:S42–S47. doi: 10.5414/cpp47042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 29.Meijer A.J., Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. doi:10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Meijer A.J., Codogno P. Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. doi:10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S., Rubinsztein D.C. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. doi:10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H., Cicchetti G., Onda H., Koon H.B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C.L., Kwiatkowski D.J. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Martínez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E.M., Cuervo A.M., Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. doi:10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri R., Suzuki T., Yamakawa K., Ganesh S. Hyperphosphorylation and aggregation of Tau in laforin-deficient mice, an animal model for Lafora disease. J. Biol. Chem. 2009;284:22657–22663. doi: 10.1074/jbc.M109.009688. doi:10.1074/jbc.M109.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjørkøy G., Lamark T., Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 36.Kirin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;3:4259–4269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Korolchuk V.I., Mansilla A., Menzies F.M., Rubinsztein D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. doi:10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Wang Y., Wu C., Liu Y., Zheng P. Dimerization of laforin is required for its optimal phosphatase activity, regulation of GSK3beta phosphorylation, and Wnt signaling. J. Biol. Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. doi:10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- 39.Kondomerkos D.J., Kalamidas S.A., Kotoulas O.B., Hann A.C. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol. Histopathol. 2005;20:689–696. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 40.Ravikumar B., Berger Z., Vacher C., O'Kane C.J., Rubinsztein D.C. Rapamycin pre-treatment protects against apoptosis. Hum. Mol. Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. doi:10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 41.Narain Y., Wyttenbach A., Rankin J., Furlong R.A., Rubinsztein D.C. A molecular investigation of true dominance in Huntington's disease. J. Med. Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. doi:10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Liu Y., Wu C., Zhang H., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Liu Y., Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell. 2006;10:179–190. doi: 10.1016/j.ccr.2006.08.008. doi:10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. doi:10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 45.Esteban I., Aguado C., Sánchez M., Knecht E. Regulation of various proteolytic pathways by insulin and amino acids in human fibroblasts. FEBS Lett. 2007;581:3415–3421. doi: 10.1016/j.febslet.2007.06.043. doi:10.1016/j.febslet.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar S., Ravikumar B., Rubinsztein D.C. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. doi:10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 47.Fuertes G., Martín de Llano J.J., Villarroya A., Rivett A.J., Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. doi:10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky D.J., Elazar Z., Seglen P.O., Rubinsztein D.C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 49.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an ‘autophagomometer. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. doi:10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 50.Sarkar S., Perlstein E.O., Imarisio S., Pineau S., Cordenier A., Maglathlin R.L., Webster J.A., Lewis T.A., O'Kane C.J., Schreiber S.L., Rubinsztein D.C. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. doi:10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. doi:10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.