Abstract
A novel bioconjugate of stearic acid capped maghemite nanoparticle (γ-Fe2O3) with bovine serum albumin (BSA) was developed by taking recourse to the fatty acid binding property of the protein. From FT-IR study, it was found that conjugation took place covalently between the amine group of protein molecule and carboxyl group of stearic acid capped maghemite nanoparticle. TEM study further signified the morphology of the proposed nanobioconjuagte. The binding constant of nanoparticle with protein molecule was evaluated from the optical property studies. Also, magnetic measurement (M–H) showed retaining of magnetic property by significant values of saturation magnetization and other hysteretic parameters.
Keywords: Bioconjugate, Maghemite nanoparticle, Bovine serum albumin, Covalent interaction, Fatty acid binding domain
Introduction
Bioconjugation of magnetic nanoparticles is basically done to make it compatible for numerous biomedical applications such as MRI contrast enhancement, drug delivery, detoxification of biological fluids, immunoassay, tissue repair, hyperthermia etc. [1-6]. Besides these, bioconjugate systems are also being applied in various large-scale bioprocesses such as nucleic acid detachment, protein separation, magnetic biosensor etc. [7-9]. All these biorelated applications require the use of magnetic nanoparticles that should have size smaller than 10 nm with overall narrow particle size distribution, so that the particles have uniform and unique properties [10]. This is mainly because of the fact that particles at this size range have the advantage of showing well-established magnetic properties which reduces the possibility of particle aggregation upon magnetic attraction in a magnetic dispersion [11]. For fabricating a bioconjugate, it generally involves lots of surface chemistry work [12]. Normally, these are synthesized by modifying the nanoparticle surface with some chemical linker molecule, so that it can further interact with next incoming bio molecular entity with the help of free functional group of linker molecule. This procedure was already well established but supposed to be having some difficulty in the sense of stability of linker molecule due to various biochemical events. This might occur mostly under in vivo condition when applying the bioconjugate system in the targeted delivery of neoplastic compounds to tumor cell.
To overcome this difficulty, we proposed some biologically evolved linker moiety (fatty acid binding domain) to fabricate a novel bioconjugate in covalent fashion by utilizing the molecular recognition property of bio molecular system, such as bovine serum albumin (BSA) protein. It is supposed to be an important substitute over the synthetic linker in designing the bioconjugate covalently so that it can be applied gently under preceding condition. This can also be called as natural anchor molecule that is functional in several of its biological activities. Here, BSA is chosen as the material of interest for the bioconjugation purpose because of the fatty acid binding domain of BSA, which helps in the conjugation of stearic acid capped nanoparticle with the protein moiety [13]. Also, serum albumin is the major vehicle for transport of nonesterified fatty acids in the circulation [14]. Magnetic bioconjugate of stearic acid capped ultrafine maghemite nanoparticle with BSA molecule is quite advantageous in case of stability because of the molecular recognition ability of the BSA molecule towards fatty acid itself and its well stability under physiological condition (pH = 7.4).
Experimental
High purity (>99%) iron (III) nitrate [Fe(NO3)3 · 9H2O], stearic acid [C18H36O2], and tetrahydrofuran (THF) were used for the synthesis of maghemite nanoparticle. The albumin protein required for the bioconjugation purpose was also of high purity from Spectrochem India Pvt. Ltd.
Ultrafine maghemite nanoparticles (γ-Fe2O3) were synthesized through a gentle chemistry route [15]. High purity iron (III) nitrate [Fe(NO3)3 · 9H2O] and stearic acid [C18H36O2 in the ratio of 1:2 were used as initial ingredients. The homogeneous solution of molten mixture was then heated at 125 °C for 11/2 h to form a reddish brown viscous mass which then subsequently treated with THF. The powdery precipitates were collected through centrifugation and dried completely in an air oven at 70 °C. The dried precipitates were further subjected to heat treatment at 250 °C for holding time of 30 min inside an electrically heating furnace to get the nanoparticles. The synthesis of the conjugate of bovine serum albumin with maghemite nanoparticles (γ-Fe2O3) was carried out by transferring 3 mmol of BSA into PBS buffer. The mixture was then kept at 4 °C and allowed to stand for 12 h, so that protein sample gets completely soluble in the PBS buffer. After this, 1 mmol of maghemite nanoparticles (γ-Fe2O3) were mixed with the BSA containing buffer and the mixture was vortexed for 1 h at room temperature. The vortexed mixture is again stored at 4 °C for 2 h for the stabilization purpose. This was basically done to control the covalent interaction taking place between stearic acid coated maghemite nanoparticles with that of protein molecule. Further, the mixture was centrifuged at high speed for 15 min to get the magnetic nanobioconjugate consisting of maghemite nanoparticles (γ-Fe2O3) and BSA. The pellet portion is collected and allowed to vacuum dried and the supernatant being kept for further characterization.
The formation of maghemite–BSA nanobioconjugate had been studied with the help of FT-IR spectrometer. The spectrum was recorded in the transmission mode on a Nicollet Impact 410-spectrometer. The dried samples of maghemite, BSA as well as the magnetic nanobioconjugate were grounded with KBr and the mixture was compressed into a pellet for characterization. Transmission electron microscopy (TEM) study for the conjugate of bovine serum albumin with maghemite nanoparticle (MNP/BSA) as well as MNP itself was carried out using a JEM-100CX model operated at 100 KV. The photoluminescence spectrum for all samples was taken with Perkin Elmer LS-55 fluorescence spectrometer. The magnetic properties of the resulting bioconjugate as well as maghemite nanoparticles had been studied with vibrating sample magnetometer (VSM, Lakeshore, 7410) for confirming the retaining of magnetic properties by the nanoparticles after the formation of nanobioconjugate.
Results and Discussion
In this work, we used a versatile technique [15] for the synthesis of maghemite nanoparticle, which was quite advantageous in the sense of not forming large aggregates, occurrence of uncontrolled oxidation, and presence of matrix etc. The procedure was characterized by a complete and homogeneous mixing of initial ingredients at molecular or atomic level. This technique ensured the presence of uniform particle size distribution as evidenced from the SAXS study [15]. We adopted the synthesis procedure for preparing magnetic nanobioconjugate system by taking the principle of covalent interaction of fatty acid binding domain of BSA molecule with stearic acid. As the synthesized magnetic nanoparticle was capped with stearic acid molecule, it can effectively help in the formation of bioconjugate with protein molecule. The reaction was carried out at 4 °C by keeping the pH of the reaction medium constant at 7.4. The precaution was taken with a motivation to keep the structure of the albumin molecule in intact from. This was primarily because of the fact that, at neutral pH(=7.4), a net charge of −10, −8 & 0 for domain I, II & III for BSA had been obtained. So, to keep the various domains of the molecule stable, primarily the domain III; the reaction had been carried out at neutral pH in PBS buffer. Serum albumin also undergoes reversible conformational changes with the changes in pH. In this regard, the % of helix transformation tends to be normal (55%) at the pH range 4.3–8. The schematic diagram of the synthesis of magnetic nanobioconjugate is shown in Fig. 1. Though bioconjugation of BSA on nanosized magnetic nanoparticles (Fe3O4) as well as semiconductor nanocrystals (CdTe) had already been done by different approaches [16,17], even using oleic acid coated magnetite nanoparticles [18].In these published reports, some of the surface functionalization reactions to conjugate BSA molecules onto the support particle surface were very complicated in comparison to the process described in this paper.
Figure 1.
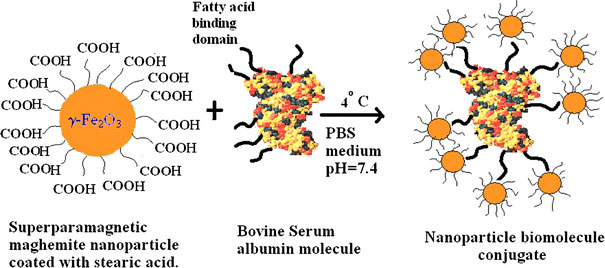
The conjugation scheme of maghemite nanoparticle with bovine serum albumin
We performed the FTIR study to primarily confirm the feasibility of the conjugation process. The FTIR spectra of maghemite/BSA nanobioconjuagte along with maghemite nanoparticle and BSA were shown in Fig. 2. The FTIR spectra exhibited strong bands in the low frequency region due to the iron oxide skeleton. In other regions, the spectra of iron oxide had weak bands. The presence of free carboxyl group on the nanoparticle surface was further confirmed from the C=O stretching band (1,704 cm−1) as well as OH stretching band (2845.40 cm−1) of the carboxyl group on the stearic acid capped magnetic nanoparticle. Due to very strong hydrophobic character of the hydrocarbon chain of stearic acid molecule, it is very difficult for it to be exposed to the aqueous solution. So, the interaction of stearic acid molecule with the iron oxide skeleton took place through the hydrocarbon segment leaving the carboxyl group towards the aqueous solution. Comparing the spectra of maghemite nanoparticles before and after conjugation with BSA, the strong absorption bands at 536.84 and 552.88 cm−1confirmed the presence of maghemite as the main phase in both samples. The characteristics band of the BSA protein at 1,650 and 1,530 cm−1are due to C=O stretching band of carboxyl functional group on the tryptophan moiety as well as carboxylate group. The conjugation of BSA to nanoparticle surface was confirmed by the appearance of the new N–H stretching band (3398.4 ~ 3,400 cm−1) as well as vanishing of the C=O stretching band (1,704 cm−1) of carboxyl functional group on stearic acid molecule. This clearly signified the formation of the covalent bond between the amino & carboxyl functional groups of the protein molecule as well as stearic acid capped maghemite nanoparticle. The new band (1655.02 cm−1) appeared after the formation of the bioconjugate was due to C=O stretching pattern of the secondary amide linkage. On the other hand, new bands appearing at around 1,100–900 cm−1were due to the C–N stretching (1080.49 cm−1) from secondary amide linkage; C–H deformation out of plane (983.83 cm−1) from stearic acid hydrocarbon chain as well as C–H deformation of aromatic hydrocarbon (864.58 cm−1) from tryptophan amino acid present over the protein molecule. The appearance of the C–N stretching band at (1080.49 cm−1) was a strong significance that the conjugation took place in covalent manner through amide linkage.
Figure 2.
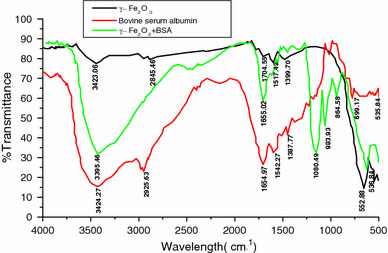
FT-IR spectra of maghemite nanoparticle before and after the conjugation with BSA
Further, to get the simplified view of the bioconjugate so prepared, we also carried out the TEM studies of the maghemite nanoparticle as well as the resulting magnetic nanobioconjugate. From the TEM micrographs, the conjugation of the nanoparticles with the protein molecule can easily be visualized as shown in Fig. 3b. It clearly illustrates the ellipsoidal pattern of BSA molecule over which the nanoparticles were conjugated and formed an assembly pattern. It is also evident that the nanoparticles are well separated from each other, i.e. aggregation has not taken place. This has occurred as a result of the perturbation of electron cloud of the bimolecular environment during the conjugation process.
Figure 3.
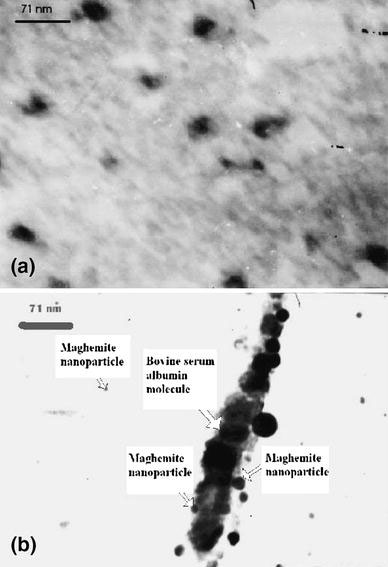
TEM micrograph of the conjugate of maghemite nanoparticle with elongated bovine serum albumin molecule
To understand the nature of interaction between magnetic nanoparticles with biomolecule, we had also studied the optical property of the resulting bioconjugate by carrying out the photoluminescence experiment. The photoluminescence pattern obtained for different samples were shown in Fig. 4. From the experimental data of the photoluminescence pattern, we calculated the binding constant of the magnetic nanoparticles with the bovine serum albumin by using the model put forward by Lehrer and Fashman [19]. According to this method, the tryptophan residue fluorescence intensity (F) scaled with the maghemite/BSA concentration through the following equation:
Figure 4.
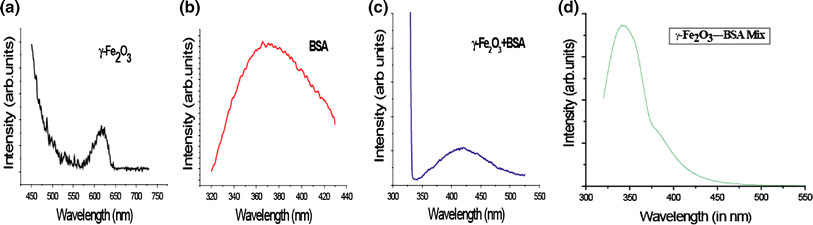
The photoluminescence spectra ofamaghemite nanoparticlebBSAcmaghemite/BSA nanobioconjugatedMaghemite/BSA mixture
where F0 and F∞ are the relative luminescence intensities of the protein alone and the protein conjugated with maghemite nanoparticles, respectively, n is the stoichiometry of the complex. The reciprocal of the Kdiss is the binding constant Kb. The intensity value (F) for the tryptophan residue was obtained from the area under the fluorescence spectra in the range of our investigation 300–500 nm on calculation, the binding constant of the nanobioconjugate has been found out to be Kb = 58.13. From the figure, it became evident that the emission wavelength of the bare maghemite nanoparticles was 617 nm. While for the standard BSA sample it was found out to be 365.5 nm. For the maghemite/BSA nanobioconjugate colloidal solution, it had been observed that there was significant shift in the emission wavelength (420.5 nm). On the other hand, in case of standard maghemite/BSA mixture the emission wavelength was found out to be 343 nm which was strictly different from that of the optical effect shown by the bioconjugate. The excitation wavelength applied in all the samples were: λEx = 320 nm for bare maghemite nanoparticle, maghemite/BSA nanobioconjuage, & maghemite/BSA mixture.On the other hand, the excitation wavelength applied in case of BSA molecule was λEx = 280 nm. The pattern obtained for bare maghemite nanoparticle was due to the intrinsic property of the electronic transition in iron oxides such as the ligand to metal charge transfer transition. This intensity was found to be low, due to the fast (picosec) overall decay as well as very efficient nonradiative relaxation raised from the dense band structure and a high density of trap states in the iron oxide skeleton as previously reported [20]. Also we had assumed that there will be substantial improvement in PL yield as no quenching phenomenon occurring in PL pattern of the bare nanoparticle [21]. On the other hand, the emission of BSA at 365.5 nm was due to luminescence originated from two aromatic tryptophan moieties present in BSA amino acid sequence: Trp 134 and Trp 214.This was further shifted to 420.5 nm after the formation of bioconjugate. This shifting might be due to the increase in the nanoparticle size during the synthesis of bioconjuagte. Also the produced BSA–NP conjugate causes effective interaction between the excited states of the biological and inorganic parts. This effect should be attributed to the spatial closeness of BSA and NP in the tightly bound covalent system [17].
For finally validating the magnetic properties of ultrafine maghemite nanoparticles before and after conjugation with bovine serum albumin molecule, we studied the same with vibrating sample magnetometer (VSM). We had mainly done it to ensure the existence of magnetic behavior of maghemite nanoparticle after the conjugation process. The magnetic behavior of the biomolecule–nanoparticles assembly depends sensitively on the morphology and the size of the nanoparticles, where the dipole coupling between the nanoparticles governs the overall magnetic behavior [22]. In conjugate of maghemite nanoparticles with BSA, we observed a formation of unusual self-alignment of nanoparticles over BSA molecules. Figure 5a, b showed the room temperature magnetization of both maghemite and maghemite/BSA nanobioconjugate. The bare maghemite nanoparticle had saturation magnetization 1.044 emu/g, whereas that of maghemite/BSA nanobioconjuagte was found to be 1.196 emu/g. Also, the retentivity (MR) and coercivity (HC) values (0.4016 emu/g as well as 25.860 Oe, respectively) of the bioconjugates were different in comparison to maghemite nanoparticles (MR = 0.2851 emu/g; HC = 23.083 Oe). The difference between these values could be attributed to the change in the microstructure of the particles due to covalent binding of BSA to the carboxyl group of stearic acid capped maghemite nanoparticle and the increase in the interparticle interactions during the bioconjugation process. This microstructural variation occurred as a result of the drastic change in particle surface effect. The particle surface effect hereby referred to the disordered alignment of surface atomic spins induced by reduced coordination and broken exchange bonds between surface spin [23]. The observed induction in saturation magnetization value could also be realized from the increase in the interparticle interactions during the bioconjugation process. It had also been found out that the obtained saturation magnetization value (1.196 emu/g) of the prepared bioconjuagte was close to the value (1.34 emu/g) obtained by Salgueirino-Maceira et al. [24] for the luminescent magnetic nanoparticle. The other hysteretic parameter, the coercivity value, also increased due to the transition of domain boundary to multidomain regime.
Figure 5.
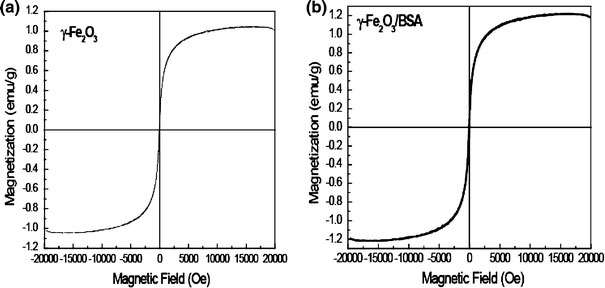
Room temperature (300 K) M–H data ofaMaghemite andbMaghemite/BSA nanobiconjugate
Conclusions
In summary, we developed a simple technique for the synthesis of bioconjugate of maghemite nanoparticles with BSA molecule by using the covalent interaction between the fatty acid binding domains of BSA molecule with stearic acid capped nanoparticles. This will lead to the development of non-toxic iron oxide nanoparticles using BSA as a biocompatible passivating agent. We confirmed the formation of the same from the FT-IR spectra as well as TEM micrograph. We also established the well retaining of magnetic property of nanoparticles after the formation of bioconjugate from M–H study. It is worth mentioning here that, this is the first report on conjugation of nanoparticles with biomolecules by utilizing biologically evolved linker moiety in covalent fashion. The designed magnetic bioconjugate seems to be applicable for targeted delivery purpose to a neoplastic cell due to the receptor action of the BSA molecule by binding to a wide variety of lipophilic compounds such as steroid present over cancer cell.
Acknowledgments
One of the author, PD, gratefully acknowledge the financial support by DAE-BRNS, Govt. of India (vide project no. 2007/20/34/04-BRNS/1865) under DAE Young Scientist Research Award. The authors would like to extend sincere thanks to CIF, IIT Guwhati, India and RSIC–NEHU for VSM and TEM facility.
References
- Kumagai M, Imai Y, Nakamura T, Yamasaki Y, Sekino M, Ueno S, Hanaoka K, Kikuchi K, Nagano T, Kaneko E, Shimokado K, Kataoka K. Colloids Surf. B: Biointerfaces. 2007. p. 174. COI number [1:CAS:528:DC%2BD2sXjslGlsLw%3D] [DOI] [PubMed]
- Gupta AK, Wells S. IEEE. 2004. p. 1536. [DOI] [PubMed]
- Kaminski MD, Rosengart AJ. J. 2005. p. 398. COI number [1:CAS:528:DC%2BD2MXktVagsL8%3D]; Bibcode number [2005JMMM..293..398K] [DOI]
- Perez JM. Nat. 2007. p. 535. COI number [1:CAS:528:DC%2BD2sXpvVOksbc%3D]; Bibcode number [2007NatNa...2..535P] [DOI] [PubMed]
- Ito A, Shinkai M, Honda H, Kobayashi T. J. 2005. p. 1. COI number [1:CAS:528:DC%2BD2MXhtVaqu7vM] [DOI] [PubMed]
- Yan S, Zhang D, Gu N, Zheng J, Ding A, Wang Z, Xing B, Ma M, Zhang Y. J. 2005. p. 1185. COI number [1:CAS:528:DC%2BD2MXnsVeltb0%3D] [DOI] [PubMed]
- Zhu N, Zhang A, He P, Fang Y. Electroanalysis. 2004. p. 1925. COI number [1:CAS:528:DC%2BD2cXhtFajt7rI] [DOI]
- Gu H, Xu K, Xu C, Xu B. Chem. 2006. p. 941. [DOI] [PubMed]
- Rossi LM, Quach AD, Rosenzweig Z. Anal. 2004. p. 606. COI number [1:CAS:528:DC%2BD2cXovV2ms7w%3D] [DOI] [PubMed]
- Gupta AK, Gupta M. Biomaterials. 2005. p. 3995. COI number [1:CAS:528:DC%2BD2MXisFWr] [DOI] [PubMed]
- Yu S, Chow GM. J. 2004. p. 2781. COI number [1:CAS:528:DC%2BD2cXntlert7k%3D] [DOI]
- Peng ZG, Hidajat K, Uddin MS. J. 2004. p. 277. COI number [1:CAS:528:DC%2BD2cXht1antLc%3D] [DOI] [PubMed]
- Reed RG. J. 1986. p. 15619. COI number [1:CAS:528:DyaL28XmtVCqur8%3D] [PubMed]
- Peters T. Adv. 1985. p. 161. COI number [1:CAS:528:DyaL2MXmtV2js70%3D] [DOI] [PubMed]
- Deb P, Basumallick A, Sen D, Mazumder S, Nath BK, Das D. Philos. 2006. p. 491. COI number [1:CAS:528:DC%2BD28XhtVCjtbnJ]; Bibcode number [2006PMagL..86..491D] [DOI]
- Samanta B, Yan H, Fischer NO, Shi J, Jerry DJ, Rotello VM. J. 2008. p. 1204. COI number [1:CAS:528:DC%2BD1cXislaku7Y%3D] [DOI] [PMC free article] [PubMed]
- Mamedova NN, Kotov NA, Rogach AL, Studer J. Nano. 2001. p. 281. COI number [1:CAS:528:DC%2BD3MXjsVOktbo%3D] [DOI]
- Tan CJ, Chua HG, Ker KH, Tong YW. Anal. 2008. p. 683. COI number [1:CAS:528:DC%2BD1cXis1ynsw%3D%3D] [DOI] [PubMed]
- Lehrer SS, Fashman GD. Biochem. 1966. p. 133. [DOI] [PubMed]
- Cherepy NJ, Liston DB, Lovejoy JA, Deng H, Zhang JZ. J. Phys. Chem. B. 1998. p. 770. COI number [1:CAS:528:DyaK1cXivFSitg%3D%3D] [DOI]
- Li Y, Yang H, He Z, Liu L, Wang W, Li F, Xu L. J. 2005. p. 2940. COI number [1:CAS:528:DC%2BD2MXht1Slt7zI]; Bibcode number [2005JMatR..20.2940L] [DOI]
- Kim T, Reis L, Rajan K, Shima M. J. 2005. p. 132. COI number [1:CAS:528:DC%2BD2MXmtlKkurk%3D]; Bibcode number [2005JMMM..295..132K] [DOI]
- Morales MP, Veintemillas-Verdaguer S, Serna CJ. J. 1999. p. 3066. COI number [1:CAS:528:DyaK1MXkslKgt70%3D]; Bibcode number [1999JMatR..14.3066M] [DOI]
- Salgueirino-Maceira V, Correa-Duarte MA, Spasova M, Liz-Marzan LM, Farle M. Adv. 2006. p. 509. COI number [1:CAS:528:DC%2BD28XislSkur8%3D] [DOI]


